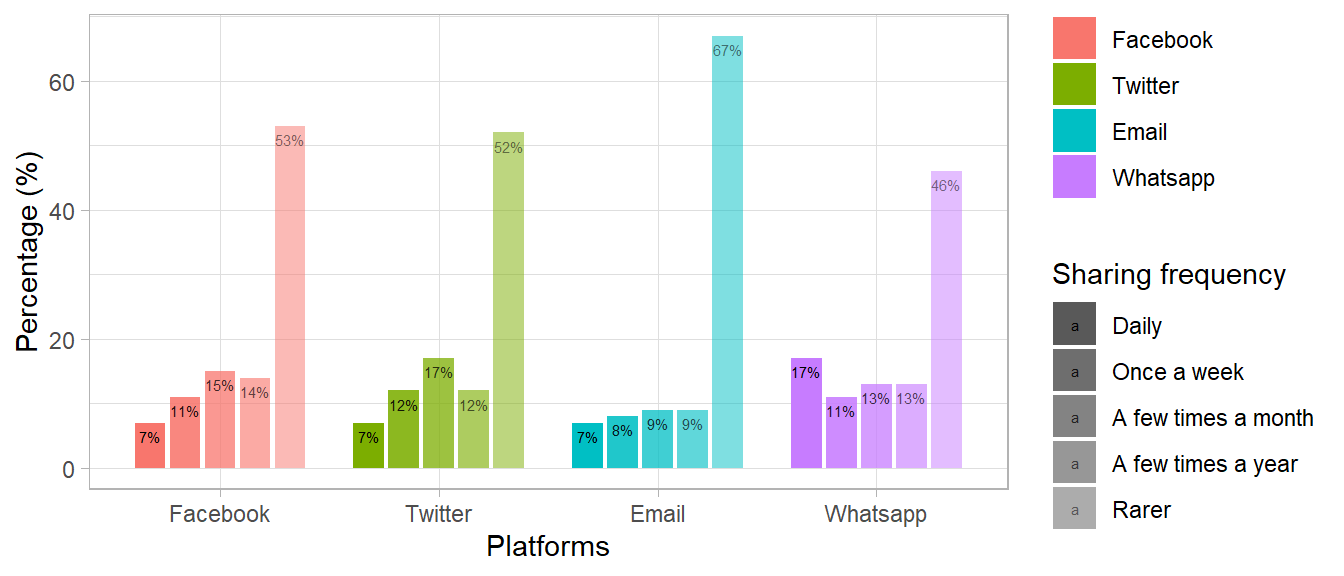
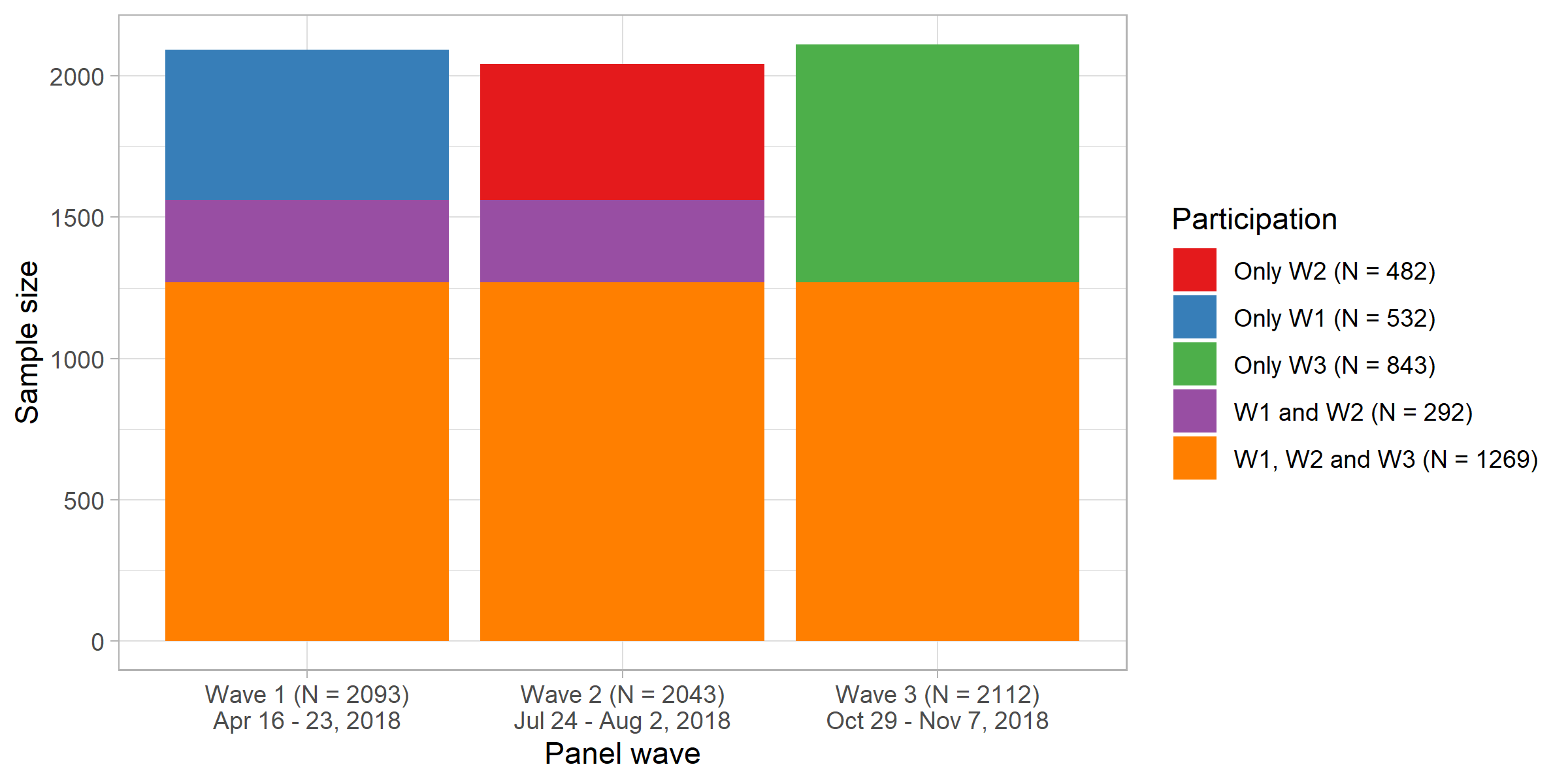library(tidyverse)
library(modelsummary)
# data_twitter_influence.csv
# data <- read_csv(sprintf("https://docs.google.com/uc?id=%s&export=download",
# "1dLSTUJ5KA-BmAdS-CHmmxzqDFm2xVfv6"))
data <- readr::read_csv("data/data_twitter_influence.csv",
col_types = cols())
# Numeric data
datasummary_skim(data, type = "numeric", output = "html")| Unique (#) | Missing (%) | Mean | SD | Min | Median | Max | ||
|---|---|---|---|---|---|---|---|---|
| n_retweets | 237 | 0 | 429.4 | 2332.7 | 0.0 | 36.0 | 48568.0 | |
| followers_count | 485 | 0 | 13184.0 | 51672.6 | 12.0 | 2647.5 | 693125.0 | |
| account_age_months | 504 | 0 | 84.1 | 40.7 | 5.0 | 87.3 | 143.6 | |
| account_age_years | 504 | 0 | 7.0 | 3.4 | 0.4 | 7.3 | 12.0 | |
| female | 2 | 0 | 0.3 | 0.5 | 0.0 | 0.0 | 1.0 |
# Categorical data (we had to create)
datasummary_skim(data %>% mutate(party = factor(party),
female = factor(female)),
type = "categorical", output = "html")| N | % | ||
|---|---|---|---|
| party | AfD | 76 | 15.1 |
| CDU_CSU | 131 | 26.0 | |
| DieLinke | 58 | 11.5 | |
| FDP | 73 | 14.5 | |
| Greens | 61 | 12.1 | |
| SPD | 105 | 20.8 | |
| party_color | black | 131 | 26.0 |
| blue | 76 | 15.1 | |
| deeppink | 58 | 11.5 | |
| gold | 73 | 14.5 | |
| green | 61 | 12.1 | |
| red | 105 | 20.8 | |
| female | 0 | 351 | 69.6 |
| 1 | 153 | 30.4 |