Capítulo 13 Variaveis Instrumentais
Motivação para usar Variáveis Instrumentais: \(\mathbb{E}(xe) \neq 0\)
Vamos ver duas formas de estuturar um modelo onde exista uma variável endógena e um ou mais instrumentos:
set.seed(1)
n= 1000
z1 = rnorm(n)
z2 = runif(n,0,1)
u = rnorm(n)
x1 = rnorm(n)
x2 = u + 1.5*z2 + 0.7*z1 + rnorm(n)
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,2)
y = X%*%beta + ucor(x2,u)## [1] 0.6290406cor(z1,u)## [1] 0.01866414cor(z1,x2)## [1] 0.4387025cor(z2,u)## [1] 0.02076497cor(z2,x2)## [1] 0.2871934Matriz de Variância e Covariância
#install.packages("mvtnorm")
library(mvtnorm) sigma = matrix(c(4,3,3,30), ncol=2)
colnames(sigma) = c("x1","x2")
rownames(sigma) = c("x1","x2")
sigma## x1 x2
## x1 4 3
## x2 3 30Normal Bivariada
n = 1000
mv = rmvnorm(n, mean=c(10,0), sigma=sigma)
dim(mv)## [1] 1000 2x1 = mv[,1]
x2 = mv[,2]
var(x1)## [1] 4.019209mean(x1)## [1] 9.918847var(x2)## [1] 31.36548mean(x2)## [1] 0.075497cov(x1,x2)## [1] 3.458533Endogeneidade
\(E(x_2u) \neq 0\)
Lembre que: \(E(x_2u) = E(x_2)E(u) + Cov(X_2,u)\), e, como \(E(u) = 0\), então \(E(x_2u) = Cov(x_2,u)\)
n = 1000
x1 = rnorm(n)
sigma = matrix(c(4,3,3,30), ncol=2)
colnames(sigma) = c("x2","u")
rownames(sigma) = c("x2","u")
sigma## x2 u
## x2 4 3
## u 3 30 mv = rmvnorm(n, mean=c(10,0), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
cov(x2,u)## [1] 2.905598Endogeneidade gera inconsistência no estimador de OLS
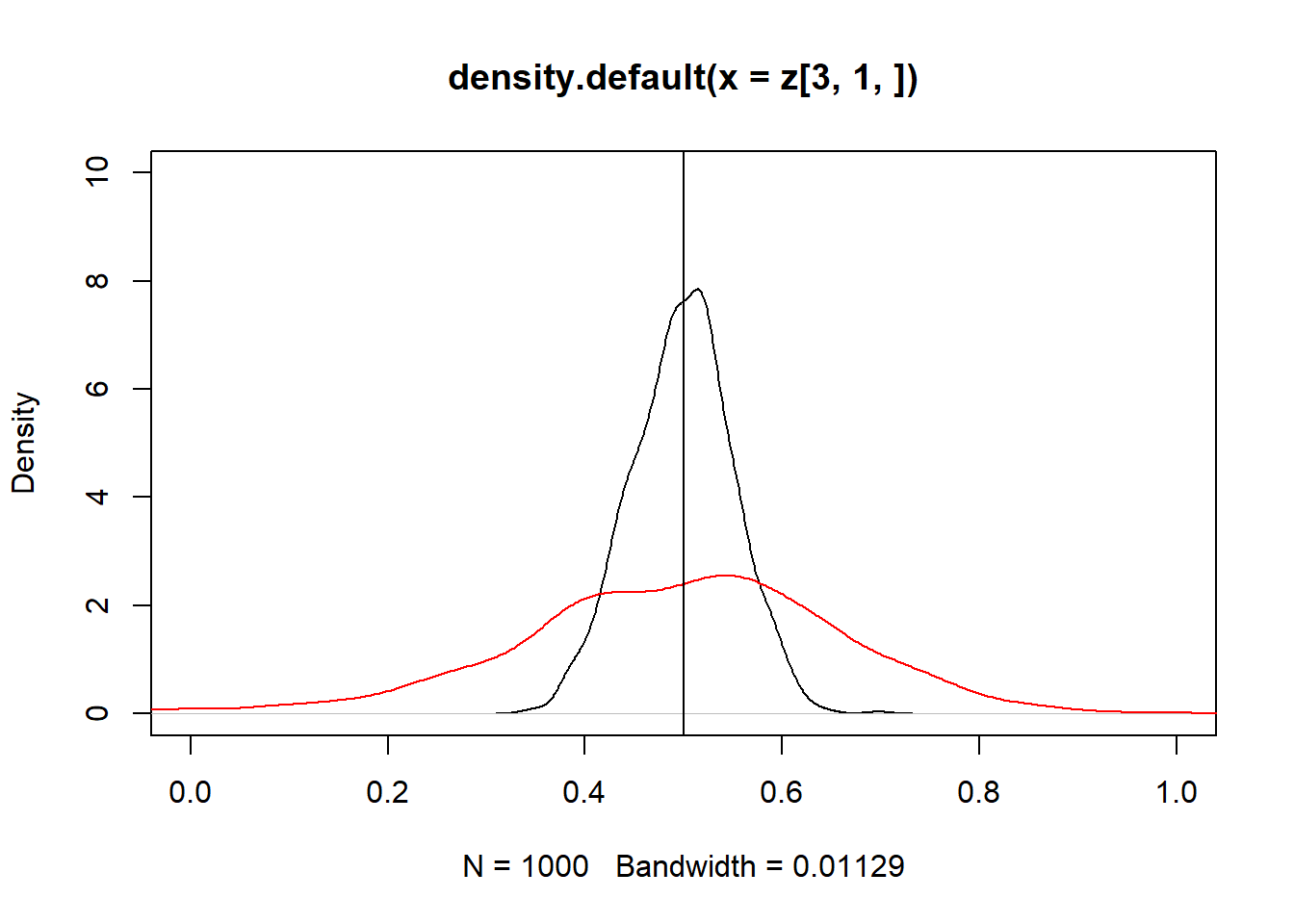
Se $Cov(x_2,u) > 0 $, então o estimador é enviesado para cima:
iv = function(n){
x1 = rnorm(n)
sigma = matrix(c(4,3,3,30), ncol=2)
colnames(sigma) = c("x2","u")
rownames(sigma) = c("x2","u")
mv = rmvnorm(n, mean=c(10,0), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
X = cbind(1,x1,x2)
beta = c(2,5,0.5)
y = X%*%beta + u
B = solve(t(X)%*%X)%*%t(X)%*%y
B
}
z = replicate(10000, expr = iv(1000))
plot(density(z[3, 1, ]), xlim = c(0,2))
abline(v = 0.5)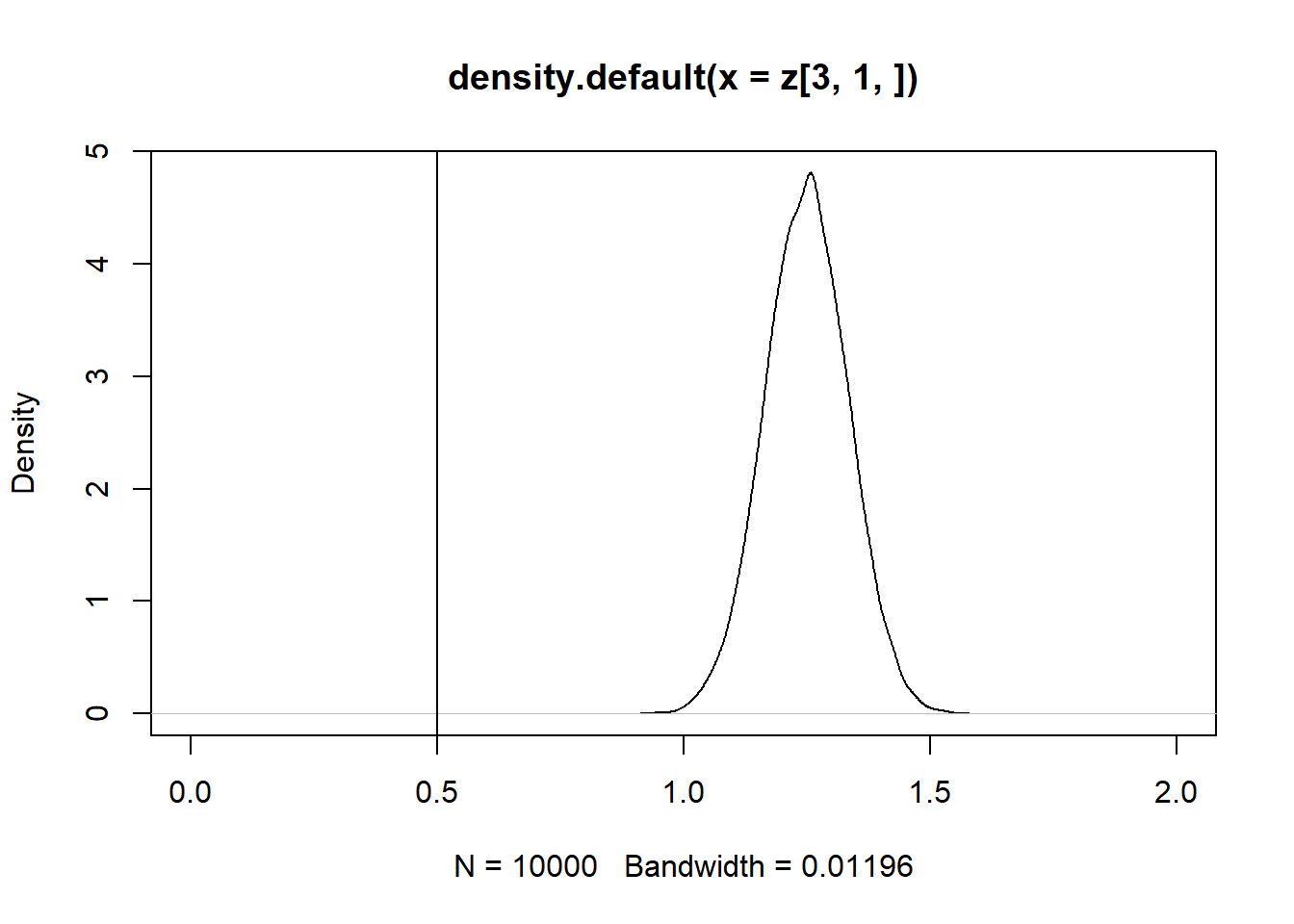
Se $Cov(x_2,u) < 0 $, então o estimador é enviesado para baixo:
iv = function(n){
x1 = rnorm(n)
sigma = matrix(c(4,-3,-3,30), ncol=2)
colnames(sigma) = c("x2","u")
rownames(sigma) = c("x2","u")
mv = rmvnorm(n, mean=c(10,0), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
X = cbind(1,x1,x2)
beta = c(2,5,0.5)
y = X%*%beta + u
B = solve(t(X)%*%X)%*%t(X)%*%y
B
}
z = replicate(10000, expr = iv(1000))
plot(density(z[3, 1, ]), xlim = c(-1,1))
abline(v = 0.5)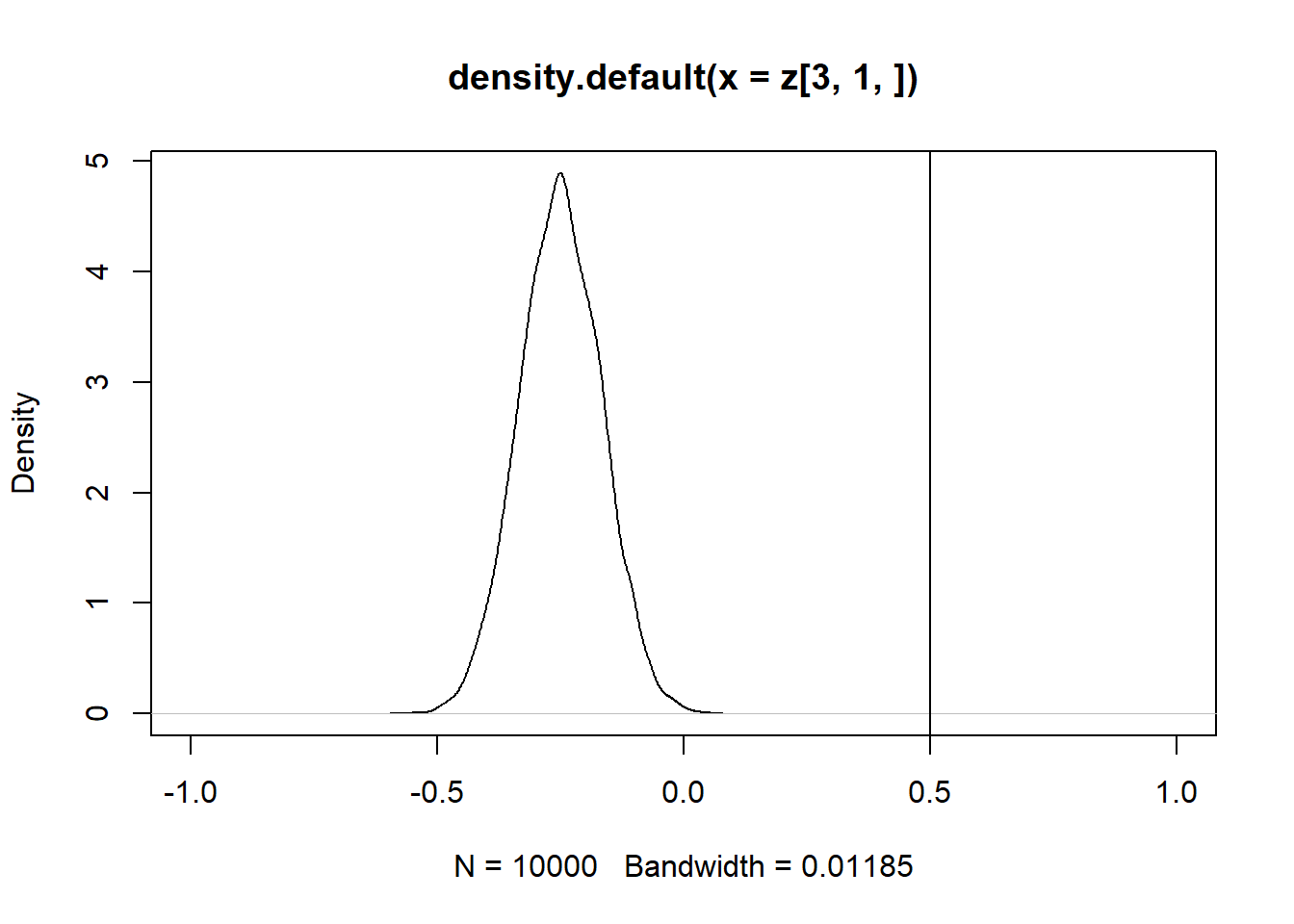
Se \(Cor(x_1,x_2) = 0\), então apenas a estimativa de \(\beta_2\) é enviesada.
iv = function(n){
x1 = rnorm(n)
sigma = matrix(c(4,-3,-3,30), ncol=2)
mv = rmvnorm(n, mean=c(10,0), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
X = cbind(1,x1,x2)
beta = c(2,5,0.5)
y = X%*%beta + u
B = solve(t(X)%*%X)%*%t(X)%*%y
B
}
z = replicate(10000, expr = iv(1000))
par(mfrow = c(1, 2))
plot(density(z[2, 1, ]), xlim = c(4.5,5.5))
abline(v = 5)
plot(density(z[3, 1, ]), xlim = c(-0.5,1))
abline(v = 0.5)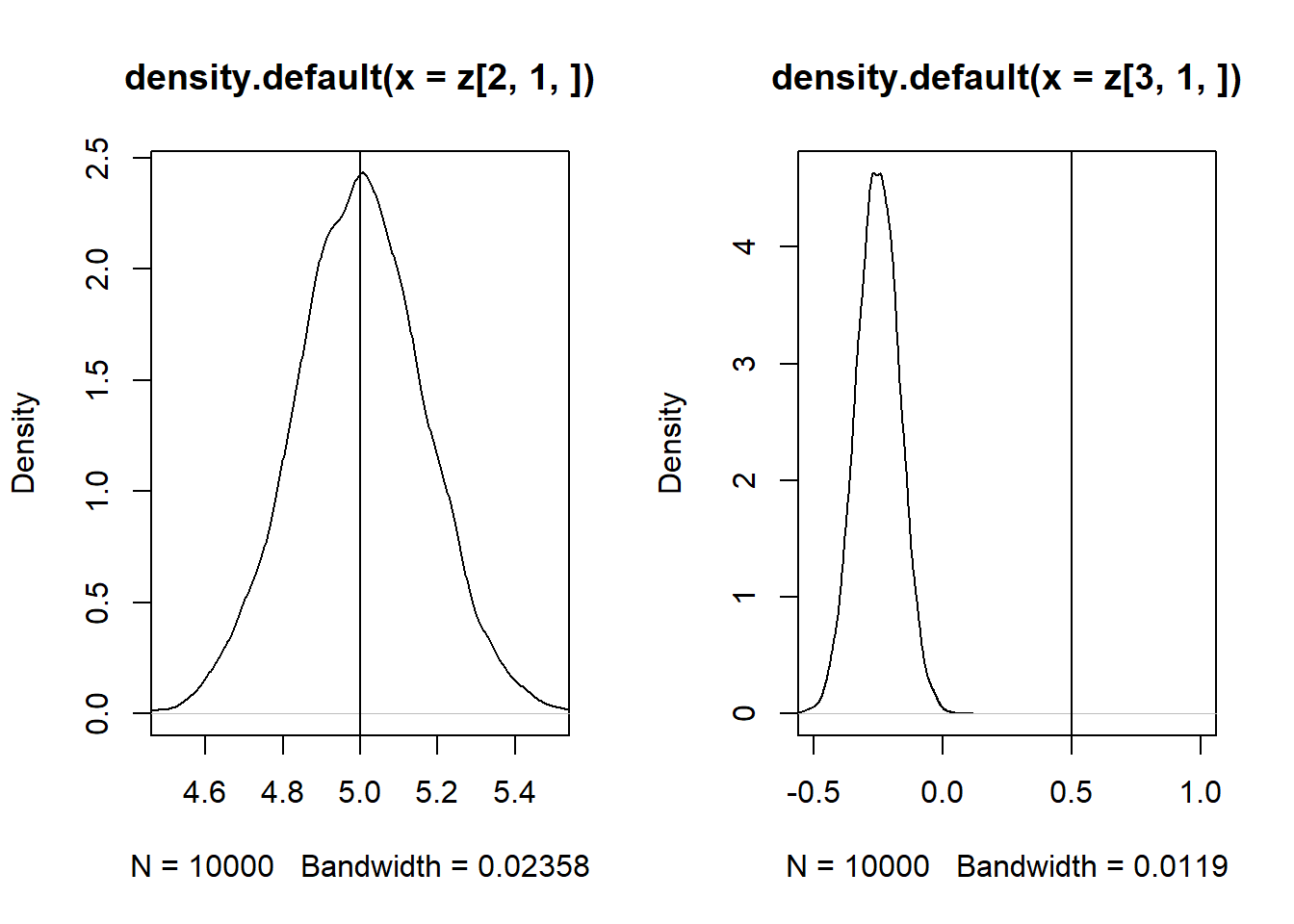
Se \(Cor(x_1,x_2) \neq 0\), então as estimativas de \(\beta_1\) e \(\beta_2\) são enviesadas.
iv = function(n){
sigma = matrix(c(4,-3,2,-3,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
x1 = mv[,3]
X = cbind(1,x1,x2)
beta = c(2,5,0.5)
y = X%*%beta + u
B = solve(t(X)%*%X)%*%t(X)%*%y
B
}
z = replicate(10000, expr = iv(1000))
par(mfrow = c(1, 2))
plot(density(z[2, 1, ]), xlim = c(4.5,6))
abline(v = 5)
plot(density(z[3, 1, ]), xlim = c(-0.5,1))
abline(v = 0.5)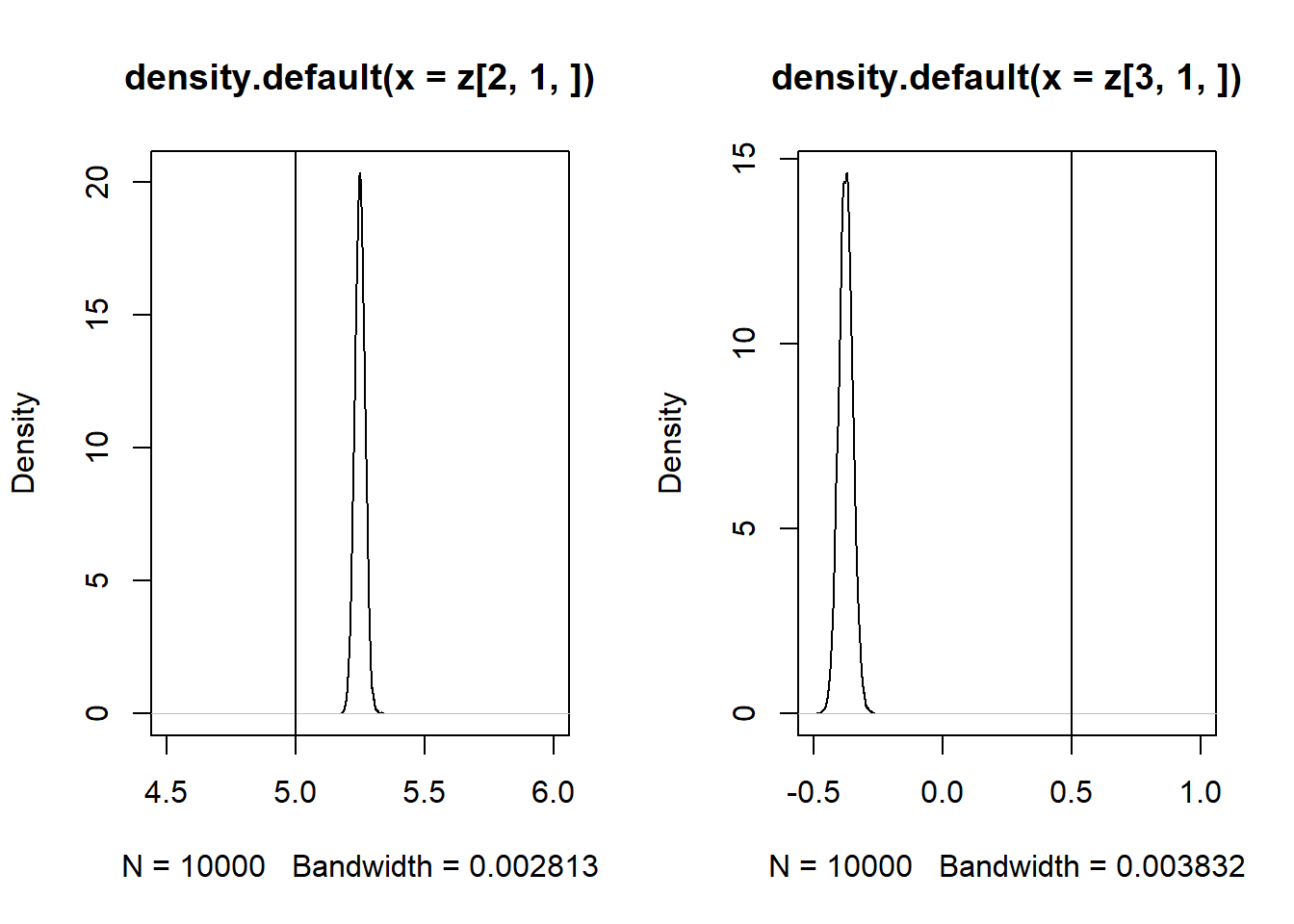
13.1 Estimador de Variáveis Instrumentais
\(\hat\beta{iv} = (Z'X)^{-1}Z'y\)
sigma = matrix(c(4,-3,2,-3,5,0,2,0,7), ncol=3)
colnames(sigma) = c("x2","u","z1")
rownames(sigma) = c("x2","u","z1")
sigma## x2 u z1
## x2 4 -3 2
## u -3 5 0
## z1 2 0 7iv = function(n){
x1 = rnorm(n)
sigma = matrix(c(4,-3,2,-3,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + u
Biv = solve(t(Z)%*%X)%*%t(Z)%*%y
Biv
}
z = replicate(10000, expr = iv(1000))
plot(density(z[3, 1, ]), xlim = c(0,1))
abline(v = 0.5)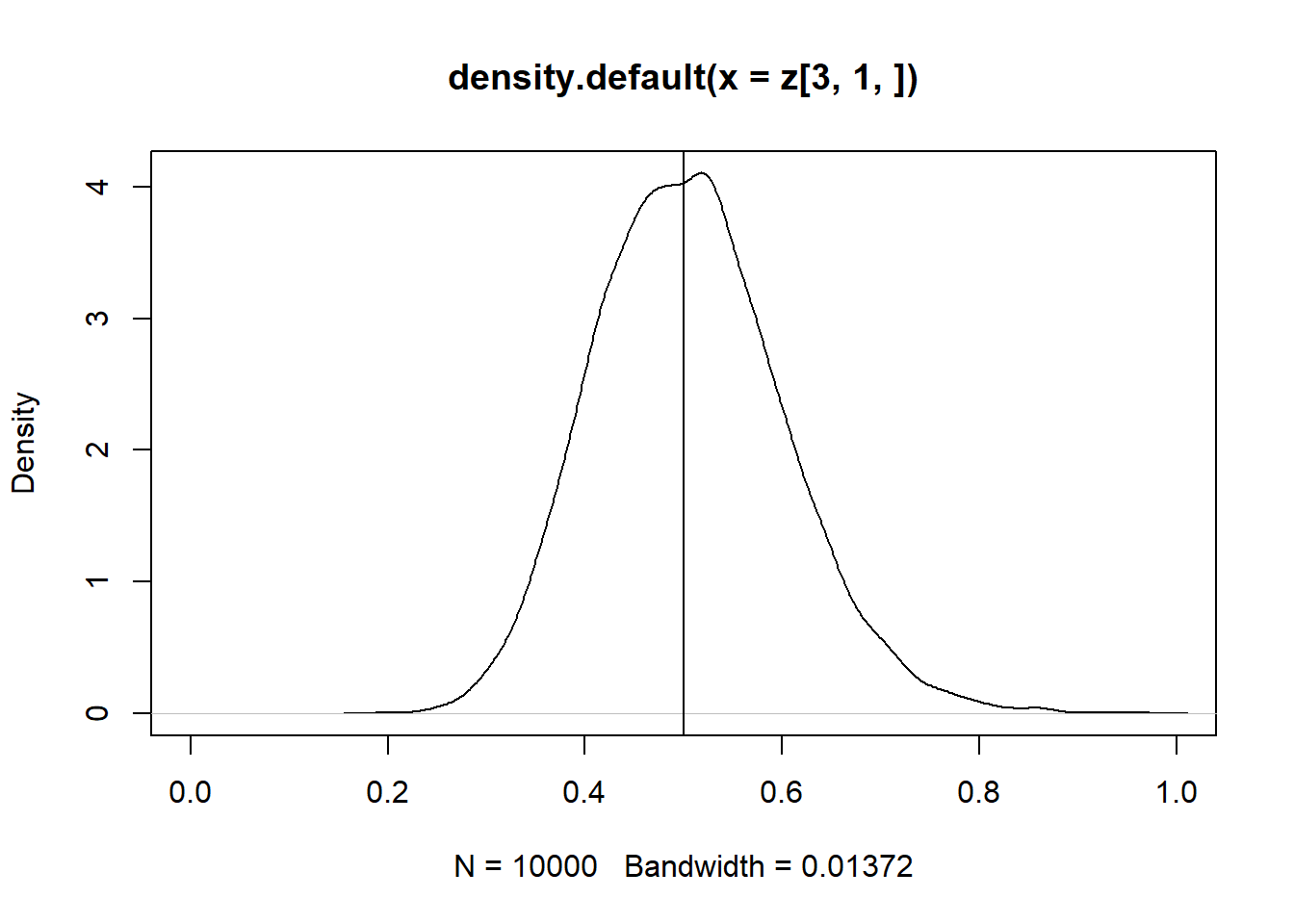
\(Cov(x_1,x_2) \ne 0\)
iv = function(n){
sigma = matrix(c(5,-3,0,0,-3,10,2,-5,0,2,4,0,0,-5,0,10), ncol=4)
mv = rmvnorm(n, mean=c(10,2,3,0), sigma=sigma)
x1 = mv[,1]
x2 = mv[,2]
u = mv[,4]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + u
Biv = solve(t(Z)%*%X)%*%t(Z)%*%y
Biv
}
z = replicate(10000, expr = iv(1000))
par(mfrow = c(1, 2))
plot(density(z[2, 1, ]), xlim = c(4.5,5.5))
abline(v = 5)
plot(density(z[3, 1, ]), xlim = c(-0.5,1))
abline(v = 0.5)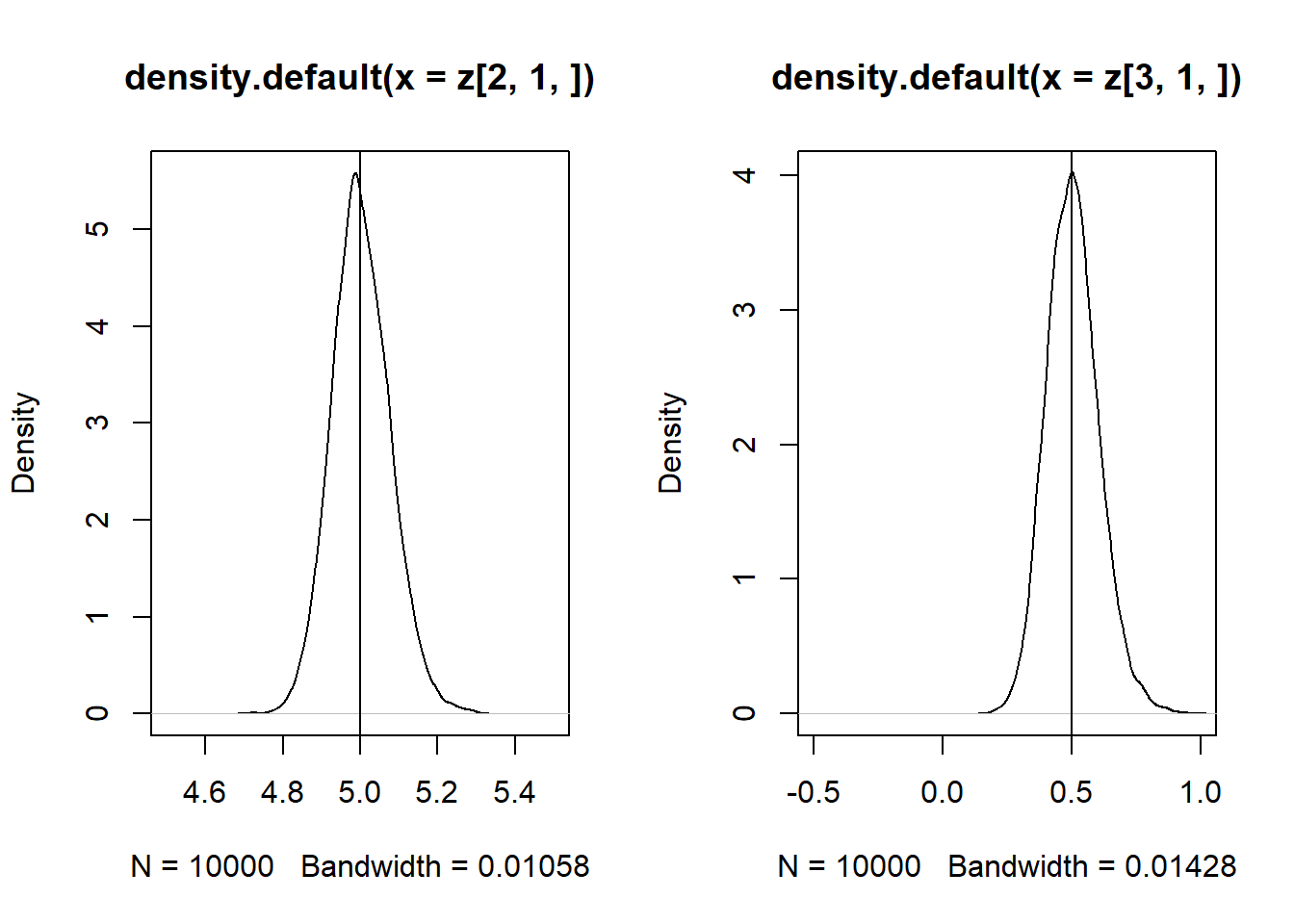
13.2 Estimador de Mínimos Quadrados em Dois Estágios - 2SLS
Utilizando apenas um instrumento: as estimativas de IV e 2SLS
\(\hat \beta_{2sls} = (X'Z(Z'Z)^{-1}Z'X)^{-1}X'Z(Z'Z)^{-1}Z'y\)
set.seed(1)
x1 = rnorm(n)
sigma = matrix(c(4,-3,2,-3,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + u
Biv = solve(t(Z)%*%X)%*%t(Z)%*%y
Biv## [,1]
## 1.1023513
## x1 4.9974349
## x2 0.5842413B2sls = solve(t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)%*%t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%y
B2sls## [,1]
## 1.1023513
## x1 4.9974349
## x2 0.5842413Por que o nome de dois estágios? Pois é possível obter as mesmas estimativas realizando um procedimento em dois estágios conforme pode ser observado abaixo.
Estimar: \(x_2 = \hat\beta_0 + \hat\beta_1 X_1 + \gamma Z_1 + e\)
set.seed(1)
n = 1000
x1 = rnorm(n)
sigma = matrix(c(4,-3,2,-3,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + u
B1 = solve(t(Z)%*%Z)%*%t(Z)%*%x2
x_hat = Z%*%B1Antes de estimar o 2º estágio, repare que o 1º estágio “limpou” a endogeneidade que existia em \(x2\).
cor(x_hat,u)## [,1]
## [1,] 0.0286093res =x2 - x_hat
cor(res,u)## [,1]
## [1,] -0.71484472º estágio: \(y = \beta_0 + \beta_1 X_1 + \beta_2 \hat{X_2} + u\)
X_hat = cbind(1,x1,x_hat)
B2sls = solve(t(X_hat)%*%X_hat)%*%t(X_hat)%*%y
B2sls## [,1]
## 1.1023513
## x1 4.9974349
## 0.5842413Comparar os resultados obtidos pelo estimador 2SLS com o procedimento em 2 estágios:
B2sls = solve(t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)%*%t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%y
B2sls## [,1]
## 1.1023513
## x1 4.9974349
## x2 0.5842413Dois Instrumentos
sigma = matrix(c(5,-2,5,3,-2,10,0,0,5,0,8,0,3,0,0,10), ncol=4)
colnames(sigma) = c("x2","z1","z2","u")
rownames(sigma) = c("x2","z1","z2","u")
sigma## x2 z1 z2 u
## x2 5 -2 5 3
## z1 -2 10 0 0
## z2 5 0 8 0
## u 3 0 0 10set.seed(1)
x1 = rnorm(n)
sigma = matrix(c(5,-2,5,3,-2,10,0,0,5,0,8,0,3,0,0,10), ncol=4)
mv = rmvnorm(n, mean=c(10,5,3,0), sigma=sigma)
x2 = mv[,1]
z1 = mv[,2]
z2 = mv[,3]
u = mv[,4]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1,z2)
beta = c(2,5,0.5)
y = X%*%beta + u
B2sls = solve(t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)%*%t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%y
B2sls## [,1]
## 2.0407802
## x1 5.0159400
## x2 0.4980277Usar dois instrumentos aumenta a eficiência, conforme veremos abaixo. Mas usando um ou dois instrumentos o estimador é consistente:
sls2 = function(n){
x1 = rnorm(n)
sigma = matrix(c(5,-2,5,3,-2,10,0,0,5,0,8,0,3,0,0,10), ncol=4)
mv = rmvnorm(n, mean=c(10,5,3,0), sigma=sigma)
x2 = mv[,1]
z1 = mv[,2]
z2 = mv[,3]
u = mv[,4]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1,z2)
beta = c(2,5,0.5)
y = X%*%beta + u
B2sls2 = solve(t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)%*%t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%y
B2sls2
}sls = function(n){
x1 = rnorm(n)
sigma = matrix(c(5,-2,5,3,-2,10,0,0,5,0,8,0,3,0,0,10), ncol=4)
mv = rmvnorm(n, mean=c(10,5,3,0), sigma=sigma)
x2 = mv[,1]
z1 = mv[,2]
z2 = mv[,3]
u = mv[,4]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + u
B2sls2 = solve(t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)%*%t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%y
B2sls2
}set.seed(10)
z = replicate(10000, expr = sls2(1000))
set.seed(10)
z1 = replicate(10000, expr = sls(1000))
par(mfrow = c(1, 2))
plot(density(z[3, 1, ]), xlim = c(0,1))
abline(v = 0.5)
plot(density(z1[3, 1, ]), xlim = c(0,1))
abline(v = 0.5)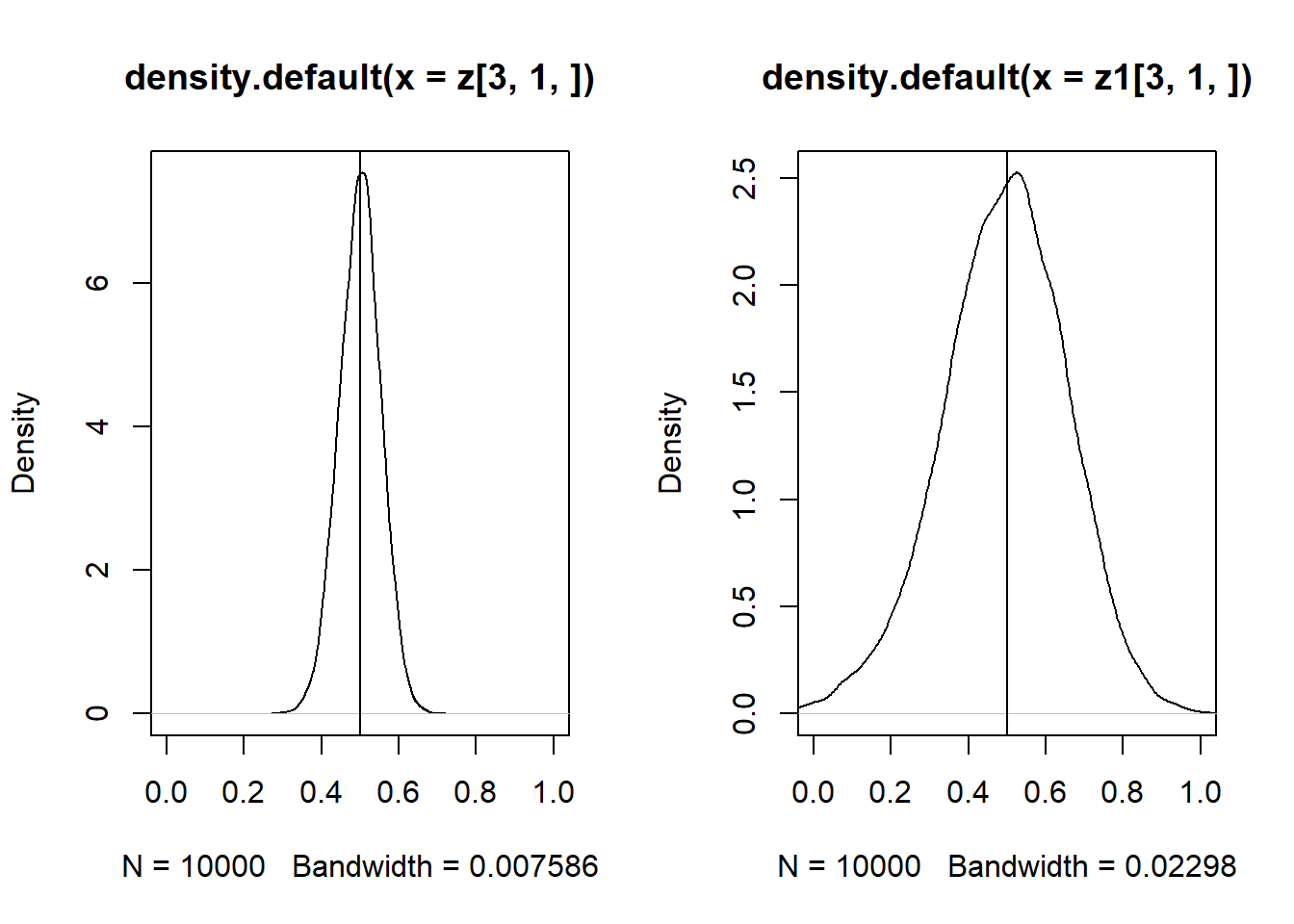
13.3 Matriz de Variânicia-Covariância de Variáveis Instrumentais
Estimador de Varáveis Instrumentais
set.seed(1)
n = 1000
x1 = rnorm(n)
sigma = matrix(c(4,-3,2,-3,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + u
Biv = solve(t(Z)%*%X)%*%t(Z)%*%y
Biv## [,1]
## 1.1023513
## x1 4.9974349
## x2 0.5842413\(Var(\hat\beta_{iv}) = \sigma_{iv}^2 (X'P_zX)^{-1}\), onde \(P_z = Z(Z'Z)^{-1}Z1\)
Portanto: \(Var(\hat\beta_{iv}) = \sigma_{iv}^2 (X'Z(Z'Z)^{-1}Z'X)^{-1}\)
e_hat_iv = y - X%*%Biv
k = ncol(X)
sigma_hat_iv = (1/(n-k)) * t(e_hat_iv)%*%e_hat_iv ## O R faz a correção do viés dividindo por (n-k) o Stata só divide por n
sigma_hat_iv## [,1]
## [1,] 5.943146V_iv = sigma_hat_iv[1,1] * solve((t(X)%*%Z)%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)
ep_iv = sqrt(diag(V_iv))
ep_iv## x1 x2
## 0.98725579 0.07471610 0.09789995Estimador de Mínimos Quadrados de Dois Estágios
set.seed(1)
x1 = rnorm(n)
sigma = matrix(c(4,-3,2,-3,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + u
B2sls = solve(t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)%*%t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%y
B2sls## [,1]
## 1.1023513
## x1 4.9974349
## x2 0.5842413e_hat_2sls = y - X%*%B2sls
k = ncol(X)
sigma_hat_2sls = (1/(n-k)) * t(e_hat_2sls)%*%e_hat_2sls ## O R faz a correção do viés dividindo por (n-k) o Stata só divide por n
sigma_hat_2sls## [,1]
## [1,] 5.943146V_2sls = sigma_hat_2sls[1,1] * solve((t(X)%*%Z)%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)
ep_2sls = sqrt(diag(V_2sls))
ep_2sls## x1 x2
## 0.98725579 0.07471610 0.09789995Estimador de Mínimos Quadrados em Dois Estágios
set.seed(1)
n = 1000
x1 = rnorm(n)
sigma = matrix(c(4,-3,2,-3,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + uB1 = solve(t(Z)%*%Z)%*%t(Z)%*%x2
x_hat = Z%*%B1
X_hat = cbind(1,x1,x_hat)
B2sls = solve(t(X_hat)%*%X_hat)%*%t(X_hat)%*%y
B2sls## [,1]
## 1.1023513
## x1 4.9974349
## 0.5842413e_hat_2sls = y - X%*%B2sls # usar o vetor original de X, e não o vetor do segundo estágio
k = ncol(X)
sigma_hat_2sls = (1/(n-k)) * t(e_hat_2sls)%*%e_hat_2sls # n-k correção de viés
sigma_hat_2sls## [,1]
## [1,] 5.943146V_2sls = sigma_hat_2sls[1,1] * solve((t(X_hat)%*%X_hat))
ep_2sls = sqrt(diag(V_2sls))
ep_2sls## x1
## 0.98725579 0.07471610 0.0978999513.4 Teste de Hausman
#install.packages("ivreg")
library(ivreg)## Warning: package 'ivreg' was built under R version 4.1.3## Registered S3 methods overwritten by 'ivreg':
## method from
## anova.ivreg AER
## hatvalues.ivreg AER
## model.matrix.ivreg AER
## predict.ivreg AER
## print.ivreg AER
## print.summary.ivreg AER
## summary.ivreg AER
## terms.ivreg AER
## update.ivreg AER
## vcov.ivreg AER##
## Attaching package: 'ivreg'## The following objects are masked from 'package:AER':
##
## ivreg, ivreg.fitModelo com Endogeneidade
set.seed(1)
n =1000
x1 = rnorm(n)
sigma = matrix(c(4,-3,2,-3,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + udf = data.frame(y,x1,x2,z1)
iv = ivreg(y ~ x1 + x2 | x1 + z1, data = df)
summary(iv)##
## Call:
## ivreg(formula = y ~ x1 + x2 | x1 + z1, data = df)
##
## Residuals:
## Min 1Q Median 3Q Max
## -7.71597 -1.63461 -0.02373 1.63873 7.22710
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.10235 0.98726 1.117 0.264
## x1 4.99743 0.07472 66.886 < 2e-16 ***
## x2 0.58424 0.09790 5.968 3.34e-09 ***
##
## Diagnostic tests:
## df1 df2 statistic p-value
## Weak instruments 1 997 170.8 <2e-16 ***
## Wu-Hausman 1 996 183.4 <2e-16 ***
## Sargan 0 NA NA NA
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 2.438 on 997 degrees of freedom
## Multiple R-Squared: 0.805, Adjusted R-squared: 0.8047
## Wald test: 2294 on 2 and 997 DF, p-value: < 2.2e-16Modelo sem Endogeneidade
set.seed(1)
n =1000
x1 = rnorm(n)
sigma = matrix(c(4,0,2,0,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + udf = data.frame(y,x1,x2,z1)
iv = ivreg(y ~ x1 + x2 | x1 + z1, data = df)
summary(iv)##
## Call:
## ivreg(formula = y ~ x1 + x2 | x1 + z1, data = df)
##
## Residuals:
## Min 1Q Median 3Q Max
## -7.96475 -1.55691 -0.01991 1.63352 7.36388
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.86049 0.90828 0.947 0.344
## x1 5.01820 0.07201 69.691 < 2e-16 ***
## x2 0.60938 0.09015 6.760 2.35e-11 ***
##
## Diagnostic tests:
## df1 df2 statistic p-value
## Weak instruments 1 997 184.298 <2e-16 ***
## Wu-Hausman 1 996 0.711 0.399
## Sargan 0 NA NA NA
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 2.347 on 997 degrees of freedom
## Multiple R-Squared: 0.8391, Adjusted R-squared: 0.8388
## Wald test: 2509 on 2 and 997 DF, p-value: < 2.2e-1613.5 Eficiência dos Estimadores
Modelo sem Endogeneidade: OLS vs IV
IV
set.seed(1)
n =1000
x1 = rnorm(n)
sigma = matrix(c(4,0,2,0,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + uBiv = solve(t(Z)%*%X)%*%t(Z)%*%y
Biv## [,1]
## 0.8604895
## x1 5.0181987
## x2 0.6093826e_hat_iv = y - X%*%Biv
k = ncol(X)
sigma_hat_iv = (1/(n-k)) * t(e_hat_iv)%*%e_hat_iv ## O R faz a correção do viés dividindo por (n-k) o Stata só divide por n
sigma_hat_iv## [,1]
## [1,] 5.507993V_iv = sigma_hat_iv[1,1] * solve((t(X)%*%Z)%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)
V_iv## x1 x2
## 0.824970876 0.0055776590 -0.0816072406
## x1 0.005577659 0.0051849073 -0.0005494873
## x2 -0.081607241 -0.0005494873 0.0081269665ep_iv = sqrt(diag(V_iv))
ep_iv## x1 x2
## 0.90827907 0.07200630 0.09014969OLS
Bols = solve(t(X)%*%X)%*%t(X)%*%y
Bols## [,1]
## 1.5605990
## x1 5.0229128
## x2 0.5396613e_hat = y - X%*%Bols
k = ncol(X)
sigma_hat = (1/(n-k)) * t(e_hat)%*%e_hat # n-k correção de viés
V = sigma_hat[1,1] * solve(t(X)%*%X)
ep = sqrt(diag(V))
ep## x1 x2
## 0.36447618 0.07165008 0.03553926iv = function(n){
x1 = rnorm(n)
sigma = matrix(c(4,0,2,0,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + u
Biv = solve(t(Z)%*%X)%*%t(Z)%*%y
Biv
}ols = function(n){
x1 = rnorm(n)
# x3 = rnorm(n,5,5)
sigma = matrix(c(4,0,2,0,5,0,2,0,7), ncol=3)
mv = rmvnorm(n, mean=c(10,0,3), sigma=sigma)
x2 = mv[,1]
u = mv[,2]
z1 = mv[,3]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + u
Bols = solve(t(X)%*%X)%*%t(X)%*%y
Bols
}set.seed(10)
z = replicate(1000, expr = iv(1000))
set.seed(10)
z1 = replicate(1000, expr = ols(1000))
plot(density(z[3, 1, ]), xlim = c(-0,1), ylim = c(0,12))
lines(density(z1[3, 1, ]), add = TRUE, col = "red")## Warning in plot.xy(xy.coords(x, y), type = type, ...): "add" não é um parâmetro
## gráficoabline(v = 0.5)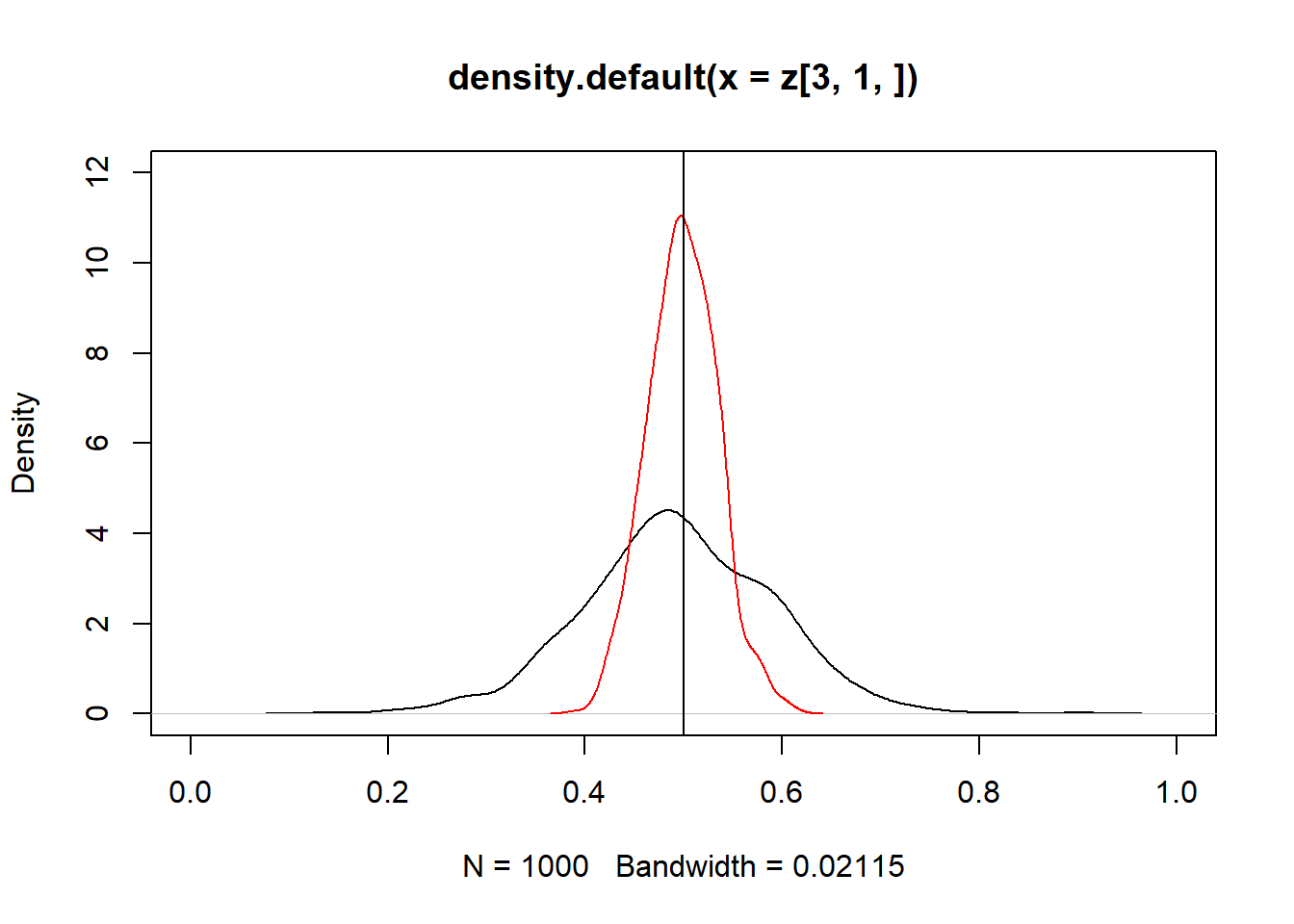
Modelo com endogeneidade: 1 instrumento vs 2 instrumentos
2 instrumentos
set.seed(1)
x1 = rnorm(n)
sigma = matrix(c(5,-2,5,3,-2,10,0,0,5,0,8,0,3,0,0,10), ncol=4)
mv = rmvnorm(n, mean=c(10,5,3,0), sigma=sigma)
x2 = mv[,1]
z1 = mv[,2]
z2 = mv[,3]
u = mv[,4]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1,z2)
beta = c(2,5,0.5)
y = X%*%beta + u
B2sls = solve(t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)%*%t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%y
B2sls## [,1]
## 2.0407802
## x1 5.0159400
## x2 0.4980277e_hat_2sls = y - X%*%B2sls
k = ncol(X)
sigma_hat_2sls = (1/(n-k)) * t(e_hat_2sls)%*%e_hat_2sls ## O R faz a correção do viés dividindo por (n-k) o Stata só divide por n
sigma_hat_2sls## [,1]
## [1,] 10.42011V_2sls = sigma_hat_2sls[1,1] * solve((t(X)%*%Z)%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)
ep_2sls = sqrt(diag(V_2sls))
ep_2sls## x1 x2
## 0.53649921 0.09868849 0.052831041 instrumento
set.seed(1)
x1 = rnorm(n)
sigma = matrix(c(5,-2,5,3,-2,10,0,0,5,0,8,0,3,0,0,10), ncol=4)
mv = rmvnorm(n, mean=c(10,5,3,0), sigma=sigma)
x2 = mv[,1]
z1 = mv[,2]
z2 = mv[,3]
u = mv[,4]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + u
B2sls = solve(t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)%*%t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%y
B2sls## [,1]
## 2.4567813
## x1 5.0166521
## x2 0.4563002e_hat_2sls = y - X%*%B2sls
k = ncol(X)
sigma_hat_2sls = (1/(n-k)) * t(e_hat_2sls)%*%e_hat_2sls ## O R faz a correção do viés dividindo por (n-k) o Stata só divide por n
sigma_hat_2sls## [,1]
## [1,] 10.69194V_2sls = sigma_hat_2sls[1,1] * solve((t(X)%*%Z)%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)
ep_2sls = sqrt(diag(V_2sls))
ep_2sls## x1 x2
## 1.57171541 0.09999934 0.15731133iv2 = function(n){
x1 = rnorm(n)
sigma = matrix(c(5,-2,5,3,-2,10,0,0,5,0,8,0,3,0,0,10), ncol=4)
mv = rmvnorm(n, mean=c(10,5,3,0), sigma=sigma)
x2 = mv[,1]
z1 = mv[,2]
z2 = mv[,3]
u = mv[,4]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1,z2)
beta = c(2,5,0.5)
y = X%*%beta + u
B2sls = solve(t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)%*%t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%y
B2sls
}iv = function(n){
x1 = rnorm(n)
sigma = matrix(c(5,-2,5,3,-2,10,0,0,5,0,8,0,3,0,0,10), ncol=4)
mv = rmvnorm(n, mean=c(10,5,3,0), sigma=sigma)
x2 = mv[,1]
z1 = mv[,2]
z2 = mv[,3]
u = mv[,4]
X = cbind(1,x1,x2)
Z = cbind(1,x1,z1)
beta = c(2,5,0.5)
y = X%*%beta + u
B2sls = solve(t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%X)%*%t(X)%*%Z%*%solve(t(Z)%*%Z)%*%t(Z)%*%y
B2sls
}set.seed(10)
z = replicate(1000, expr = iv2(1000))
set.seed(10)
z1 = replicate(1000, expr = iv(1000))
plot(density(z[3, 1, ]), xlim = c(-0,1), ylim = c(0,10))
lines(density(z1[3, 1, ]), add = TRUE, col = "red")## Warning in plot.xy(xy.coords(x, y), type = type, ...): "add" não é um parâmetro
## gráficoabline(v = 0.5)