Documento 23 Apuntes Clase
23.1 C1 - 21-02
Instalamos los siguientes paquetes:
- arules
- devtools
- https://github.com/neuroimaginador/fcaR
## [1] "ts"## Time-Series [1:144] from 1949 to 1961: 112 118 132 129 121 135 148 148 136 119 ...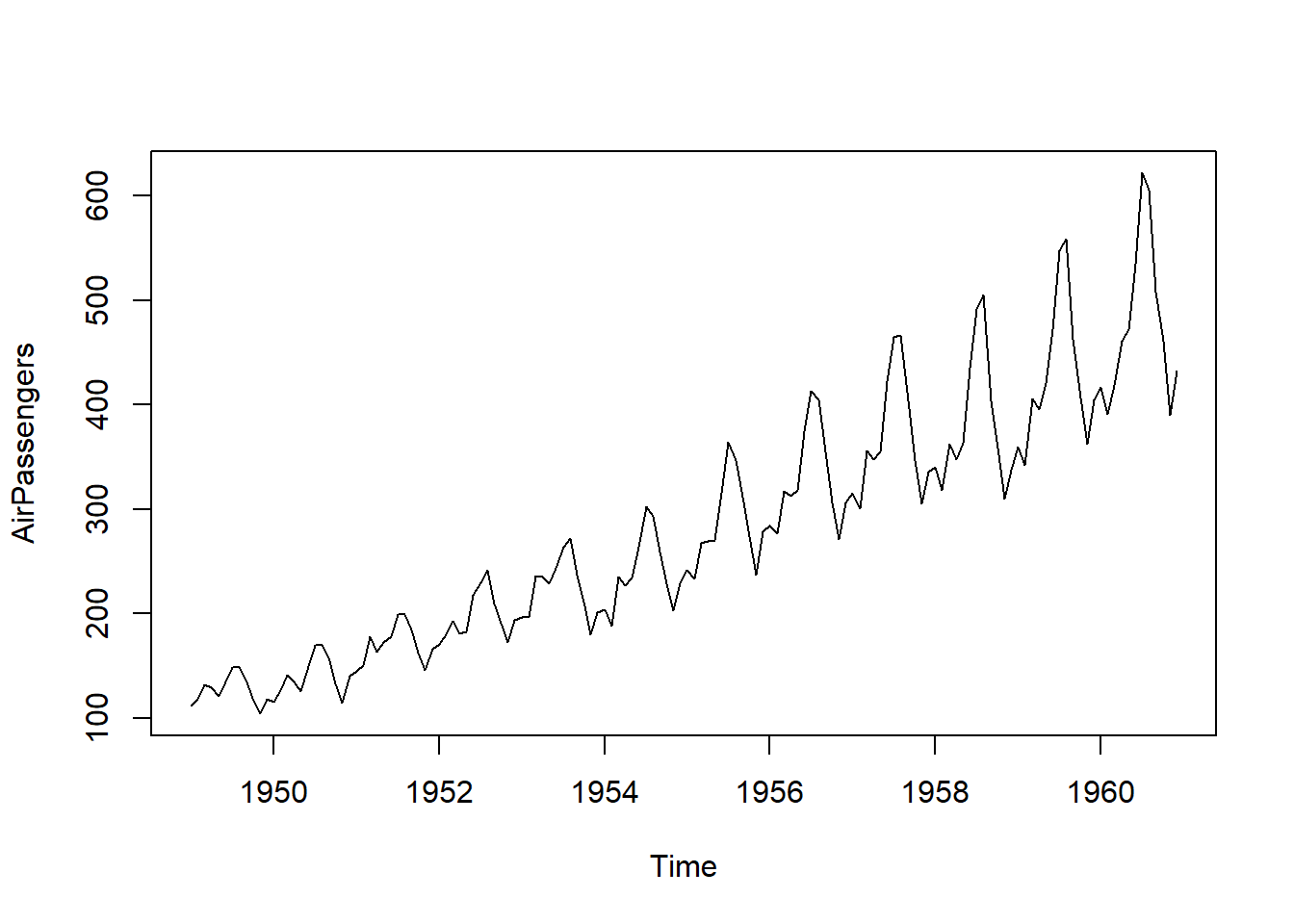
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 104.0 180.0 265.5 280.3 360.5 622.0## [1] "_book" "_bookdown.yml"
## [3] "_bookdown_files" "_output.yml"
## [5] "01-Vectores.Rmd" "02-DFyFunciones.Rmd"
## [7] "03-breastCancer.Rmd" "04-ApplyFunctions.Rmd"
## [9] "05-cuestion1.Rmd" "06-cuestion2.Rmd"
## [11] "07-DTpackage.Rmd" "08-Visualización.Rmd"
## [13] "09-Soporte_Confianza.Rmd" "10-Iris.Rmd"
## [15] "11-Online_dataset.Rmd" "12-TutorialGanter.Rmd"
## [17] "13-Groceries_Prueba.Rmd" "14-Proyecto_Futbin.Rmd"
## [19] "15-Examen_Reglas_fcaR.Rmd" "16-EjemploRegresion.Rmd"
## [21] "17-FuelEfficiency_dataset.Rmd" "18-FancyDataset.Rmd"
## [23] "19-ExamenRegresion.Rmd" "22-Homicide_PDF.Rmd"
## [25] "23-Analyze_Tweets.Rmd" "24-Tweets_tidytext.Rmd"
## [27] "30-Apuntes-Clase.Rmd" "book.bib"
## [29] "context.csv" "datasets"
## [31] "fc.rds" "homicide-pdfs"
## [33] "index.Rmd" "IPG-LabComp.Rmd"
## [35] "IPG-LabComp.Rproj" "IPG-LabComp_files"
## [37] "packages.bib" "preamble.tex"
## [39] "rsconnect" "style.css"## [1] "a1" "a2"
## [3] "Adult" "AirPassengers"
## [5] "asoc" "asociaciones"
## [7] "attr_ganter" "aux1"
## [9] "C" "cancer"
## [11] "color" "color_pal1"
## [13] "colors" "colors1"
## [15] "computer_support_confidence" "computer_suppport_confidence"
## [17] "concept1" "concept2"
## [19] "conj" "consumo.de.gas"
## [21] "context" "context.no.sparness"
## [23] "corpus" "d"
## [25] "d_" "d1"
## [27] "d2" "d3"
## [29] "dataset" "dataset_ganter"
## [31] "df.asoc" "df1"
## [33] "directorio.textos" "divido.matriz"
## [35] "docsCorpus" "dsIris1"
## [37] "dsIris2" "dsIris3"
## [39] "dtm" "duplicados"
## [41] "e1" "e2"
## [43] "e3" "e4"
## [45] "e5" "e7"
## [47] "ej20" "elems"
## [49] "encodeSource" "f"
## [51] "f1" "f2"
## [53] "famosos" "fc"
## [55] "FC" "fc_2"
## [57] "fc_ganter" "fc1"
## [59] "fc2" "fitting1"
## [61] "frecuencia_items" "frecuencias.df"
## [63] "freq" "freq2"
## [65] "freqdsIris3" "frequent_terms"
## [67] "FuelEfficiency" "g1"
## [69] "groc" "HairEyeColor"
## [71] "hollywood" "idxNoRed"
## [73] "idxRed" "ind"
## [75] "indices" "interc"
## [77] "iris" "iris2"
## [79] "iset" "label.att"
## [81] "label.obj" "landdata.states"
## [83] "landdata_states" "leaf_lhs"
## [85] "list.files" "lista.asoc"
## [87] "lista.compra.usuarios" "lista1"
## [89] "m" "M"
## [91] "m_cuad" "m1"
## [93] "M1" "M1.2"
## [95] "M1.3" "m2"
## [97] "M2" "m3"
## [99] "m4" "marketing"
## [101] "matrix_tdm" "maximo"
## [103] "meanV2" "media"
## [105] "minimo" "mis.Plantas"
## [107] "mis_reglas" "mis_tokens"
## [109] "mis_tokens_afinn" "mis_tweets"
## [111] "model_cuadratic" "model_exp"
## [113] "model_linear" "msleep"
## [115] "mtcars" "mu"
## [117] "mush" "Mushroom"
## [119] "my_corpus" "my_tdm"
## [121] "nombres" "nu"
## [123] "obj_ganter" "OjosAzules"
## [125] "online" "onlinePlot"
## [127] "ord" "palabras"
## [129] "pizzagate" "pizzagateSinRt"
## [131] "porc" "putOnes"
## [133] "putZeros" "r"
## [135] "r1" "r1.noredunt"
## [137] "r1.ordenado" "r1.sort"
## [139] "r2" "r3"
## [141] "r4" "rbv"
## [143] "redundantes" "reg1"
## [145] "reglas" "reglas_redundantes"
## [147] "reglas_redundantes2" "reglas2"
## [149] "regs" "res"
## [151] "rh" "rInterseccion"
## [153] "rules.ord" "rules.sub"
## [155] "rules.sub_2" "rules.sub_3"
## [157] "rules2" "rules3"
## [159] "rUnion" "s"
## [161] "S" "sentiment_mis_tokens_bing"
## [163] "sentiment_mis_tokens_nrc" "sihayNA"
## [165] "simbols" "sizes"
## [167] "sleep" "slope"
## [169] "stop_words" "subreticulo"
## [171] "subset.matrix" "sumV2"
## [173] "tabla_tweets" "termino"
## [175] "test" "Textprocessing"
## [177] "texts" "Tlista.compra.usuarios"
## [179] "to_TDM" "toSpace"
## [181] "toString" "TSS"
## [183] "tweets" "UKgas"
## [185] "v2" "v3"
## [187] "v4" "v5"
## [189] "v7.2" "vals1"
## [191] "vals2" "vals3"
## [193] "vals4" "vals5"
## [195] "vals6" "vals7"
## [197] "ventas" "ventas_pol2"
## [199] "ventas_youtube" "ventas2"
## [201] "vRed"Ejemplo de función:
arearectangulo <- function(base, altura){
res <- base*altura
return(res)
} #end function
area = arearectangulo(5,4)
area## [1] 20Asignación de variables y operadores:
## [1] "integer"## [1] "numeric"## [1] 6## [1] "integer"## [1] NaN## [1] Inf## [1] 2.5## [1] NA## [1] NA# Si no sabes un dato no lo puedes sumar o hacer cualquier operación que dependa de ella
mean(c(1,2,3,4,NA), na.rm = TRUE)## [1] 2.5Vectores:
## [1] "character"# Carácter tiene más prioridad que numérico
# Convierte todos los valores del vector a carácter
v1 <- c(1,2,3)
n1 <- c("a1", "a2", "a3")
names(v1) <- n1
v1## a1 a2 a3
## 1 2 3## a2
## 2## a2
## 2## <NA>
## NA## [1] 7 6 5 0 7 6 5 NA## [1] 7 6 5 6 7 6 5## [1] TRUEReciclaje
## [1] 5 4 7 6## [1] 1 4 5 1# Dentro del corchete me permite crear un vector de los elementos a los que quiero acceder
v6 <- 1:10
v6 [-c(1, length(v6))]## [1] 2 3 4 5 6 7 8 9## [1] 3 6 9 12 1523.2 C2 - 26-02
## [1] 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0 1.1 1.2 1.3 1.4
## [16] 1.5 1.6 1.7 1.8 1.9 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9
## [31] 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 3.9 4.0 4.1 4.2 4.3 4.4
## [46] 4.5 4.6 4.7 4.8 4.9 5.0 5.1 5.2 5.3 5.4 5.5 5.6 5.7 5.8 5.9
## [61] 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 6.8 6.9 7.0 7.1 7.2 7.3 7.4
## [76] 7.5 7.6 7.7 7.8 7.9 8.0 8.1 8.2 8.3 8.4 8.5 8.6 8.7 8.8 8.9
## [91] 9.0 9.1 9.2 9.3 9.4 9.5 9.6 9.7 9.8 9.9 10.0## [1] 11 11 11 11 11 11 11 11 11 11## [1] 10 18 24 28 30 30 28 24 18 10## [1] FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE## [1] TRUE## [1] 4## [1] NA 1 4 9 16 25 36 49 64 81 100 150## [1] TRUE## [1] TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE## [1] 1## [1] TRUE## [1] TRUE## [1] FALSEFactor: vector que además de los valores, almacena qué posibles posibles valores puede tener ese valores.
## [1] "Bueno" "Muy bueno" "Regular"## [1] Malo Muy malo Regular
## Levels: Muy malo Malo Regular Bien Muy bien## Warning in Ops.factor(v1[1], v1[2]): '<' not meaningful for factors## [1] NA# No tiene sentido usar menor que o similares en factors
v1 <- ordered(c("Malo", "Regular"), levels = c("Muy malo", "Malo", "Regular", "Bien", "Muy bien"))
v1## [1] Malo Regular
## Levels: Muy malo < Malo < Regular < Bien < Muy bien## [1] TRUELISTAS
## [1] 1 2 3 4## [[1]]
## [1] 1
##
## [[2]]
## [1] 2
##
## [[3]]
## [[3]][[1]]
## [1] 3
##
## [[3]][[2]]
## [1] 4# Si hago v1$ no me sale nada porque no tengo un nombre asociado
v1 <- list(n1=1,n2=2, lista1=list(3,4))
v1## $n1
## [1] 1
##
## $n2
## [1] 2
##
## $lista1
## $lista1[[1]]
## [1] 3
##
## $lista1[[2]]
## [1] 4## [1] 1## [[1]]
## [1] 3
##
## [[2]]
## [1] 4## [1] 3## [[1]]
## [1] 3## num 3## List of 1
## $ : num 3Datasets:
## [1] "data.frame"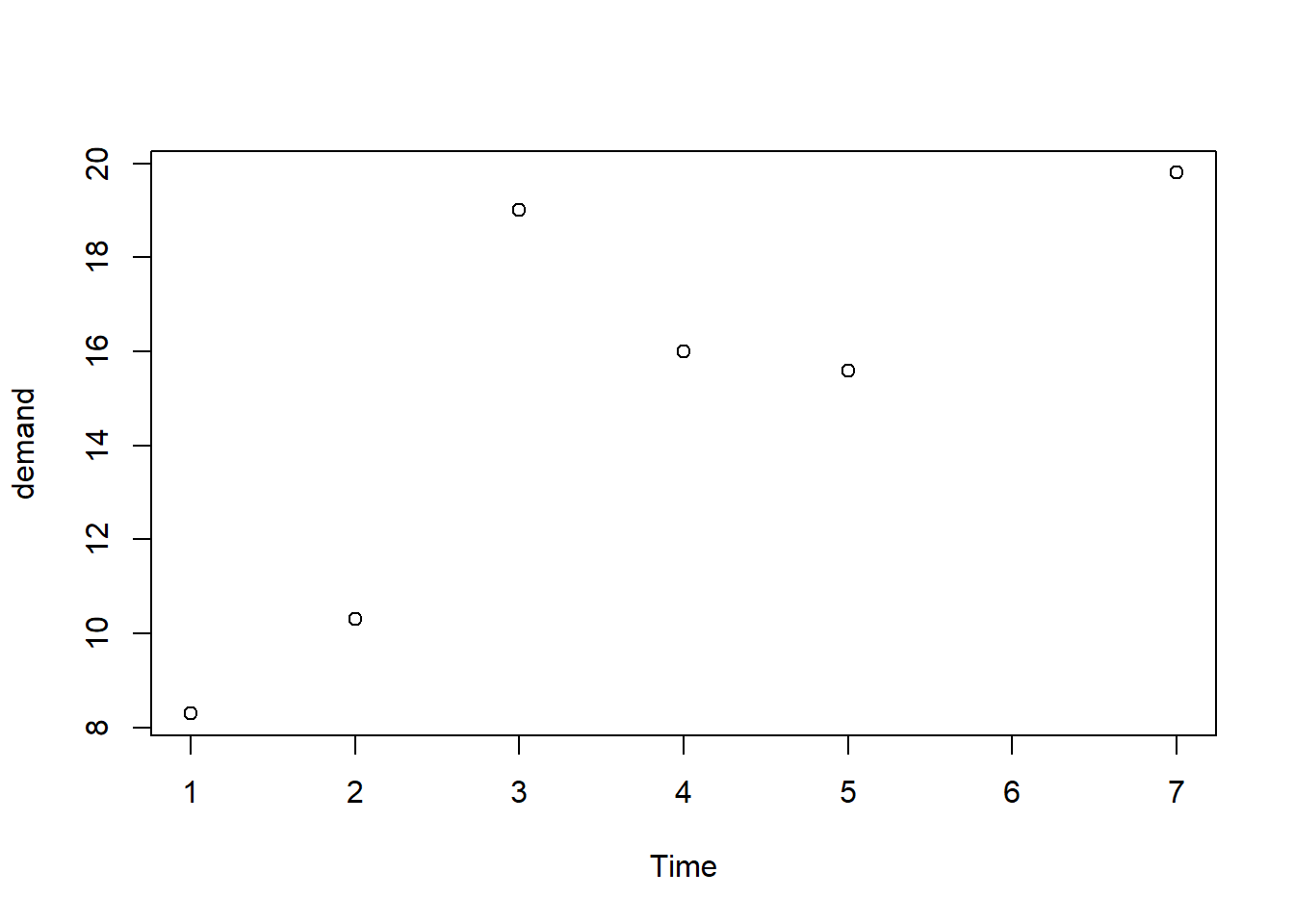
#Hacemos una regresión
f1 <- lm(demand~Time, data = BOD) # Variable y frente la x (Demand frente Time)
f1##
## Call:
## lm(formula = demand ~ Time, data = BOD)
##
## Coefficients:
## (Intercept) Time
## 8.521 1.721## List of 12
## $ coefficients : Named num [1:2] 8.52 1.72
## ..- attr(*, "names")= chr [1:2] "(Intercept)" "Time"
## $ residuals : Named num [1:6] -1.943 -1.664 5.314 0.593 -1.529 ...
## ..- attr(*, "names")= chr [1:6] "1" "2" "3" "4" ...
## $ effects : Named num [1:6] -36.334 8.315 5.857 0.943 -1.371 ...
## ..- attr(*, "names")= chr [1:6] "(Intercept)" "Time" "" "" ...
## $ rank : int 2
## $ fitted.values: Named num [1:6] 10.2 12 13.7 15.4 17.1 ...
## ..- attr(*, "names")= chr [1:6] "1" "2" "3" "4" ...
## $ assign : int [1:2] 0 1
## $ qr :List of 5
## ..$ qr : num [1:6, 1:2] -2.449 0.408 0.408 0.408 0.408 ...
## .. ..- attr(*, "dimnames")=List of 2
## .. .. ..$ : chr [1:6] "1" "2" "3" "4" ...
## .. .. ..$ : chr [1:2] "(Intercept)" "Time"
## .. ..- attr(*, "assign")= int [1:2] 0 1
## ..$ qraux: num [1:2] 1.41 1.18
## ..$ pivot: int [1:2] 1 2
## ..$ tol : num 1e-07
## ..$ rank : int 2
## ..- attr(*, "class")= chr "qr"
## $ df.residual : int 4
## $ xlevels : Named list()
## $ call : language lm(formula = demand ~ Time, data = BOD)
## $ terms :Classes 'terms', 'formula' language demand ~ Time
## .. ..- attr(*, "variables")= language list(demand, Time)
## .. ..- attr(*, "factors")= int [1:2, 1] 0 1
## .. .. ..- attr(*, "dimnames")=List of 2
## .. .. .. ..$ : chr [1:2] "demand" "Time"
## .. .. .. ..$ : chr "Time"
## .. ..- attr(*, "term.labels")= chr "Time"
## .. ..- attr(*, "order")= int 1
## .. ..- attr(*, "intercept")= int 1
## .. ..- attr(*, "response")= int 1
## .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
## .. ..- attr(*, "predvars")= language list(demand, Time)
## .. ..- attr(*, "dataClasses")= Named chr [1:2] "numeric" "numeric"
## .. .. ..- attr(*, "names")= chr [1:2] "demand" "Time"
## $ model :'data.frame': 6 obs. of 2 variables:
## ..$ demand: num [1:6] 8.3 10.3 19 16 15.6 19.8
## ..$ Time : num [1:6] 1 2 3 4 5 7
## ..- attr(*, "terms")=Classes 'terms', 'formula' language demand ~ Time
## .. .. ..- attr(*, "variables")= language list(demand, Time)
## .. .. ..- attr(*, "factors")= int [1:2, 1] 0 1
## .. .. .. ..- attr(*, "dimnames")=List of 2
## .. .. .. .. ..$ : chr [1:2] "demand" "Time"
## .. .. .. .. ..$ : chr "Time"
## .. .. ..- attr(*, "term.labels")= chr "Time"
## .. .. ..- attr(*, "order")= int 1
## .. .. ..- attr(*, "intercept")= int 1
## .. .. ..- attr(*, "response")= int 1
## .. .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
## .. .. ..- attr(*, "predvars")= language list(demand, Time)
## .. .. ..- attr(*, "dataClasses")= Named chr [1:2] "numeric" "numeric"
## .. .. .. ..- attr(*, "names")= chr [1:2] "demand" "Time"
## - attr(*, "class")= chr "lm"## (Intercept) Time
## 8.521429 1.721429## 1 2 3 4 5 6
## -1.9428571 -1.6642857 5.3142857 0.5928571 -1.5285714 -0.7714286## Named num 8.52
## - attr(*, "names")= chr "(Intercept)"## num 8.52NOTA: Comando unlist aplana todo
MATRICES
## [,1]
## [1,] 1
## [2,] 2
## [3,] 3
## [4,] 4
## [5,] 5
## [6,] 6
## [7,] 7
## [8,] 8
## [9,] 9
## [10,] 10
## [11,] 11
## [12,] 12## [,1] [,2] [,3] [,4]
## [1,] 1 4 7 10
## [2,] 2 5 8 11
## [3,] 3 6 9 12## [,1] [,2] [,3] [,4]
## [1,] 1 4 7 10
## [2,] 2 5 8 11
## [3,] 3 6 9 12## Warning in matrix(1:13, nrow = 3): la longitud de los datos [13] no es un
## submúltiplo o múltiplo del número de filas [3] en la matriz## [,1] [,2] [,3] [,4] [,5]
## [1,] 1 4 7 10 13
## [2,] 2 5 8 11 1
## [3,] 3 6 9 12 2## Warning in matrix(1:13, nrow = 3, byrow = TRUE): la longitud de los datos [13]
## no es un submúltiplo o múltiplo del número de filas [3] en la matriz## [,1] [,2] [,3] [,4] [,5]
## [1,] 1 2 3 4 5
## [2,] 6 7 8 9 10
## [3,] 11 12 13 1 2## [1] 6## [1] 8## [1] 10## [1] 2 10 5## [,1] [,2]
## [1,] 2 3
## [2,] 12 13## [,1] [,2] [,3] [,4] [,5]
## [1,] 1 2 3 4 5
## [2,] 6 7 8 9 10
## [3,] 11 12 13 1 2## [,1] [,2] [,3] [,4] [,5]
## [1,] FALSE FALSE FALSE FALSE FALSE
## [2,] FALSE FALSE FALSE TRUE TRUE
## [3,] TRUE TRUE TRUE FALSE FALSE## [1] 11 12 13 9 10## [1] 3 6 9 11 14## [1] 11 12 13 9 10Operando con DATA FRAMES
## Time demand
## 1 1 8.3
## 2 2 10.3
## 3 3 19.0
## 4 4 16.0
## 5 5 15.6
## 6 7 19.8## [1] 1 2 3 4 5 7## [1] 8.3 10.3 19.0 16.0 15.6 19.8## [1] 3## [1] 3## [1] 8.3## NULL## Time demand
## 1 1 8.3## [1] "data.frame"## [1] "numeric"## [1] 1 2 3 4 5 7## [1] 1 2 3 4 5 7## Time
## 1 1
## 2 2
## 3 3
## 4 4
## 5 5
## 6 7## [1] "data.frame"## Time demand
## 1 1 8.3
## 2 2 10.3
## 3 3 19.0
## 4 4 16.0
## 5 5 15.6## Time demand
## 1 1 8.3
## 2 2 10.3
## 3 3 19.0
## 6 7 19.8## Time demand
## 1 1 8.3
## 2 2 10.3
## 3 3 19.0
## 4 4 16.0
## 5 5 15.6
## 6 7 19.8## demand
## 1 8.3
## 2 10.3
## 3 19.0
## 4 16.0
## 5 15.6
## 6 19.8# Por defecto, cuando tú le metes un vector de caracteres
# a un dataframe te lo convierte a factora
# Para evitar esto, hay que usar:
# data.frame(stringsAsFactors = )
data(BOD)
f1 <- BOD[1:2, ]
f1## Time demand
## 1 1 8.3
## 2 2 10.3## Time demand
## 2 2 10.3
## 3 3 19.0
## 4 4 16.0
## 5 5 15.6
## 6 7 19.8## Time demand
## 1 1 8.3
## 2 2 10.3
## 21 2 10.3
## 3 3 19.0
## 4 4 16.0
## 5 5 15.6
## 6 7 19.8# Tabla de contingencias: posibles valores, frecuencias absolutas y relativas,...
# table()
# USO DEL FOR
v <- (1:10)^2+3
for(k in 1:length(v)){
print(v[k])
}## [1] 4
## [1] 7
## [1] 12
## [1] 19
## [1] 28
## [1] 39
## [1] 52
## [1] 67
## [1] 84
## [1] 103## NULL
## NULL## [1] 0## integer(0)## NULL
## NULL## [1] "v1" "v2" "v3"## [1] 1 2 3# Nos dice las posiciones de los elementos
# USAMOS EL FOR ASÍ, NO DA PROBLEMAS
v <- (1:10)^2+3
for(k in seq_along(v)){
print(v[k])
}## [1] 4
## [1] 7
## [1] 12
## [1] 19
## [1] 28
## [1] 39
## [1] 52
## [1] 67
## [1] 84
## [1] 10323.3 C3 04-03
Para crear un .Rmd
New rmarkdown - presentattion - pdf(beamer)
Hemos empezado la clase haciendo la P1(voluntaria), nos ha enseñado a utilizar books, ahí tenemos que poner la información de cada temas + prácticas
La teoría es LCC_presentatiomn: Github/ejemplo
23.4 C4 06-03
Intalamos dplyr y readr, que están dentro de tidyverse
dplyr::filter Se usa para del paquete dplyr ejecutar la función filter.
Existen 5 funciones clave en dplyr:
- filter()
- arrange()
- select()
- mutate()
- summarise()
Cargamos los paquetes y el dataset:
FILTER
## # A tibble: 6 x 13
## name height mass hair_color skin_color eye_color birth_year gender homeworld
## <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
## 1 Chew~ 228 112 brown unknown blue 200 male Kashyyyk
## 2 Han ~ 180 80 brown fair brown 29 male Corellia
## 3 Jek ~ 180 110 brown fair blue NA male Bestine ~
## 4 Qui-~ 193 89 brown fair blue 92 male <NA>
## 5 Tarf~ 234 136 brown brown blue NA male Kashyyyk
## 6 Raym~ 188 79 brown light brown NA male Alderaan
## # ... with 4 more variables: species <chr>, films <list>, vehicles <list>,
## # starships <list>## # A tibble: 6 x 13
## name height mass hair_color skin_color eye_color birth_year gender homeworld
## <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
## 1 Chew~ 228 112 brown unknown blue 200 male Kashyyyk
## 2 Han ~ 180 80 brown fair brown 29 male Corellia
## 3 Jek ~ 180 110 brown fair blue NA male Bestine ~
## 4 Qui-~ 193 89 brown fair blue 92 male <NA>
## 5 Tarf~ 234 136 brown brown blue NA male Kashyyyk
## 6 Raym~ 188 79 brown light brown NA male Alderaan
## # ... with 4 more variables: species <chr>, films <list>, vehicles <list>,
## # starships <list>## # A tibble: 44 x 13
## name height mass hair_color skin_color eye_color birth_year gender
## <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr>
## 1 Dart~ 202 136 none white yellow 41.9 male
## 2 Leia~ 150 49 brown light brown 19 female
## 3 Owen~ 178 120 brown, gr~ light blue 52 male
## 4 Beru~ 165 75 brown light blue 47 female
## 5 Bigg~ 183 84 black light brown 24 male
## 6 Anak~ 188 84 blond fair blue 41.9 male
## 7 Chew~ 228 112 brown unknown blue 200 male
## 8 Han ~ 180 80 brown fair brown 29 male
## 9 Jabb~ 175 1358 <NA> green-tan~ orange 600 herma~
## 10 Wedg~ 170 77 brown fair hazel 21 male
## # ... with 34 more rows, and 5 more variables: homeworld <chr>, species <chr>,
## # films <list>, vehicles <list>, starships <list>## # A tibble: 73 x 13
## name height mass hair_color skin_color eye_color birth_year gender
## <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr>
## 1 Luke~ 172 77 blond fair blue 19 male
## 2 C-3PO 167 75 <NA> gold yellow 112 <NA>
## 3 R2-D2 96 32 <NA> white, bl~ red 33 <NA>
## 4 Dart~ 202 136 none white yellow 41.9 male
## 5 Leia~ 150 49 brown light brown 19 female
## 6 Owen~ 178 120 brown, gr~ light blue 52 male
## 7 Beru~ 165 75 brown light blue 47 female
## 8 R5-D4 97 32 <NA> white, red red NA <NA>
## 9 Bigg~ 183 84 black light brown 24 male
## 10 Obi-~ 182 77 auburn, w~ fair blue-gray 57 male
## # ... with 63 more rows, and 5 more variables: homeworld <chr>, species <chr>,
## # films <list>, vehicles <list>, starships <list>Operador %>%
Viene de los pipelines de linux, que sirven para conectar dispositivos
d1 <- starwars %>% filter(!(mass > 78 | hair_color == "brown")) # Equivalente a (4)
# En visualización trabajar así.
# Si falla el comando %>%: library(magrittr)
# Ahora combinamos select
d1 <- starwars %>%
filter(!(mass > 78 | hair_color == "brown")) %>%
select(name, height, mass)
d1## # A tibble: 18 x 3
## name height mass
## <chr> <int> <dbl>
## 1 Luke Skywalker 172 77
## 2 Obi-Wan Kenobi 182 77
## 3 Yoda 66 17
## 4 Palpatine 170 75
## 5 Nien Nunb 160 68
## 6 Jar Jar Binks 196 66
## 7 Sebulba 112 40
## 8 Ayla Secura 178 55
## 9 Dud Bolt 94 45
## 10 Ben Quadinaros 163 65
## 11 Adi Gallia 184 50
## 12 Luminara Unduli 170 56.2
## 13 Barriss Offee 166 50
## 14 Zam Wesell 168 55
## 15 Ratts Tyerell 79 15
## 16 Wat Tambor 193 48
## 17 Shaak Ti 178 57
## 18 Sly Moore 178 48## # A tibble: 18 x 5
## name height mass hair_color skin_color
## <chr> <int> <dbl> <chr> <chr>
## 1 Luke Skywalker 172 77 blond fair
## 2 Obi-Wan Kenobi 182 77 auburn, white fair
## 3 Yoda 66 17 white green
## 4 Palpatine 170 75 grey pale
## 5 Nien Nunb 160 68 none grey
## 6 Jar Jar Binks 196 66 none orange
## 7 Sebulba 112 40 none grey, red
## 8 Ayla Secura 178 55 none blue
## 9 Dud Bolt 94 45 none blue, grey
## 10 Ben Quadinaros 163 65 none grey, green, yellow
## 11 Adi Gallia 184 50 none dark
## 12 Luminara Unduli 170 56.2 black yellow
## 13 Barriss Offee 166 50 black yellow
## 14 Zam Wesell 168 55 blonde fair, green, yellow
## 15 Ratts Tyerell 79 15 none grey, blue
## 16 Wat Tambor 193 48 none green, grey
## 17 Shaak Ti 178 57 none red, blue, white
## 18 Sly Moore 178 48 none pale- Otras: Mutate, Transmute, Arrange,
Top_n
Ordenar por grados y quiero mostrar los x primeros (ej diapositivas)
dplyr devuelve tibble filas x cols, que es un derivado de dataframes.
En este formato, en la salida te devuelve a parte del nombre, el tipo de la columna justo debajo
Ahora, decargamos los ejercicios de dplyr y realizamos dpylr_breast-cancer guion
23.5 C5 11-03
Explicación de las funciones apply. URL: Dealing with apply function
- Comparativa con apply()
¿Cuánto se puede reducir el tiempo de ejecución entre usar una y otra? Entre 2 y 3 veces en muchos casos.
Paquete profvis: sirve para analizar cuellos de botella.
Ejemplo funciones Apply:
## [,1] [,2] [,3]
## [1,] 1 4 7
## [2,] 2 5 8
## [3,] 3 6 9## [1] 6 7 8## [1] 6 7 8# Aquí vemos que la función se puede definir externa o internamente
ratings <- 1:10
result <- lapply(ratings, mean)
result## [[1]]
## [1] 1
##
## [[2]]
## [1] 2
##
## [[3]]
## [1] 3
##
## [[4]]
## [1] 4
##
## [[5]]
## [1] 5
##
## [[6]]
## [1] 6
##
## [[7]]
## [1] 7
##
## [[8]]
## [1] 8
##
## [[9]]
## [1] 9
##
## [[10]]
## [1] 10# Hace la media de cada número y te lo devuelve en forma de lista
# Si tuviéramos una lista de vectores si resultaría útil
calif <- list(as1=c(3,7,9,10),
as2=c(4,7,8,6))
result <- lapply(calif, mean)
result## $as1
## [1] 7.25
##
## $as2
## [1] 6.25Diferencia entre sapply y lapply? Lo que devuelven.
## as1 as2
## 7.25 6.25## as1
## 7.25Hacer lapply exercises.
23.6 C6 18-03
Filosofia para realización de graficos: http://motioninsocial.com/tufte/
Enlace para observar graficas de R sobre el COVID 19 https://elpais.com/sociedad/2020/03/17/actualidad/1584436648_230452.html
Visualizacion Buscar ‘Boxplot lenguaje R’, sirve para mostrar distriucion de valores, así como la media y valores anómalos. Tambien podemos realizar directamente summary().
Como analizar los valores: repetidos, como se comportan, si se distribuyen con una normal - EXAMEN
Funcion curve() para poder representar funciones matematicas.
R por defecto trae el paquete plot para representaciones graficas, pero nosotros usaremos el paquete ggplot2. A continuacion un ejemplo de ggplot:
library(gcookbook)
library(ggplot2)
g1 <- ggplot(BOD, aes(x=Time, y=demand)) + geom_point(aes(color = demand))
g1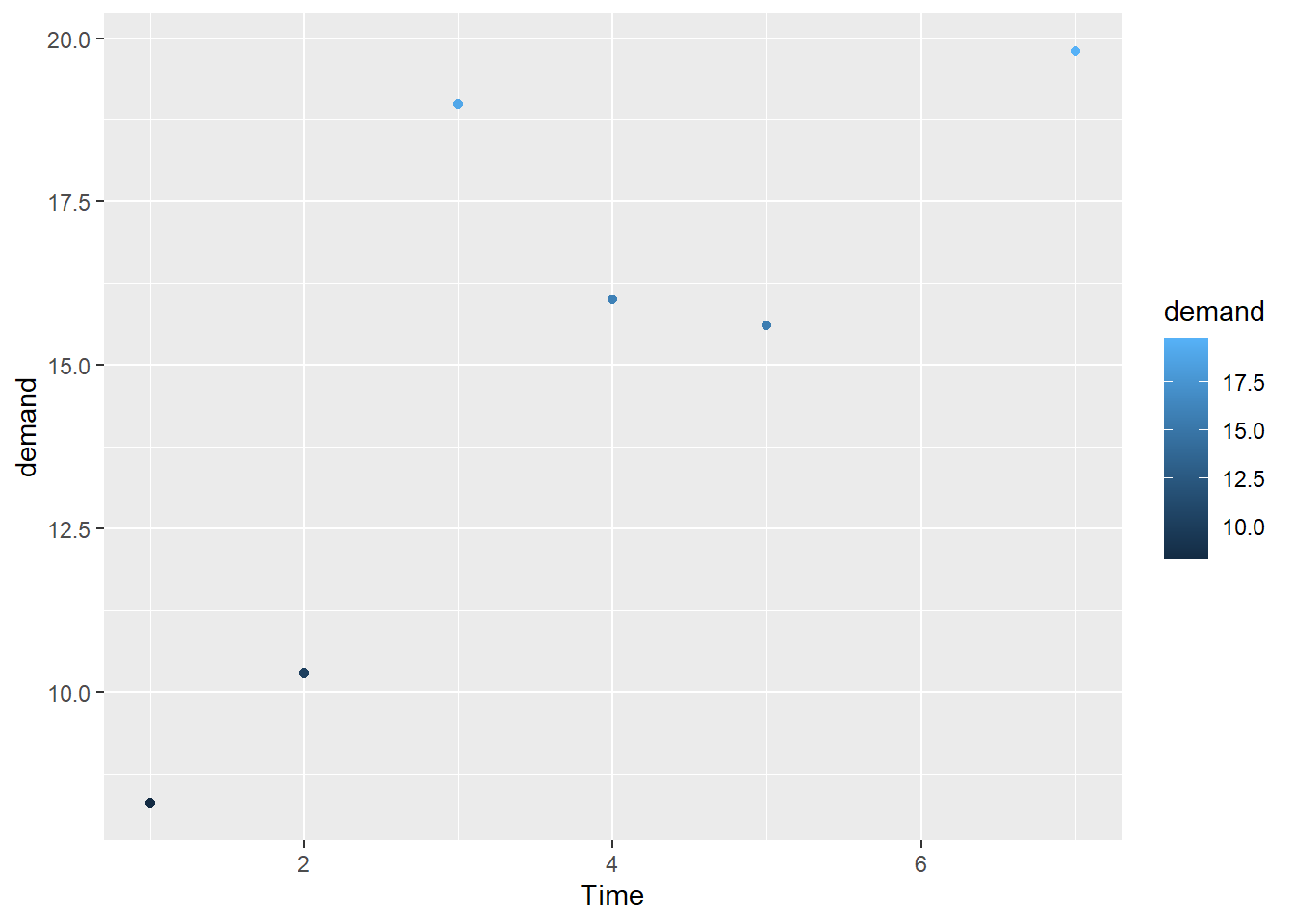
Con la funcion aes() defines lo que puedes ver, es decir, lo que va en el eje X, el eje Y, la forma, el tamaño, el color…
library(magrittr)
library(dplyr)
landdata_states <- read.csv("Datasets/landdata-states.csv")
hp2001Q1 <- landdata_states %>%
filter(Date == 2001.25)
p1 <- ggplot(hp2001Q1, aes(x = log(Land.Value), y = Structure.Cost))
p1 + geom_point(aes(color=Home.Value)) + geom_smooth()## `geom_smooth()` using method = 'loess' and formula 'y ~ x'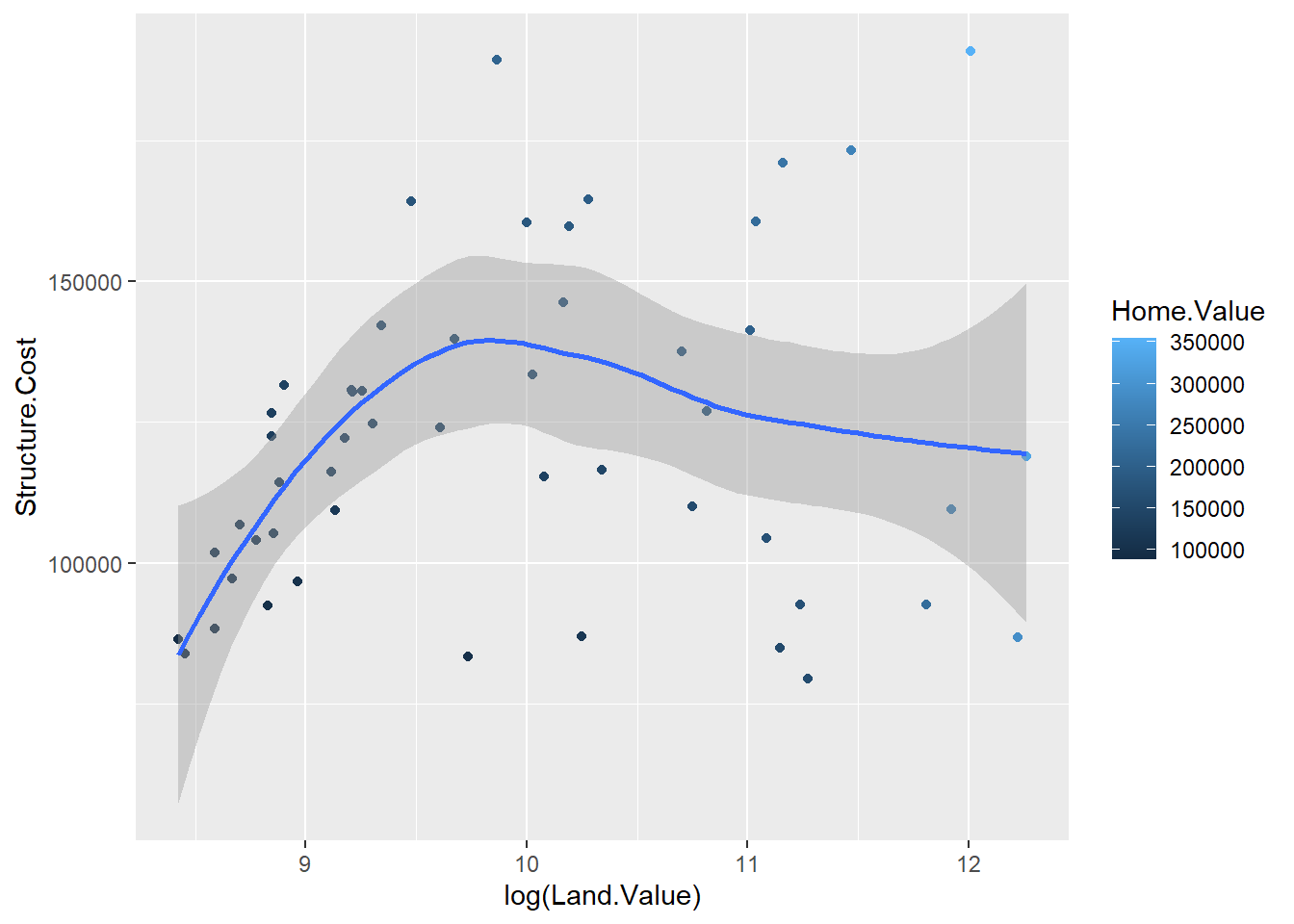
## Warning: Removed 1 rows containing missing values (geom_point).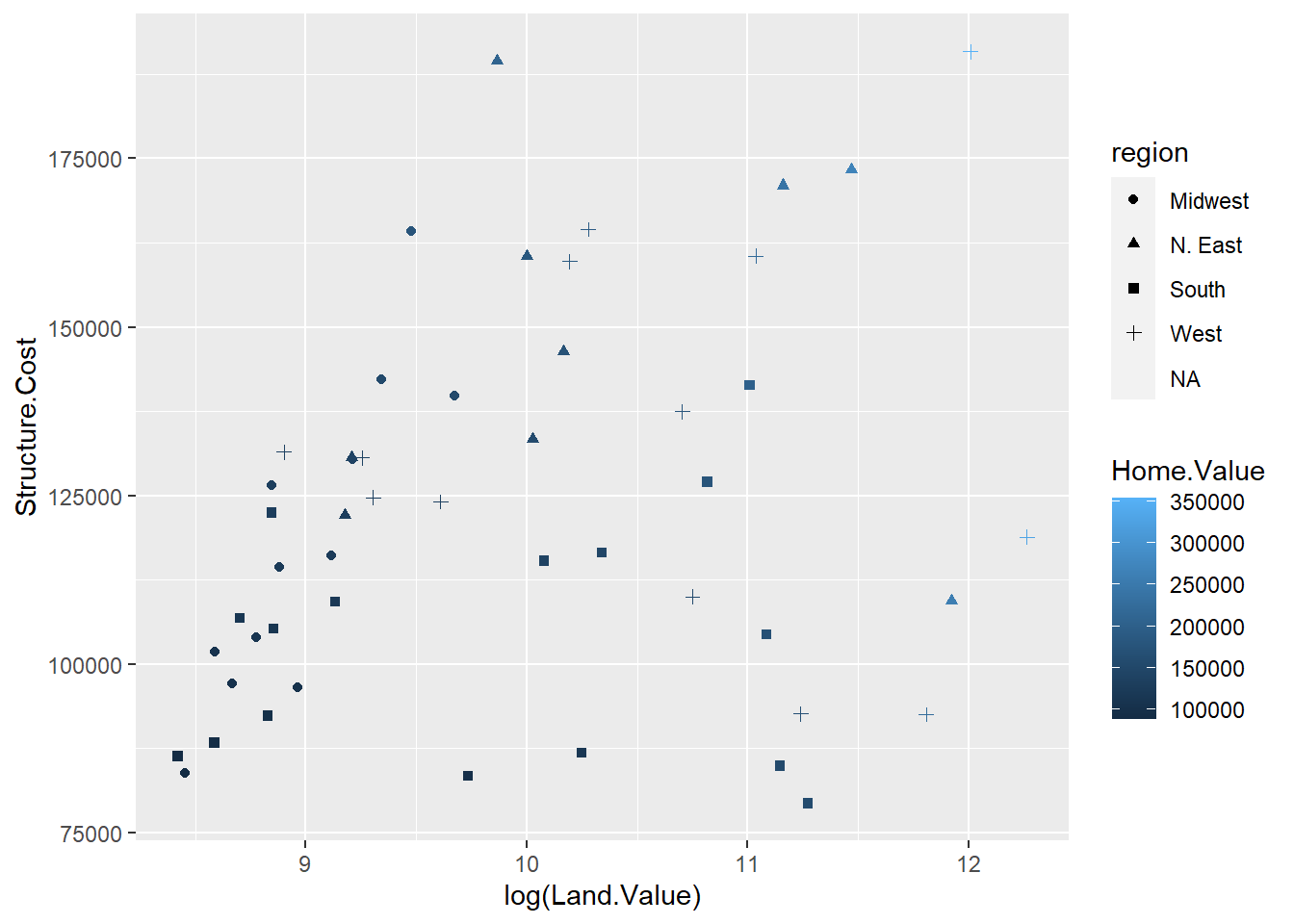
log sirve para linealizar los valores.
Importante: Hay ciertos problemas con los graficos de barra si la variable no es continua, por ejemplo, una vairable de numeros reales, porque podria haber infinitos valores, por lo que hay que convertir dichas variables en Factor, y luego usar stat=“identity” dentro de geom_bar: x=factor(Time) y geom_bar(stat=“identity”). Transparencia 22.
Aplicacion shiny con R detras: https://scitilab.shinyapps.io/Covid19/
aggregate permite la agregacion de una variable respecto de otra aplicando una funcion determinada.
23.7 C8 20-03
Soporte y Confianza:
Soporte de leche y pan
soporte(leche, pan) = casos favorables/casos posi = 2/5 = 0.4
soporte(leche,pan,mantequilla) = casos favorables/casos posi = 1/5 = 0.2
confianza (leche pan –> mantequilla) = 0.2/0.4 = 0.5
soporte(leche,pan,mantequilla) = 4/5 MUY FRECUENTE leche, pan MUY FRECUENTE
Cualquier subconjunto de muy frecuente será muy frecuente
Usaremos el paquete arules: apriori function
# install.packages("arules")
library(arules)
# inspect para ver reglas, no se puede hacer un view
data("Groceries")
reglas <- apriori(Groceries)## Apriori
##
## Parameter specification:
## confidence minval smax arem aval originalSupport maxtime support minlen
## 0.8 0.1 1 none FALSE TRUE 5 0.1 1
## maxlen target ext
## 10 rules FALSE
##
## Algorithmic control:
## filter tree heap memopt load sort verbose
## 0.1 TRUE TRUE FALSE TRUE 2 TRUE
##
## Absolute minimum support count: 983
##
## set item appearances ...[0 item(s)] done [0.00s].
## set transactions ...[169 item(s), 9835 transaction(s)] done [0.01s].
## sorting and recoding items ... [8 item(s)] done [0.00s].
## creating transaction tree ... done [0.00s].
## checking subsets of size 1 2 done [0.00s].
## writing ... [0 rule(s)] done [0.00s].
## creating S4 object ... done [0.00s].## Apriori
##
## Parameter specification:
## confidence minval smax arem aval originalSupport maxtime support minlen
## 0.3 0.1 1 none FALSE TRUE 5 0.1 1
## maxlen target ext
## 10 rules FALSE
##
## Algorithmic control:
## filter tree heap memopt load sort verbose
## 0.1 TRUE TRUE FALSE TRUE 2 TRUE
##
## Absolute minimum support count: 983
##
## set item appearances ...[0 item(s)] done [0.00s].
## set transactions ...[169 item(s), 9835 transaction(s)] done [0.00s].
## sorting and recoding items ... [8 item(s)] done [0.00s].
## creating transaction tree ... done [0.00s].
## checking subsets of size 1 2 done [0.00s].
## writing ... [0 rule(s)] done [0.00s].
## creating S4 object ... done [0.00s].SOBRE TRABAJO EN GRUPO
Análisis de datos -> proyecto kaggle
Buscar info también de competición de covid.
Hacer trabajovisualización, tipo covid.
Tutorial de ggplot aplicando nuevas técnicas.
23.8 Apuntes pre-examen (1)
## [1] "ts"# View(UKgas)
# vemos que es una ts (serie temporal), queremos un vector
consumo.de.gas <- as.vector(UKgas)
class(consumo.de.gas)## [1] "numeric"## [1] 160.1 129.7 84.8 120.1 160.1 124.9 84.8 116.9 169.7 140.9
## [11] 89.7 123.3 187.3 144.1 92.9 120.1 176.1 147.3 89.7 123.3
## [21] 185.7 155.3 99.3 131.3 200.1 161.7 102.5 136.1 204.9 176.1
## [31] 112.1 140.9 227.3 195.3 115.3 142.5 244.9 214.5 118.5 153.7
## [41] 244.9 216.1 188.9 142.5 301.0 196.9 136.1 267.3 317.0 230.5
## [51] 152.1 336.2 371.4 240.1 158.5 355.4 449.9 286.6 179.3 403.4
## [61] 491.5 321.8 177.7 409.8 593.9 329.8 176.1 483.5 584.3 395.4
## [71] 187.3 485.1 669.2 421.0 216.1 509.1 827.7 467.5 209.7 542.7
## [81] 840.5 414.6 217.7 670.8 848.5 437.0 209.7 701.2 925.3 443.4
## [91] 214.5 683.6 917.3 515.5 224.1 694.8 989.4 477.1 233.7 730.0
## [101] 1087.0 534.7 281.8 787.6 1163.9 613.1 347.4 782.8## [1] 129.7 120.1 124.9 84.8 116.9 169.7 140.9 89.7 123.3 187.3 144.1 92.9
## [13] 120.1 176.1 147.3 89.7 123.3 185.7 155.3 99.3 131.3 200.1 161.7 102.5
## [25] 136.1 204.9 176.1 112.1 140.9 227.3 195.3 115.3 142.5 244.9 214.5 118.5
## [37] 153.7 244.9 216.1 188.9 142.5 301.0 196.9 136.1 267.3 317.0 230.5## [1] FALSE## [1] FALSE FALSE FALSE FALSE## [1] TRUE FALSE FALSE## [1] 0## [1] 2## [1] TRUE## Warning in matrix(1:10, 3, 3, byrow = TRUE): la longitud de los datos [10] no es
## un submúltiplo o múltiplo del número de filas [3] en la matriz## [,1] [,2] [,3]
## [1,] 1 2 3
## [2,] 4 5 6
## [3,] 7 8 9## X1 X2 X3
## 12 15 18## [1] 6 15 24## X1 X2 X3
## 4 5 6## [1] 0 25 50 75 100## [1] 1 2 1 2 1 2## [1] 0 1 4 9 16 25 36 49 64 81 100 1000## [1] NA NA 1 4 9 16 25 36 49 64 81 100 150## [1] TRUE## [1] FALSE## [1] 1 2## [1] 1.0 3.0 1.2 2.0 1.0 2.0 5.0 6.0 3.0 2.0## [1] 1.0 1.0 1.2 2.0 2.0 2.0 3.0 3.0 5.0 6.0## [1] 2.0 3.0 6.0 5.0 2.0 1.0 2.0 1.2 3.0 1.0## [1] 1.0 3.0 1.2 2.0 5.0 6.0## [1] 2.0 -1.8 0.8 -1.0 1.0 3.0 1.0 -3.0 -1.0## [1] 1 5 3 4 6 10 2 9 7 8## [1] 1 2 3 5 4## [1] 1 2 3 5 4## [1] 1 2 3 4 5# x2[is.na(x2)] <- 0 # Sustituimos NA por 0. CUIDADO
v1 <- 1:10
# M1 <- append(M1,c(0,0,0), after=0)
# M1 <- matrix(M1, 4,3, byrow = TRUE)
# M1
v1 <- append(v1,5.5,after=2)
v1## [1] 1.0 2.0 5.5 3.0 4.0 5.0 6.0 7.0 8.0 9.0 10.0alumno1 <- list(nombre = "Luis", no.asignaturas = 4,
nombre.asignaturas = c("Lab1", "Lab2", "Lab3"))
L <- list(list(c(1,2,3), 4), 5)
unlist(L)## [1] 1 2 3 4 5## [,1] [,2] [,3]
## f1 1 2 3
## f2 4 5 6
## f3 7 8 9## f1 f2 f3
## [1,] 1 4 7
## [2,] 2 5 8
## [3,] 3 6 9## alumnos edad
## 1 Luis 21
## 2 Antonio 20
## 3 Daniel 22## alumnos ciudad
## 1 Luis Madrid
## 2 Daniel Córdoba
## 3 Ángel Málaga## alumnos edad ciudad
## 1 Daniel 22 Córdoba
## 2 Luis 21 Madrid## [,1] [,2] [,3]
## [1,] 1 4 7
## [2,] 2 5 8
## [3,] 3 6 9## [1] 4 5 6## [1] "numeric"## [1] 6 15 24## [1] "integer"result <- apply(m1,1,cumsum) #cumulative sum of elements for each row
result #by default column-wise order## [,1] [,2] [,3]
## [1,] 1 2 3
## [2,] 5 7 9
## [3,] 12 15 18## [1] "matrix"## [,1] [,2] [,3]
## [1,] 1 5 12
## [2,] 2 7 15
## [3,] 3 9 18#user defined function
check<-function(x){
return(x[x>5])
}
result <- apply(m1,1,check) #user defined function as an argument
result## [[1]]
## [1] 7
##
## [[2]]
## [1] 8
##
## [[3]]
## [1] 6 9## [1] "list"#case 2. data frame as an input
ratings <- c(4.2, 4.4, 3.4, 3.9, 5, 4.1, 3.2, 3.9, 4.6, 4.8, 5, 4, 4.5, 3.9, 4.7, 3.6)
employee.mat <- matrix(ratings,byrow=TRUE,nrow=4,dimnames = list(c("Quarter1","Quarter2","Quarter3","Quarter4"),c("Hari","Shri","John","Albert")))
employee <- as.data.frame(employee.mat)
employee## Hari Shri John Albert
## Quarter1 4.2 4.4 3.4 3.9
## Quarter2 5.0 4.1 3.2 3.9
## Quarter3 4.6 4.8 5.0 4.0
## Quarter4 4.5 3.9 4.7 3.6## Hari Shri John Albert
## 18.3 17.2 16.3 15.4## [1] "numeric"result <- apply(employee,1,cumsum) #cumulative sum of elements for each row
result #by default column-wise order## Quarter1 Quarter2 Quarter3 Quarter4
## Hari 4.2 5.0 4.6 4.5
## Shri 8.6 9.1 9.4 8.4
## John 12.0 12.3 14.4 13.1
## Albert 15.9 16.2 18.4 16.7## [1] "matrix"#user defined function
check<-function(x){
return(x[x>4.2])
}
result <- apply(employee,2,check) #user defined function as an argument
result## $Hari
## Quarter2 Quarter3 Quarter4
## 5.0 4.6 4.5
##
## $Shri
## Quarter1 Quarter3
## 4.4 4.8
##
## $John
## Quarter3 Quarter4
## 5.0 4.7
##
## $Albert
## named numeric(0)## [1] "list"