Documento 12 Formal Context Ganter dataset
12.1 Primera parte
library(readr)
library(knitr)
library(fcaR)
library(rmarkdown)
dataset_ganter <- read.csv("datasets/contextformal_tutorialGanter.csv", sep=";")
paged_table(dataset_ganter)Los nombres de las filas aparecen como primera columna. Trasladamos el nombre de las filas a la variable rownames(dataset_ganter)
Eliminamos la primera columna
- Introduce el dataset anterior en un contexto formal de nombre fc_ganter usando el paquete fcaR.
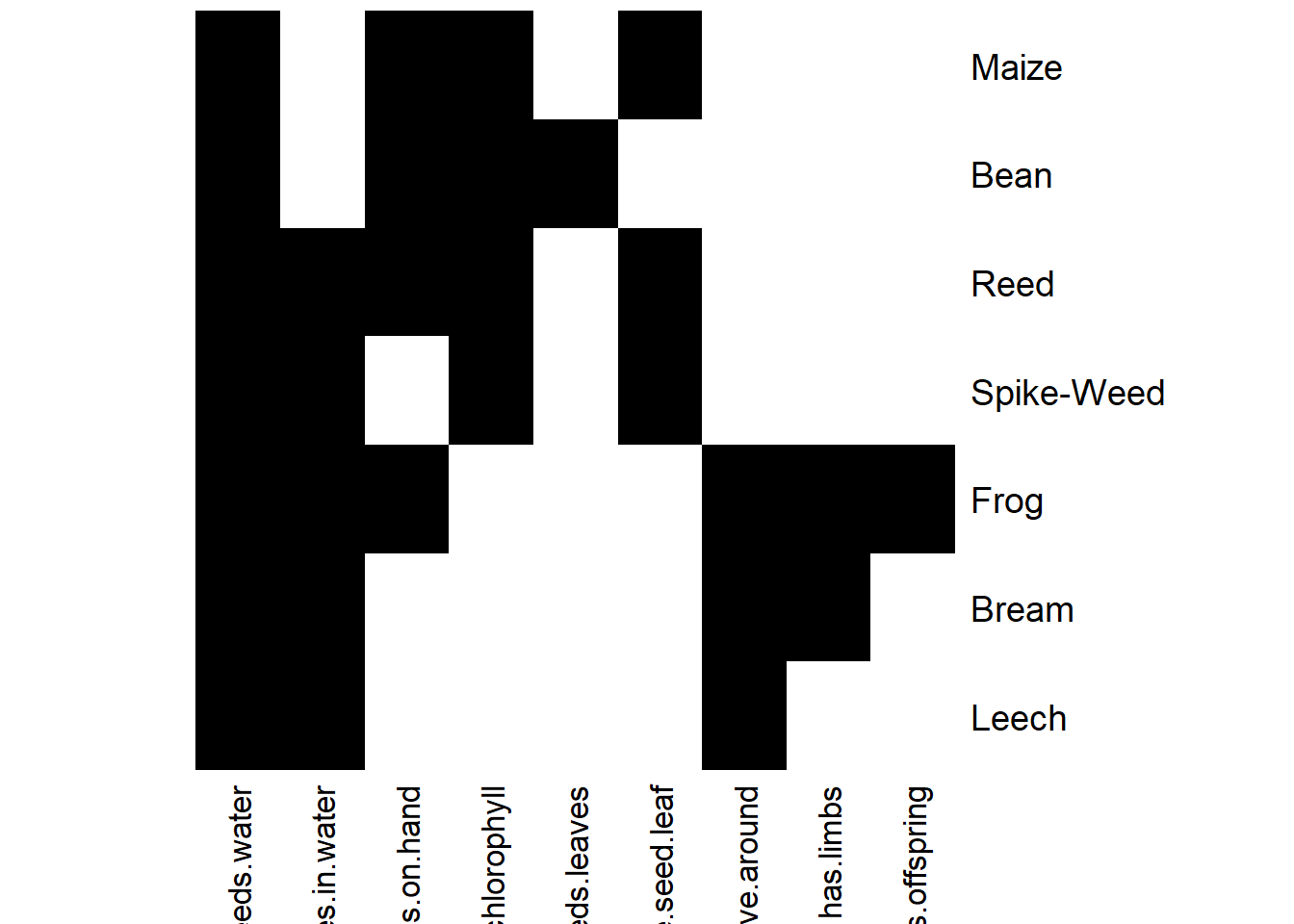
- Imprime el contexto formal (print). Haz plot también del contexto formal.
## Warning: Too many attributes, output will be truncated.## FormalContext with 7 objects and 9 attributes.
## Attributes' names are: needs.water, lives.in.water, lives.on.hand,
## needs.chlorophyll, two.seeds.leaves, one.seed.leaf, ...
## Matrix:
## needs.water lives.in.water lives.on.hand needs.chlorophyll
## Leech 1 1 0 0
## Bream 1 1 0 0
## Frog 1 1 1 0
## Spike-Weed 1 1 0 1
## Reed 1 1 1 1
## Bean 1 0 1 1
## two.seeds.leaves one.seed.leaf can.move.around
## Leech 0 0 1
## Bream 0 0 1
## Frog 0 0 1
## Spike-Weed 0 1 0
## Reed 0 1 0
## Bean 1 0 0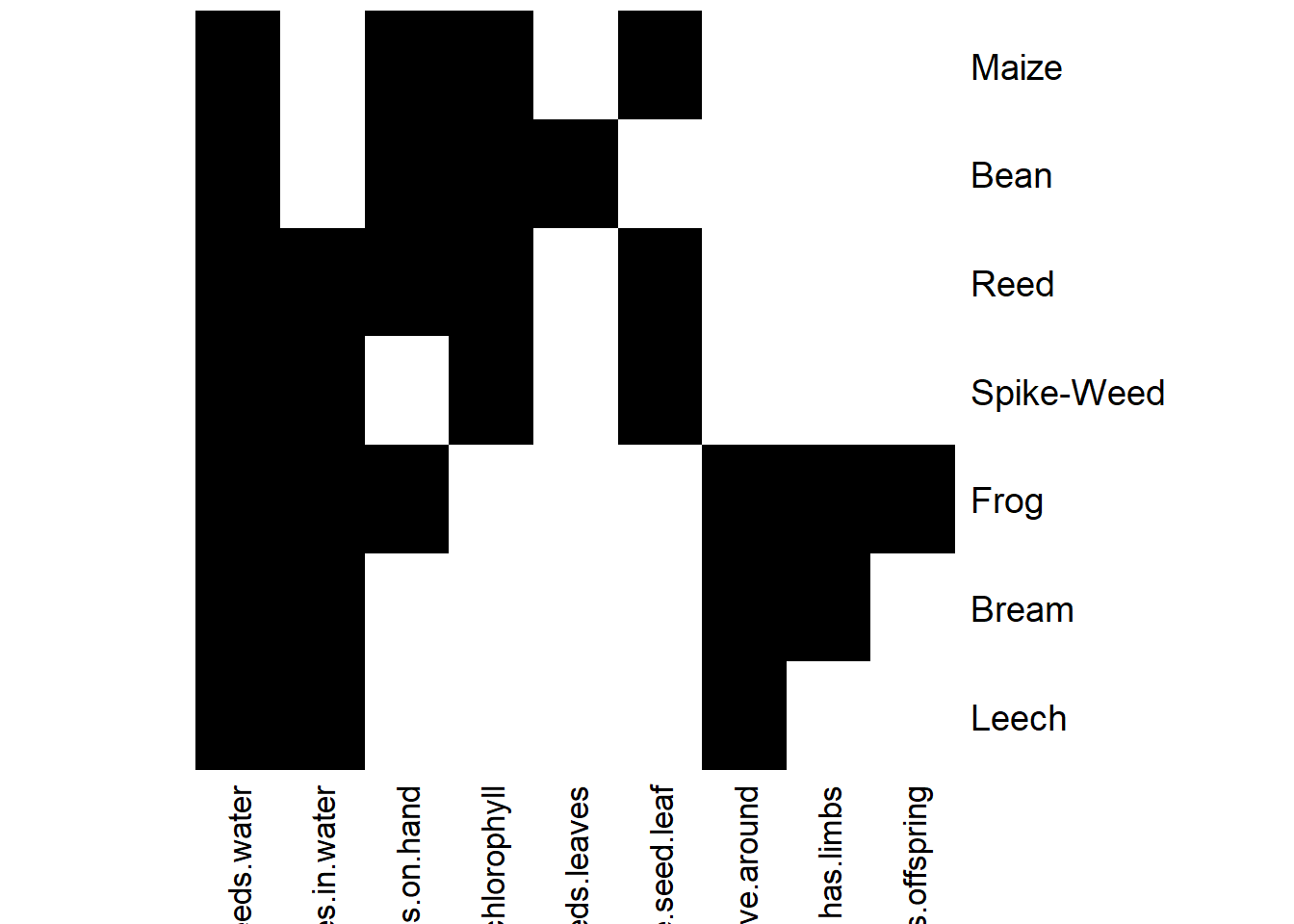
Convierte a latex el contexto formal. En el Rmd introduce el código latex del contexto formal para visualizarlo.
## \begin{table} \centering
## \begin{tabular}{lrrrrrrrrr}
## \toprule
## & needs.water & lives.in.water & lives.on.hand & needs.chlorophyll & two.seeds.leaves & one.seed.leaf & can.move.around & has.limbs & suckles.its.offspring\\
## \midrule
## Leech & 1 & 1 & 0 & 0 & 0 & 0 & 1 & 0 & 0\\
## Bream & 1 & 1 & 0 & 0 & 0 & 0 & 1 & 1 & 0\\
## Frog & 1 & 1 & 1 & 0 & 0 & 0 & 1 & 1 & 1\\
## Spike-Weed & 1 & 1 & 0 & 1 & 0 & 1 & 0 & 0 & 0\\
## Reed & 1 & 1 & 1 & 1 & 0 & 1 & 0 & 0 & 0\\
## Bean & 1 & 0 & 1 & 1 & 1 & 0 & 0 & 0 & 0\\
## Maize & 1 & 0 & 1 & 1 & 0 & 1 & 0 & 0 & 0\\
## \bottomrule
## \end{tabular} \caption{\label{}} \end{table}Guarda todos los atributos en una variable attr_ganter usando los comandos del paquete.
## [1] "needs.water" "lives.in.water" "lives.on.hand"
## [4] "needs.chlorophyll" "two.seeds.leaves" "one.seed.leaf"
## [7] "can.move.around" "has.limbs" "suckles.its.offspring"Guarda todos los objetos en una variable obj_ganter usando los comandos del paquete fCar.
## [1] "Leech" "Bream" "Frog" "Spike-Weed" "Reed"
## [6] "Bean" "Maize"¿De que tipo es la variable attr_ganter?
## [1] "character"¿De que tipo es la variable attr_objetos?
## [1] "character"Visualizando el contexto formal y utilizando los operadores de derivación, calcula dos conceptos sin usar el método que calcula todos los conceptos.
#Definimos un conjunto de atributos
concept1 <- SparseSet$new(attributes = fc_ganter$objects)
concept1$assign(Reed = 1, Frog = 1)
concept1## {Frog, Reed}## {needs.water, lives.in.water, lives.on.hand}## -----------------------------------------------------------------------------
# Define a set of objects
concept2 <- SparseSet$new(attributes = fc_ganter$attributes)
concept2$assign(needs.water = 1)
concept2## {needs.water}## {Leech, Bream, Frog, Spike-Weed, Reed, Bean, Maize}Usar método de fcaR para calcular todos los conceptos.
## A set of 15 concepts:
## 1: ({Leech, Bream, Frog, Spike-Weed, Reed, Bean, Maize}, {needs.water})
## 2: ({Spike-Weed, Reed, Bean, Maize}, {needs.water, needs.chlorophyll})
## 3: ({Spike-Weed, Reed, Maize}, {needs.water, needs.chlorophyll, one.seed.leaf})
## 4: ({Frog, Reed, Bean, Maize}, {needs.water, lives.on.hand})
## 5: ({Reed, Bean, Maize}, {needs.water, lives.on.hand, needs.chlorophyll})
## 6: ({Reed, Maize}, {needs.water, lives.on.hand, needs.chlorophyll, one.seed.leaf})
## 7: ({Bean}, {needs.water, lives.on.hand, needs.chlorophyll, two.seeds.leaves})
## 8: ({Leech, Bream, Frog, Spike-Weed, Reed}, {needs.water, lives.in.water})
## 9: ({Leech, Bream, Frog}, {needs.water, lives.in.water, can.move.around})
## 10: ({Bream, Frog}, {needs.water, lives.in.water, can.move.around, has.limbs})
## 11: ({Spike-Weed, Reed}, {needs.water, lives.in.water, needs.chlorophyll, one.seed.leaf})
## 12: ({Frog, Reed}, {needs.water, lives.in.water, lives.on.hand})
## 13: ({Frog}, {needs.water, lives.in.water, lives.on.hand, can.move.around, has.limbs, suckles.its.offspring})
## 14: ({Reed}, {needs.water, lives.in.water, lives.on.hand, needs.chlorophyll, one.seed.leaf})
## 15: ({}, {needs.water, lives.in.water, lives.on.hand, needs.chlorophyll, two.seeds.leaves, one.seed.leaf, can.move.around, has.limbs, suckles.its.offspring})¿Cuantos conceptos hemos calculado a partir del contexto formal?
15
Muestra los 10 primeros conceptos.
## ({Leech, Bream, Frog, Spike-Weed, Reed, Bean, Maize}, {needs.water})
## ({Spike-Weed, Reed, Bean, Maize}, {needs.water, needs.chlorophyll})
## ({Spike-Weed, Reed, Maize}, {needs.water, needs.chlorophyll, one.seed.leaf})
## ({Frog, Reed, Bean, Maize}, {needs.water, lives.on.hand})
## ({Reed, Bean, Maize}, {needs.water, lives.on.hand, needs.chlorophyll})
## ({Reed, Maize}, {needs.water, lives.on.hand, needs.chlorophyll, one.seed.leaf})
## ({Bean}, {needs.water, lives.on.hand, needs.chlorophyll, two.seeds.leaves})
## ({Leech, Bream, Frog, Spike-Weed, Reed}, {needs.water, lives.in.water})
## ({Leech, Bream, Frog}, {needs.water, lives.in.water, can.move.around})
## ({Bream, Frog}, {needs.water, lives.in.water, can.move.around, has.limbs})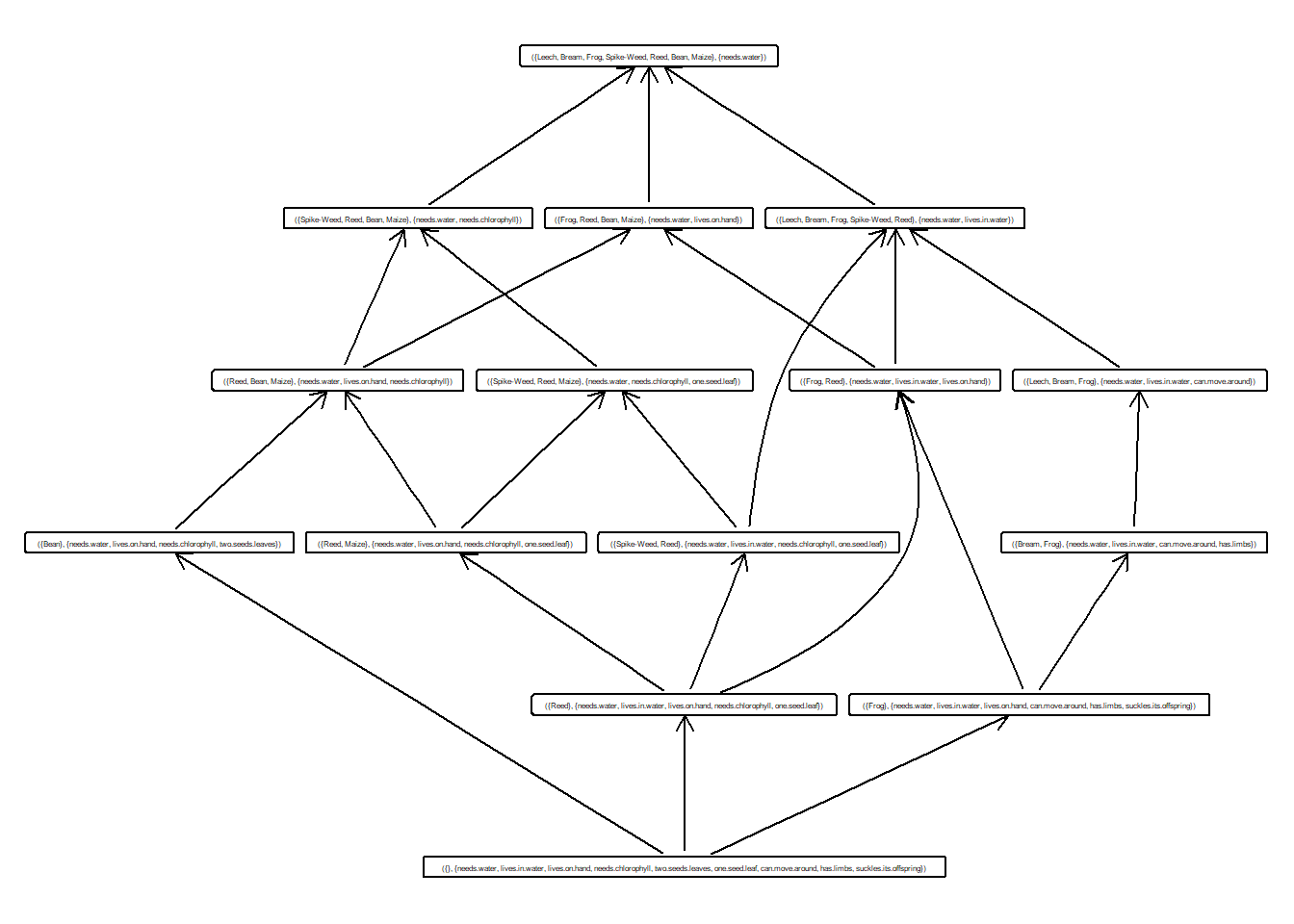
Calcular y guardar en una variable el subretículo con soporte mayor que 0.3.
## [1] 1.0000000 0.5714286 0.4285714 0.5714286 0.4285714 0.2857143 0.1428571
## [8] 0.7142857 0.4285714 0.2857143 0.2857143 0.2857143 0.1428571 0.1428571
## [15] 0.0000000## [1] 1 2 3 4 5 8 9## A set of 13 concepts:
## 1: ({Leech, Bream, Frog, Spike-Weed, Reed, Bean, Maize}, {needs.water})
## 2: ({Spike-Weed, Reed, Bean, Maize}, {needs.water, needs.chlorophyll})
## 3: ({Spike-Weed, Reed, Maize}, {needs.water, needs.chlorophyll, one.seed.leaf})
## 4: ({Frog, Reed, Bean, Maize}, {needs.water, lives.on.hand})
## 5: ({Reed, Bean, Maize}, {needs.water, lives.on.hand, needs.chlorophyll})
## 6: ({Reed, Maize}, {needs.water, lives.on.hand, needs.chlorophyll, one.seed.leaf})
## 7: ({Leech, Bream, Frog, Spike-Weed, Reed}, {needs.water, lives.in.water})
## 8: ({Leech, Bream, Frog}, {needs.water, lives.in.water, can.move.around})
## 9: ({Spike-Weed, Reed}, {needs.water, lives.in.water, needs.chlorophyll, one.seed.leaf})
## 10: ({Frog, Reed}, {needs.water, lives.in.water, lives.on.hand})
## 11: ({Frog}, {needs.water, lives.in.water, lives.on.hand, can.move.around, has.limbs, suckles.its.offspring})
## 12: ({Reed}, {needs.water, lives.in.water, lives.on.hand, needs.chlorophyll, one.seed.leaf})
## 13: ({}, {needs.water, lives.in.water, lives.on.hand, needs.chlorophyll, two.seeds.leaves, one.seed.leaf, can.move.around, has.limbs, suckles.its.offspring})Dibujar dicho subretículo.
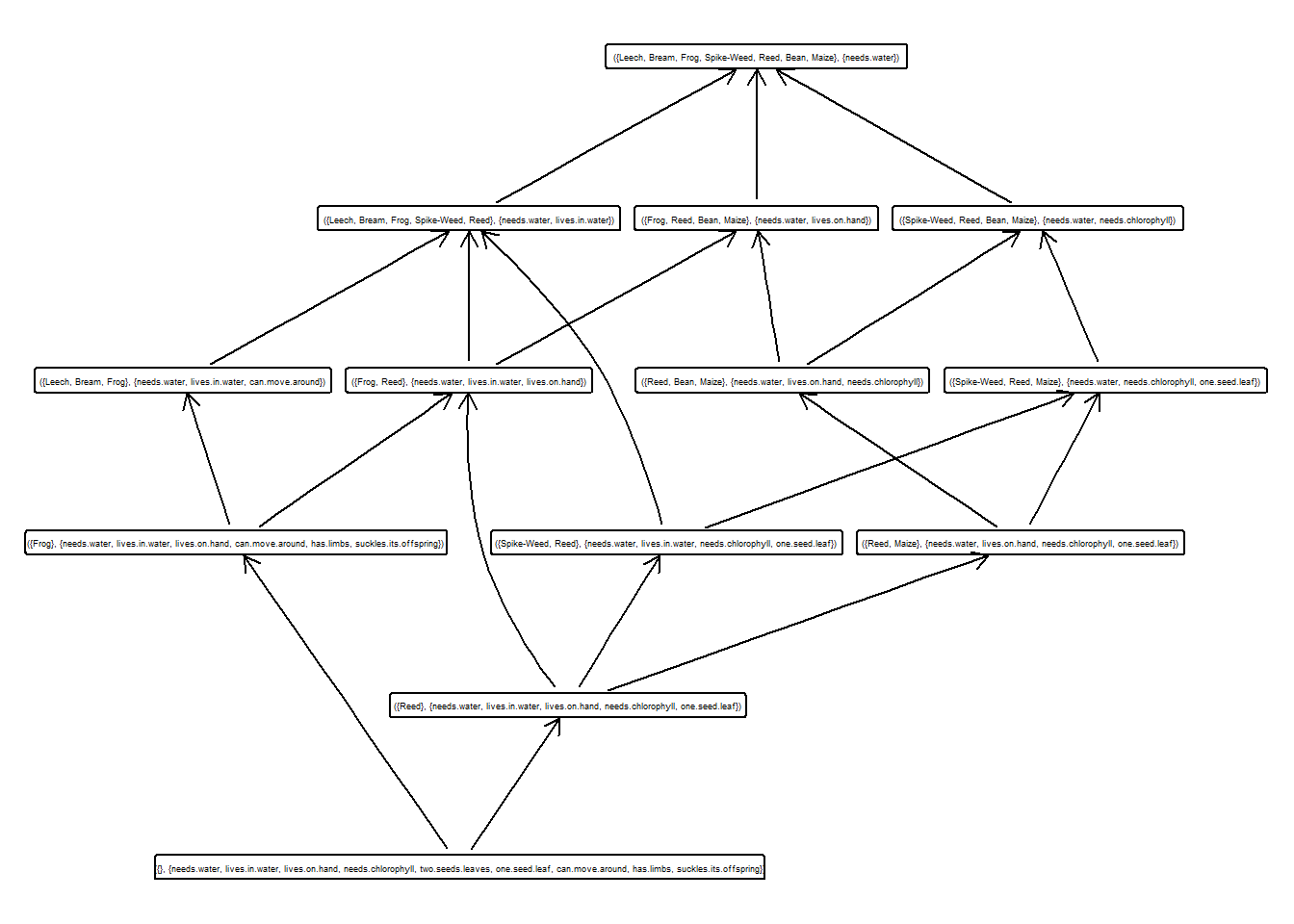
¿De que tipo es el subretículo obtenido?.
## [1] "ConceptLattice" "R6"¿Qué metodos puedes usar con el subretículo calculado? Pon ejemplos y explica.
Clone, extents, infimum, initialize, intents, is_empty, join_irreducibles, lower_neighbours, meet_irreducibles, plot, print, size, subconcepts, sublattice, superconcepts, support, supremum, to_latex, upper_neighbours
Calcula el superior y el infimo de los conceptos calculados para fc_ganter y lo mismo para el subretículo anterior. Visualizalos.
## ({Leech, Bream, Frog, Spike-Weed, Reed}, {needs.water, lives.in.water})## ({Bream, Frog}, {needs.water, lives.in.water, can.move.around, has.limbs})## ({Leech, Bream, Frog, Spike-Weed, Reed, Bean, Maize}, {needs.water})## ({Spike-Weed, Reed, Maize}, {needs.water, needs.chlorophyll, one.seed.leaf})Grabar el objeto fc_ganter en un fichero fc_ganter.rds.
Elimina la variable fc_ganter. Carga otra vez en la variable del fichero anterior y comprueba que tenemos toda la información: atributos, conceptos, etc.
fc2 <- FormalContext$new()
fc2$load("./fc.rds")
#Comprobamos que se ha cargado correctamente
fc2$plot()
Calcula lo siguientes conjuntos usando los métodos del paquete fcaR:
{Bean}′ {livesonland}′ {twoseedleaves}′ {Frog,Maize}′ {needschlorophylltoproducefood,canmovearound}′ {livesinwater,livesonland}′ {needschlorophylltoproducefood,canmovearound}′
Únicamente calculamos el primero ya que todos siguen el mismo proceso:
## {needs.water, lives.on.hand, needs.chlorophyll, two.seeds.leaves}12.2 Segunda parte, clase 06/05/20
Calcula las implicaciones del contexto
Muestra las implicaciones en pantalla
## Implication set with 10 implications.
## Rule 1: {} -> {needs.water}
## Rule 2: {needs.water, suckles.its.offspring} -> {lives.in.water,
## lives.on.hand, can.move.around, has.limbs}
## Rule 3: {needs.water, has.limbs} -> {lives.in.water, can.move.around}
## Rule 4: {needs.water, can.move.around} -> {lives.in.water}
## Rule 5: {needs.water, one.seed.leaf} -> {needs.chlorophyll}
## Rule 6: {needs.water, two.seeds.leaves} -> {lives.on.hand,
## needs.chlorophyll}
## Rule 7: {needs.water, lives.on.hand, needs.chlorophyll,
## two.seeds.leaves, one.seed.leaf} -> {lives.in.water, can.move.around,
## has.limbs, suckles.its.offspring}
## Rule 8: {needs.water, lives.in.water, needs.chlorophyll} ->
## {one.seed.leaf}
## Rule 9: {needs.water, lives.in.water, needs.chlorophyll,
## one.seed.leaf, can.move.around} -> {lives.on.hand, two.seeds.leaves,
## has.limbs, suckles.its.offspring}
## Rule 10: {needs.water, lives.in.water, lives.on.hand, can.move.around}
## -> {has.limbs, suckles.its.offspring}¿Cuantas implicaciones se han extraido?
## [1] 10Calcula el tamaño de las implicaciones y la media de la parte y derecha de dichas implicaciones.
## LHS RHS
## 2.7 2.2Aplica las reglas de la lógica de simplificación. ¿Cuántas implicaciones han aparecido tras aplicar la lógica?
fc_ganter$implications$apply_rules(rules = c("simplification"),
reorder = FALSE,
parallelize = FALSE)## Processing batch## --> simplification: from 10 to 10 in 0.14 secs.## Batch took 0.14 secs.## [1] 10## Implication set with 10 implications.
## Rule 1: {} -> {needs.water}
## Rule 2: {suckles.its.offspring} -> {lives.in.water, lives.on.hand,
## can.move.around, has.limbs}
## Rule 3: {has.limbs} -> {lives.in.water, can.move.around}
## Rule 4: {can.move.around} -> {lives.in.water}
## Rule 5: {one.seed.leaf} -> {needs.chlorophyll}
## Rule 6: {two.seeds.leaves} -> {lives.on.hand, needs.chlorophyll}
## Rule 7: {two.seeds.leaves, one.seed.leaf} -> {lives.in.water,
## can.move.around, has.limbs, suckles.its.offspring}
## Rule 8: {lives.in.water, needs.chlorophyll} -> {one.seed.leaf}
## Rule 9: {one.seed.leaf, can.move.around} -> {lives.on.hand,
## two.seeds.leaves, has.limbs, suckles.its.offspring}
## Rule 10: {lives.on.hand, can.move.around} -> {has.limbs,
## suckles.its.offspring}## LHS RHS
## 1.3 2.2Eliminar la redundancia en el conjunto de implicaciones. ¿Cuantas implicaciones han aparecido tras aplicar la lógica?
fc_ganter$implications$apply_rules(rules = c("composition",
"generalization",
"simplification"),
parallelize = FALSE)## Processing batch## --> composition: from 10 to 10 in 0 secs.## --> generalization: from 10 to 10 in 0.03 secs.## --> simplification: from 10 to 10 in 0.06 secs.## Batch took 0.09 secs.## Implication set with 10 implications.
## Rule 1: {} -> {needs.water}
## Rule 2: {suckles.its.offspring} -> {lives.in.water, lives.on.hand,
## can.move.around, has.limbs}
## Rule 3: {has.limbs} -> {lives.in.water, can.move.around}
## Rule 4: {can.move.around} -> {lives.in.water}
## Rule 5: {one.seed.leaf} -> {needs.chlorophyll}
## Rule 6: {two.seeds.leaves} -> {lives.on.hand, needs.chlorophyll}
## Rule 7: {two.seeds.leaves, one.seed.leaf} -> {lives.in.water,
## can.move.around, has.limbs, suckles.its.offspring}
## Rule 8: {lives.in.water, needs.chlorophyll} -> {one.seed.leaf}
## Rule 9: {one.seed.leaf, can.move.around} -> {lives.on.hand,
## two.seeds.leaves, has.limbs, suckles.its.offspring}
## Rule 10: {lives.on.hand, can.move.around} -> {has.limbs,
## suckles.its.offspring}## [1] 10Calcular el cierre de los atributos needs.water, one.seed.leaf.
## {needs.water, one.seed.leaf}## $closure
## {needs.water, needs.chlorophyll, one.seed.leaf}Copia (clona) el conjunto fc_ganter en una variable fc1.
Elimina la implicación que está en la primera posición:
## Implication set with 10 implications.
## Rule 1: {} -> {needs.water}
## Rule 2: {suckles.its.offspring} -> {lives.in.water, lives.on.hand,
## can.move.around, has.limbs}
## Rule 3: {has.limbs} -> {lives.in.water, can.move.around}
## Rule 4: {can.move.around} -> {lives.in.water}
## Rule 5: {one.seed.leaf} -> {needs.chlorophyll}
## Rule 6: {two.seeds.leaves} -> {lives.on.hand, needs.chlorophyll}
## Rule 7: {two.seeds.leaves, one.seed.leaf} -> {lives.in.water,
## can.move.around, has.limbs, suckles.its.offspring}
## Rule 8: {lives.in.water, needs.chlorophyll} -> {one.seed.leaf}
## Rule 9: {one.seed.leaf, can.move.around} -> {lives.on.hand,
## two.seeds.leaves, has.limbs, suckles.its.offspring}
## Rule 10: {lives.on.hand, can.move.around} -> {has.limbs,
## suckles.its.offspring}Extrae de todas las implicaciones la que tengan en el lado izquierdo de la implicación el atributo one.seed.leaf.
## Implication set with 9 implications.
## Rule 1: {suckles.its.offspring} -> {lives.in.water, lives.on.hand,
## can.move.around, has.limbs}
## Rule 2: {has.limbs} -> {lives.in.water, can.move.around}
## Rule 3: {can.move.around} -> {lives.in.water}
## Rule 4: {one.seed.leaf} -> {needs.chlorophyll}
## Rule 5: {two.seeds.leaves} -> {lives.on.hand, needs.chlorophyll}
## Rule 6: {two.seeds.leaves, one.seed.leaf} -> {lives.in.water,
## can.move.around, has.limbs, suckles.its.offspring}
## Rule 7: {lives.in.water, needs.chlorophyll} -> {one.seed.leaf}
## Rule 8: {one.seed.leaf, can.move.around} -> {lives.on.hand,
## two.seeds.leaves, has.limbs, suckles.its.offspring}
## Rule 9: {lives.on.hand, can.move.around} -> {has.limbs,
## suckles.its.offspring}## Implication set with 3 implications.
## Rule 1: {one.seed.leaf} -> {needs.chlorophyll}
## Rule 2: {two.seeds.leaves, one.seed.leaf} -> {lives.in.water,
## can.move.around, has.limbs, suckles.its.offspring}
## Rule 3: {one.seed.leaf, can.move.around} -> {lives.on.hand,
## two.seeds.leaves, has.limbs, suckles.its.offspring}## Implication set with 9 implications.
## Rule 1: {suckles.its.offspring} -> {lives.in.water, lives.on.hand,
## can.move.around, has.limbs}
## Rule 2: {has.limbs} -> {lives.in.water, can.move.around}
## Rule 3: {can.move.around} -> {lives.in.water}
## Rule 4: {one.seed.leaf} -> {needs.chlorophyll}
## Rule 5: {two.seeds.leaves} -> {lives.on.hand, needs.chlorophyll}
## Rule 6: {two.seeds.leaves, one.seed.leaf} -> {lives.in.water,
## can.move.around, has.limbs, suckles.its.offspring}
## Rule 7: {lives.in.water, needs.chlorophyll} -> {one.seed.leaf}
## Rule 8: {one.seed.leaf, can.move.around} -> {lives.on.hand,
## two.seeds.leaves, has.limbs, suckles.its.offspring}
## Rule 9: {lives.on.hand, can.move.around} -> {has.limbs,
## suckles.its.offspring}Obtén los atributos que aparezcan en las implicaciones.
## [1] "needs.water" "lives.in.water" "lives.on.hand"
## [4] "needs.chlorophyll" "two.seeds.leaves" "one.seed.leaf"
## [7] "can.move.around" "has.limbs" "suckles.its.offspring"Calcula el soporte de la implicación 3.
## [1] 0.1111111 0.1111111 0.3333333 0.3333333 0.2222222 0.1111111 0.1111111
## [8] 0.1111111 0.1111111## [1] 0.3333333