5.3 Worked Example
Helene Wagner and Max Hesselbarth
1. Overview of Worked Example
5.3.0.1 a. Background on spatial packages in R
There has been a lot of development recently in R regarding object types for geospatial data. We are currently in a transition period, where some packages still expect the “old” object types (based e.g. on packages raster and sp), others expect the “new” object types (based e.g. on packages terra and sf), and some accept both. This vignette uses the newer packages terra and sf. The bonus vignette includes code for converting between sf and sp, and between terra and raster.
Further resources:
Intro GIS with R: to learn about GIS functionality using R, https://bookdown.org/michael_bcalles/gis-crash-course-in-r/data.html#vector-data
R Tutorial: For an introductory tutorial on spatial data analysis with R, which explains object types in more detail than this worked example, see: https://rpubs.com/jguelat/fdsfd
Advanced: For a thorough resource on spatial data analysis with R, see this excellent Gitbook: https://geocompr.robinlovelace.net/index.html.
b. Goals
This worked example shows:
- How to import spatial coordinates and site attributes as spatially referenced data.
- How to plot raster data in R and overlay sampling locations.
- How to calculate landscape metrics.
- How to extract landscape data at sampling locations and within a buffer around them.
Try modifying the code to import your own data!
c. Data set
This code builds on data and code from the GeNetIt package by Jeff Evans and Melanie Murphy. Landscape metrics will be calculated with the landscapemetrics package described in: Hesselbarth et al. (2019), Ecography 42: 1648-1657.
This code uses landscape data and spatial coordinates from 30 locations where Colombia spotted frogs (Rana luteiventris) were sampled for the full data set analyzed by Funk et al. (2005) and Murphy et al. (2010). Please see the separate introduction to the data set.
ralu.site:sfobject with UTM coordinates (zone 11) and 17 site variables for 31 sites. The data are included in the ‘GeNetIt’ package, for meta data type: ?ralu.site
We will extract values at sampling point locations and within a local neighborhood (buffer) from six raster maps (see Murphy et al. 2010 for definitions), which are included with the GeNetIt package as a SpatialPixelsDataFrame called ‘rasters’:
- cti: Compound Topographic Index (“wetness”)
- err27: Elevation Relief Ratio
- ffp: Frost Free Period
- gsp: Growing Season Precipitation
- hli: Heat Load Index
- nlcd: USGS Landcover (categorical map)
2. Import site data from .csv file
a. Import data into an sf object
The site data are already in an sf object named ralu.site that comes with the package GeNetIt. Use data(ralu.site) to load it. This will create an object ralu.site.
## [1] "sf" "data.frame"To demonstrate how to create an sf object from two data frames (one with the coordinates and one with the attribute data), we’ll extract these data frames from the sf object ralu.site and then recreate the sf object (we’ll call it Sites) from the two data frames.
We can extract the coordinates with the function st_coordinates:
## X Y
## [1,] 688816.6 5003207
## [2,] 688494.4 4999093
## [3,] 687938.4 5000223
## [4,] 689732.8 5002522
## [5,] 690104.0 4999355
## [6,] 688742.5 4997481Question: What are the variable names for the spatial coordinates?
Similarly, we can drop the geometry (spatial information) from the sf object to reduce it to a data frame:
## [1] "data.frame"Now we can create an sf object again. Here, we:
- combine the two data frames
DataandCoordinatesinto a single data frame with functiondata.frame, - use the function
st_as_sffrom thesfpackage to convert this data frame to ansfobject, - tell R that the variables with the coordinates are called “X” and “Y”.
Sites <- data.frame(Data, Coordinates)
Sites.sf <- st_as_sf(Sites, coords=c("X", "Y"))
head(Sites.sf)## Simple feature collection with 6 features and 17 fields
## Geometry type: POINT
## Dimension: XY
## Bounding box: xmin: 687938.4 ymin: 4997481 xmax: 690104 ymax: 5003207
## CRS: NA
## SiteName Drainage Basin Substrate
## 1 AirplaneLake ShipIslandCreek Sheepeater Silt
## 2 BachelorMeadow WilsonCreek Skyhigh Silt
## 3 BarkingFoxLake WaterfallCreek Terrace Silt
## 4 BirdbillLake ClearCreek Birdbill Sand
## 5 BobLake WilsonCreek Harbor Silt
## 6 CacheLake WilsonCreek Skyhigh Silt
## NWI AREA_m2 PERI_m Depth_m TDS FISH ACB AUC
## 1 Lacustrine 62582.2 1142.8 21.64 2.5 1 0 0.411
## 2 Riverine_Intermittent_Streambed 225.0 60.0 0.40 0.0 0 0 0.000
## 3 Lacustrine 12000.0 435.0 5.00 13.8 1 0 0.300
## 4 Lacustrine 12358.6 572.3 3.93 6.4 1 0 0.283
## 5 Palustrine 4600.0 321.4 2.00 14.3 0 0 0.000
## 6 Palustrine 2268.8 192.0 1.86 10.9 0 0 0.000
## AUCV AUCC AUF AWOOD AUFV geometry
## 1 0 0.411 0.063 0.063 0.464 POINT (688816.6 5003207)
## 2 0 0.000 1.000 0.000 0.000 POINT (688494.4 4999093)
## 3 0 0.300 0.700 0.000 0.000 POINT (687938.4 5000223)
## 4 0 0.283 0.717 0.000 0.000 POINT (689732.8 5002522)
## 5 0 0.000 0.500 0.000 0.500 POINT (690104 4999355)
## 6 0 0.000 0.556 0.093 0.352 POINT (688742.5 4997481)Question: Where and how are the spatial coordinates shown in the sf object Sites.sf?
To illustrate importing spatial data from Excel, here we export the combined data frame as a csv file, import it again as a data frame, then convert it to an sf object. First we create a folder output if it does not yet exist.
Note: to run the code below, remove all the hashtags # at the beginning of the lines to un-comment them. This part assumes that you have writing permission on your computer. Alternatively, try setting up your R project folder on an external drive where you have writing permission.
The code below does the following:
- Line 1: Load package here that helps with file paths.
- Line 2: Check if folder output exists, and if not, create it.
- Line 3: Export the combined data frame as a .csv file.
- Line 4: Re-imports the .csv file as a
data.frameobjectSites. - Line 5: Create
sfobjectSites.sffrom df`.
#require(here)
#if(!dir.exists(here("output"))) dir.create(here("output"))
#write.csv(data.frame(Data, Coordinates), file=here("output/ralu.site.csv"), quote=FALSE, row.names=FALSE)
#Sites <- read.csv(here("output/ralu.site.csv"), header=TRUE)
#Sites.sf <- st_as_sf(df, coords=c("X", "Y"))The sf object Sites.sf contains 17 attribute variables and one variable geometry that contains the spatial information. Now R knows these are spatial data and knows how to handle them.
b. Add spatial reference data
Before we can combine the sampling locations with other spatial datasets, such as raster data, we need to tell R where on Earth these locations are (georeferencing). This is done by specifying the “Coordinate Reference System” (CRS).
For a great explanation of projections, see: https://michaelminn.net/tutorials/gis-projections/index.html
For more information on CRS, see: https://www.nceas.ucsb.edu/~frazier/RSpatialGuides/OverviewCoordinateReferenceSystems.pdf
We know that these coordinates are UTM zone 11 (Northern hemisphere) coordinates. We define it here by its EPSG code (32611). You can search for EPSG codes here: https://epsg.io/32611.
Here we call the function and the package simultaneously (this is good practice, as it helps keep track of where the functions in your code come from).
Question: Print Sites.sf and check what CRS is listed in the header.
Important: the code above, using function st_crs, only declares the existing projection, it does not change the coordinates to that projection!
However, the ralu.site dataset uses a slightly different definition of the projection (the difference is in the datum argument WGS84 vs. NAD83). Hence it may be safer to simply copy the crs information from ralu.site to Sites.sf. Again, this does not change the projection but declares that Sites.sf has the same projection as ralu.site. R even prints a warning to remind us of this:
## Warning: st_crs<- : replacing crs does not reproject data; use st_transform for
## thatc. Change projection
In case we needed to transform the projection, e.g., from UTM zone 11 to longitude/latitude (EPSG code: 4326), we could create a new sf object Sites.sf.longlat. We use the function st_transform to change the projection from the projection of the old object Sites.sf to the “longlat” coordinate system, which we define by the argument crs.
With
## Simple feature collection with 6 features and 17 fields
## Geometry type: POINT
## Dimension: XY
## Bounding box: xmin: -114.61 ymin: 45.1056 xmax: -114.5828 ymax: 45.15708
## Geodetic CRS: WGS 84
## SiteName Drainage Basin Substrate
## 1 AirplaneLake ShipIslandCreek Sheepeater Silt
## 2 BachelorMeadow WilsonCreek Skyhigh Silt
## 3 BarkingFoxLake WaterfallCreek Terrace Silt
## 4 BirdbillLake ClearCreek Birdbill Sand
## 5 BobLake WilsonCreek Harbor Silt
## 6 CacheLake WilsonCreek Skyhigh Silt
## NWI AREA_m2 PERI_m Depth_m TDS FISH ACB AUC
## 1 Lacustrine 62582.2 1142.8 21.64 2.5 1 0 0.411
## 2 Riverine_Intermittent_Streambed 225.0 60.0 0.40 0.0 0 0 0.000
## 3 Lacustrine 12000.0 435.0 5.00 13.8 1 0 0.300
## 4 Lacustrine 12358.6 572.3 3.93 6.4 1 0 0.283
## 5 Palustrine 4600.0 321.4 2.00 14.3 0 0 0.000
## 6 Palustrine 2268.8 192.0 1.86 10.9 0 0 0.000
## AUCV AUCC AUF AWOOD AUFV geometry
## 1 0 0.411 0.063 0.063 0.464 POINT (-114.5977 45.15708)
## 2 0 0.000 1.000 0.000 0.000 POINT (-114.6034 45.12016)
## 3 0 0.300 0.700 0.000 0.000 POINT (-114.61 45.13047)
## 4 0 0.283 0.717 0.000 0.000 POINT (-114.5864 45.15067)
## 5 0 0.000 0.500 0.000 0.500 POINT (-114.5828 45.12208)
## 6 0 0.000 0.556 0.093 0.352 POINT (-114.6008 45.1056)Question: What has changed in the summary?
d. Map sampling sites on world map
Where on earth is this? You could enter the coordinates from the “longlat” projection in Google maps. Note that Google expects the Latitude (Y coordinate) first, then the Longitude (X coordinate). In the variable geometry, longitude (e.g., -114.5977 for the first data point) is listed before latitude (45.15708 for the first data point). Thus, to locate the first site in Google maps, you will need to enter 45.15708, -114.5977.
There is a much easier way to find out where on Earth the sampling points are located. And we don’t even need to change the coordinates to longitude/latitude - R will do this for us internally.
- We load the R package
tmap(which stands for “thematic maps”) - With
tmap_mode("view"), we indicate that we want to create an interactive map. - With
tm_shape(Sites.sf)we select the data set (Sites.sf`) to be displayed. For now, we only plot the location, no attribute information. - With
tm_sf, we specify how the points should be displayed (color, size).
## tmap mode set to interactive viewingQuestion: Try changing the basemap by clicking on the Layers symbol and selecting a different map. Which map is most informative for these data? Zoom in and out to find out where on Earth the sampling points are located.
Note: see this week’s bonus materials to learn more about creating maps with tmap.
3. Display raster data and overlay sampling locations, extract data
a. Display raster data
The raster data for this project are already available in the package GeNetIt. They are stored as a SpatRaster object from package terra.
## [1] "SpatRaster"
## attr(,"package")
## [1] "terra"Printing the name of the SpatRaster object displays a summary. A few explanations:
- dimensions: number of rows (nrow), number of columns (ncol), number of layers (nlyr). So we see there are 6 layers (i.e., variables).
- resolution: cell size is 30 m both in x and y directions (typical for Landsat-derived remote sensing data)
- extent: the bounding box that defines the spatial extent of the raster data.
- coord.ref: projected in UTM zone 11, with ‘datum’ (NAD83).
- source: the file from which the data were imported.
- names, min values, max values: summary of the layers (variables).
## class : SpatRaster
## dimensions : 426, 358, 6 (nrow, ncol, nlyr)
## resolution : 30, 30 (x, y)
## extent : 683282.5, 694022.5, 4992833, 5005613 (xmin, xmax, ymin, ymax)
## coord. ref. : NAD83 / UTM zone 11N (EPSG:26911)
## source : covariates.tif
## names : cti, err27, ffp, gsp, hli, nlcd
## min values : 0.8429851, 0.03906551, 0, 227.0000, 1014, 11
## max values : 23.7147598, 0.76376426, 51, 338.0697, 9263, 95Now we can use plot, which knows what to do with a raster SpatRaster object.
Note: layer nlcd is a categorical map of land cover types. See this week’s bonus materials for how to better display a categorical map in R.
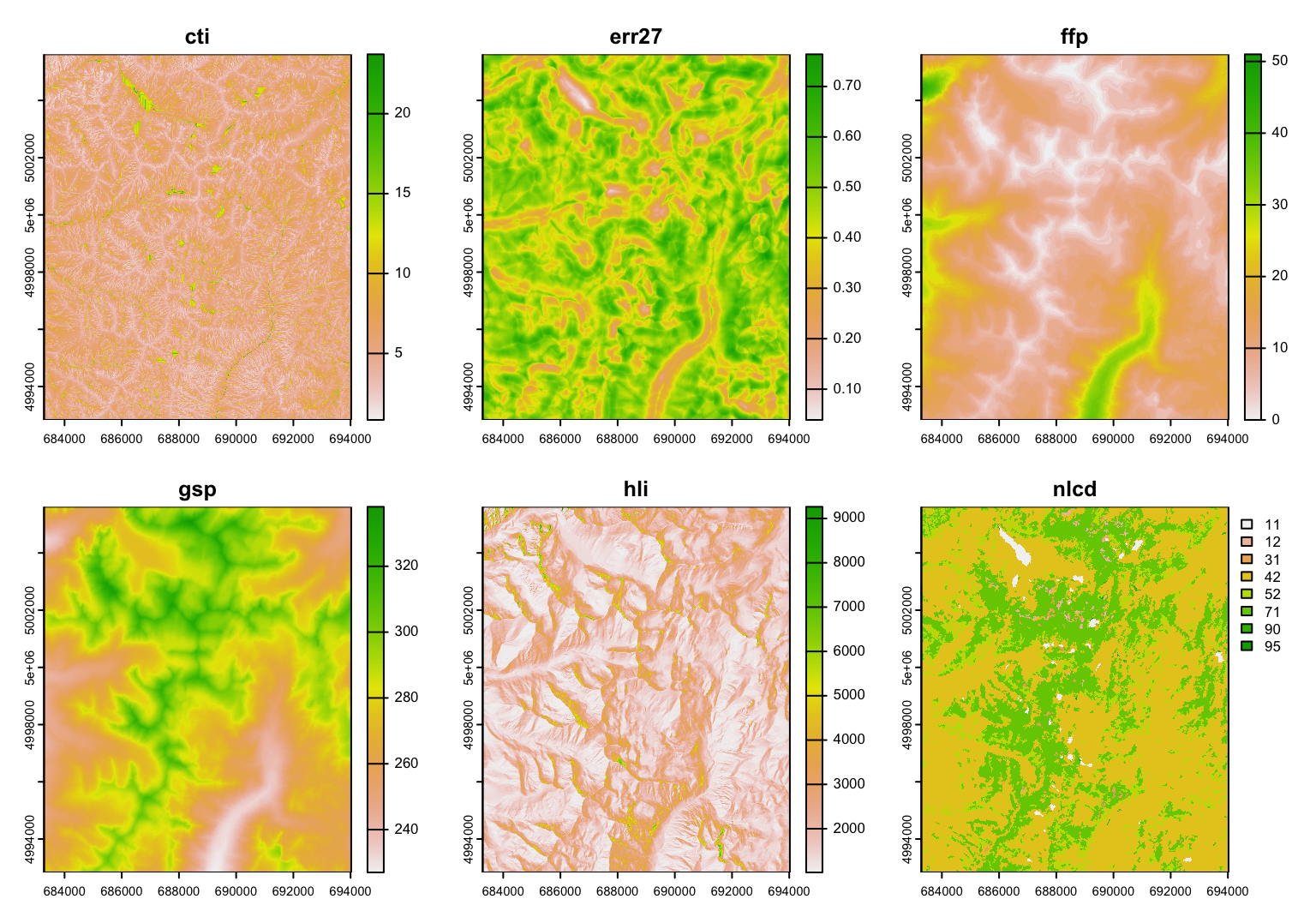
Some layers seem to show a similar pattern. It is easy to calculate the correlation between quantitative raster layers. Here, the last layer ncld, is in fact categorical (land cover type), and it’s correlation here is meaningless.
## NULLQuestions:
- What does each value in this table represent?
- Which layers are most strongly correlated?
- What do these variables (layers) represent?
- Is the correlation positive or negative?
b. Change color ramp, add sampling locations
We can specify a color ramp by setting the col argument. The default is terrain.colors(255). Here we change it to rainbow(9), a rainbow color palette with 9 color levels.
Note: To learn about options for the plot function for SpatRaster objects, access the help file by typing ?plot and select Make a map.
We can add the sampling locations (if we plot only a single raster layer). Here we use rev to reverse the color ramp for plotting raster layer ffp, and add the sites as white circles with black outlines.
par(mar=c(3,3,1,2))
plot(RasterMaps, "ffp", col=rev(rainbow(9)))
points(Sites.sf, pch=21, col="black", bg="white")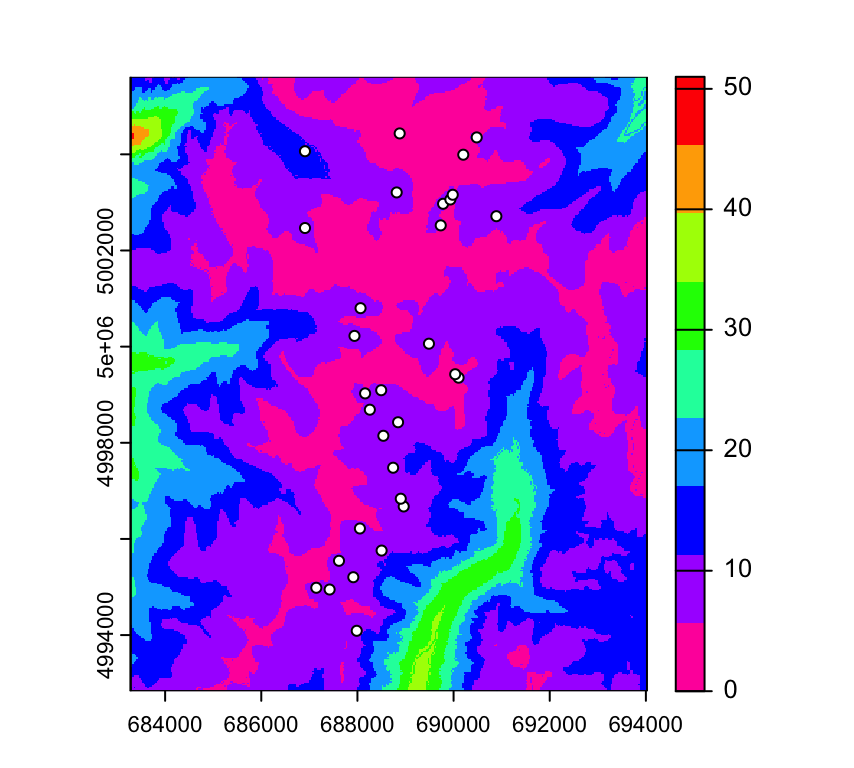
Question: Recall that ‘ffp’ stands for frost free period (in days). What do you think is the average length of the frost free period at theses sampling sites?
c. Extract raster values at sampling locations
The following code adds six variables to Sites.sf. However, it automatically changes the object type to a SpatVector object, which we’ll call Sites.terra to remind us of the object type. Technically we combine the columns of the existing data frame Sites.sf with the new columns in a new data frame with the same name.
We can convert back to an sf object:
## Simple feature collection with 31 features and 23 fields
## Geometry type: POINT
## Dimension: XY
## Bounding box: xmin: 686908.5 ymin: 4994089 xmax: 690890.1 ymax: 5004435
## Projected CRS: PROJCRS["unknown",
## BASEGEOGCRS["unknown",
## DATUM["North American Datum 1983",
## ELLIPSOID["GRS 1980",6378137,298.257222101,
## LENGTHUNIT["metre",1]],
## ID["EPSG",6269]],
## PRIMEM["Greenwich",0,
## ANGLEUNIT["degree",0.0174532925199433],
## ID["EPSG",8901]]],
## CONVERSION["UTM zone 11N",
## METHOD["Transverse Mercator",
## ID["EPSG",9807]],
## PARAMETER["Latitude of natural origin",0,
## ANGLEUNIT["degree",0.0174532925199433],
## ID["EPSG",8801]],
## PARAMETER["Longitude of natural origin",-117,
## ANGLEUNIT["degree",0.0174532925199433],
## ID["EPSG",8802]],
## PARAMETER["Scale factor at natural origin",0.9996,
## SCALEUNIT["unity",1],
## ID["EPSG",8805]],
## PARAMETER["False easting",500000,
## LENGTHUNIT["metre",1],
## ID["EPSG",8806]],
## PARAMETER["False northing",0,
## LENGTHUNIT["metre",1],
## ID["EPSG",8807]],
## ID["EPSG",16011]],
## CS[Cartesian,2],
## AXIS["(E)",east,
## ORDER[1],
## LENGTHUNIT["metre",1,
## ID["EPSG",9001]]],
## AXIS["(N)",north,
## ORDER[2],
## LENGTHUNIT["metre",1,
## ID["EPSG",9001]]]]
## First 10 features:
## SiteName Drainage Basin Substrate
## 1 AirplaneLake ShipIslandCreek Sheepeater Silt
## 2 BachelorMeadow WilsonCreek Skyhigh Silt
## 3 BarkingFoxLake WaterfallCreek Terrace Silt
## 4 BirdbillLake ClearCreek Birdbill Sand
## 5 BobLake WilsonCreek Harbor Silt
## 6 CacheLake WilsonCreek Skyhigh Silt
## 7 DoeLake WilsonCreek Skyhigh Silt
## 8 EggWhiteLake WilsonCreek Skyhigh Silt
## 9 ElenasLake ShipIslandCreek Sheepeater Sand
## 10 FawnLake WilsonCreek Skyhigh Silt
## NWI AREA_m2 PERI_m Depth_m TDS FISH ACB AUC
## 1 Lacustrine 62582.2 1142.8 21.64 2.5 1 0 0.411
## 2 Riverine_Intermittent_Streambed 225.0 60.0 0.40 0.0 0 0 0.000
## 3 Lacustrine 12000.0 435.0 5.00 13.8 1 0 0.300
## 4 Lacustrine 12358.6 572.3 3.93 6.4 1 0 0.283
## 5 Palustrine 4600.0 321.4 2.00 14.3 0 0 0.000
## 6 Palustrine 2268.8 192.0 1.86 10.9 0 0 0.000
## 7 Lacustrine 13034.9 463.2 6.03 10.0 1 0 0.415
## 8 Palustrine 4544.5 291.9 3.30 2.4 0 0 0.000
## 9 Palustrine 0.0 0.0 0.00 0.0 0 0 0.000
## 10 Palustrine 3865.9 237.7 1.98 3.6 0 0 0.000
## AUCV AUCC AUF AWOOD AUFV cti err27 ffp gsp hli nlcd
## 1 0.000 0.411 0.063 0.063 0.464 6.551698 0.2211860 8 286.0000 1438 42
## 2 0.000 0.000 1.000 0.000 0.000 7.257386 0.3399709 8 286.0000 1718 71
## 3 0.000 0.300 0.700 0.000 0.000 7.587409 0.2636720 9 283.0000 1308 42
## 4 0.000 0.283 0.717 0.000 0.000 4.864036 0.3027736 5 294.1377 1416 12
## 5 0.000 0.000 0.500 0.000 0.500 10.962278 0.5830295 6 290.0000 1685 42
## 6 0.000 0.000 0.556 0.093 0.352 7.817918 0.4895256 11 274.0000 1277 42
## 7 0.171 0.585 0.341 0.000 0.073 6.940942 0.1684043 11 273.0000 1306 42
## 8 0.047 0.047 0.686 0.209 0.058 6.560551 0.4499388 10 279.0000 1244 42
## 9 0.000 0.000 0.000 0.000 0.000 4.031181 0.3834549 2 308.0000 1169 71
## 10 0.000 0.000 1.000 0.000 0.000 10.988009 0.1625656 11 273.0000 1296 42
## geometry
## 1 POINT (688816.6 5003207)
## 2 POINT (688494.4 4999093)
## 3 POINT (687938.4 5000223)
## 4 POINT (689732.8 5002522)
## 5 POINT (690104 4999355)
## 6 POINT (688742.5 4997481)
## 7 POINT (688962.4 4996675)
## 8 POINT (688539.3 4998146)
## 9 POINT (688878.7 5004435)
## 10 POINT (688901.5 4996837)Let’s calculate the mean length of the frost free period for these sites:
## [1] 8.0963What land cover type is assigned to the most sampling units? Let’s tabulate them.
##
## 11 12 42 52 71 90
## 3 1 21 1 4 1Note: the land cover types are coded by numbers. Check here what the numbers mean: https://www.mrlc.gov/data/legends/national-land-cover-database-2016-nlcd2016-legend
Question: A total of 21 sites are classified as 42. What is this most frequent land cover type?
4. Calculate landscape metrics
We are going to use the package landscapemetrics. It is an R package to calculate landscape metrics in a tidy workflow (for more information about tidy data see here). landscapemetrics is basically a reimplementation of ‘FRAGSTATS’, which allows an integration into larger workflows within the R environment. The core of the package are functions to calculate landscape metrics, but also several auxiliary functions exist.
To facilitate an integration into larger workflows, landscapemetrics is based on the terra and stars packages. It expects a raster with integers that represent land cover classes. To check if a raster is suitable for landscapemetrics, run the check_landscape() function first. The function checks the coordinate reference system (and mainly if units are in meters) and if the raster values are discrete classes. If the check fails, the calculation of metrics is still possible, however, especially metrics that are based on area and distances must be used with caution.
## Warning: Caution: Land-cover classes must be decoded as integer values.## Warning: Caution: More than 30 land cover-classes - Please check if discrete
## land-cover classes are present.## Warning: Caution: Land-cover classes must be decoded as integer values.## Warning: Caution: More than 30 land cover-classes - Please check if discrete
## land-cover classes are present.## Warning: Caution: Land-cover classes must be decoded as integer values.## Warning: Caution: More than 30 land cover-classes - Please check if discrete
## land-cover classes are present.## Warning: Caution: Land-cover classes must be decoded as integer values.## Warning: Caution: More than 30 land cover-classes - Please check if discrete
## land-cover classes are present.
## Warning: Caution: More than 30 land cover-classes - Please check if discrete
## land-cover classes are present.## layer crs units class n_classes OK
## 1 1 projected m non-integer 150973 ✖
## 2 2 projected m non-integer 151161 ✖
## 3 3 projected m non-integer 37266 ✖
## 4 4 projected m non-integer 79335 ✖
## 5 5 projected m integer 4776 ❓
## 6 6 projected m integer 8 ✔Question: Which raster layer(s) are suitable for calculating landscape metrics? Why are the others not suitable?
There are three different levels of landscape metrics. Firstly, metrics can be calculated for each single patch (a patch is defined as neighbouring cells of the same class). Secondly, metrics can be calculated for a certain class (i.e. all patches belonging to the same class) and lastly for the whole landscape. All these levels are implemented and easily accessible in landscapemetrics.
All functions to calculate metrics start with lsm_ (for landscapemetrics). The second part of the name specifies the level (patch - p, class - c or landscape - l). Lastly, the final part of the function name is the abbreviation of the corresponding metric (e.g. enn for the Euclidean nearest-neighbor distance). To list all available metrics, you can use the list_lsm() function. The function also allows to show metrics filtered by level, type or metric name. For more information about the metrics, please see either the corresponding helpfile(s) or https://r-spatialecology.github.io/landscapemetrics.
List available diversity metrics:
## # A tibble: 9 × 5
## metric name type level function_name
## <chr> <chr> <chr> <chr> <chr>
## 1 msidi modified simpson's diversity index diversity metric land… lsm_l_msidi
## 2 msiei modified simpson's evenness index diversity metric land… lsm_l_msiei
## 3 pr patch richness diversity metric land… lsm_l_pr
## 4 prd patch richness density diversity metric land… lsm_l_prd
## 5 rpr relative patch richness diversity metric land… lsm_l_rpr
## 6 shdi shannon's diversity index diversity metric land… lsm_l_shdi
## 7 shei shannon's evenness index diversity metric land… lsm_l_shei
## 8 sidi simpson's diversity index diversity metric land… lsm_l_sidi
## 9 siei simspon's evenness index diversity metric land… lsm_l_sieiList available area metrics:
## # A tibble: 7 × 5
## metric name type level function_name
## <chr> <chr> <chr> <chr> <chr>
## 1 area patch area area and edge metric patch lsm_p_area
## 2 area_cv patch area area and edge metric class lsm_c_area_cv
## 3 area_mn patch area area and edge metric class lsm_c_area_mn
## 4 area_sd patch area area and edge metric class lsm_c_area_sd
## 5 area_cv patch area area and edge metric landscape lsm_l_area_cv
## 6 area_mn patch area area and edge metric landscape lsm_l_area_mn
## 7 area_sd patch area area and edge metric landscape lsm_l_area_sdList available aggregation metrics:
landscapemetrics::list_lsm(level = c("class", "landscape"), type = "aggregation metric",
simplify = TRUE)## [1] "lsm_c_ai" "lsm_c_clumpy" "lsm_c_cohesion" "lsm_c_division"
## [5] "lsm_c_enn_cv" "lsm_c_enn_mn" "lsm_c_enn_sd" "lsm_c_iji"
## [9] "lsm_c_lsi" "lsm_c_mesh" "lsm_c_nlsi" "lsm_c_np"
## [13] "lsm_c_pd" "lsm_c_pladj" "lsm_c_split" "lsm_l_ai"
## [17] "lsm_l_cohesion" "lsm_l_contag" "lsm_l_division" "lsm_l_enn_cv"
## [21] "lsm_l_enn_mn" "lsm_l_enn_sd" "lsm_l_iji" "lsm_l_lsi"
## [25] "lsm_l_mesh" "lsm_l_np" "lsm_l_pd" "lsm_l_pladj"
## [29] "lsm_l_split"a. Calculate patch-, class- and landscape level landscape metrics
Note: This section explains different ways of calculating a selection of landscape metrics from a raster map with ‘landscapemetrics’. If this seems too technical for a first go, you may jump to section 4b.
To calculate a single metric, just use the corresponding function. The result of all landscape metric functions is always an identically structured tibble (i.e. an advanced data.frame). The first coloumn is the layer id (only interesting for e.g. a RasterStack). The second coloumn specifies the level (‘patch’, ‘class’ or ‘landscape’). The third coloumn is the class ID (NA on landscape level) and the fourth coloumn is the patch ID (NA on class- and landscape level). Lastly, The fith coloumn is the abbreviation of the metric and finally the corresponding value in the last coloumn.
## c.lculate percentage of landscape of class
percentage_class <- lsm_c_pland(landscape = RasterMaps$nlcd)
percentage_class## # A tibble: 8 × 6
## layer level class id metric value
## <int> <chr> <int> <int> <chr> <dbl>
## 1 1 class 11 NA pland 0.948
## 2 1 class 12 NA pland 0.441
## 3 1 class 31 NA pland 0.394
## 4 1 class 42 NA pland 59.1
## 5 1 class 52 NA pland 11.0
## 6 1 class 71 NA pland 28.1
## 7 1 class 90 NA pland 0.0557
## 8 1 class 95 NA pland 0.0210Questions:
- What percentage of the overall landscape (total map) is evergreen forest (class 42)?
- What percentage of the landscape is classified as wetlands (classes 90 and 95)?
Because the resulting tibble is type stable, you can easily row-bind (rbind) different metrics (even of different levels):
metrics <- rbind(
landscapemetrics::lsm_c_pladj(RasterMaps$nlcd),
landscapemetrics::lsm_l_pr(RasterMaps$nlcd),
landscapemetrics::lsm_l_shdi(RasterMaps$nlcd)
)
metrics## # A tibble: 10 × 6
## layer level class id metric value
## <int> <chr> <int> <int> <chr> <dbl>
## 1 1 class 11 NA pladj 77.9
## 2 1 class 12 NA pladj 52.5
## 3 1 class 31 NA pladj 48.8
## 4 1 class 42 NA pladj 89.9
## 5 1 class 52 NA pladj 58.6
## 6 1 class 71 NA pladj 81.6
## 7 1 class 90 NA pladj 41.8
## 8 1 class 95 NA pladj 32.8
## 9 1 landscape NA NA pr 8
## 10 1 landscape NA NA shdi 1.01To calculate a larger set of landscape metrics, you can just use the wrapper calculate_lsm(). The arguments are similar to list_lsm(), e.g. you can specify the level or the type of metrics to calculate. Alternatively, you can also provide a vector with the function names of metrics to calculate to the what argument.
However, watch out, for large rasters and many metrics, this can be rather slow (set progress = TRUE to get an progress report on the console). Also, we suggest to not just calculate all available metrics, but rather think about which ones might be actually suitable for your research question.
Calculate all patch-level metrics using wrapper:
nlcd_patch <- landscapemetrics::calculate_lsm(landscape = RasterMaps$nlcd,
level = "patch")
nlcd_patch## # A tibble: 19,776 × 6
## layer level class id metric value
## <int> <chr> <int> <int> <chr> <dbl>
## 1 1 patch 11 1 area 0.45
## 2 1 patch 11 2 area 0.45
## 3 1 patch 11 3 area 41.7
## 4 1 patch 11 4 area 0.72
## 5 1 patch 11 5 area 6.12
## 6 1 patch 11 6 area 0.9
## 7 1 patch 11 7 area 0.9
## 8 1 patch 11 8 area 1.89
## 9 1 patch 11 9 area 0.09
## 10 1 patch 11 10 area 1.17
## # ℹ 19,766 more rowsShow abbreviation of all calculated metrics:
## [1] "area" "cai" "circle" "contig" "core" "enn" "frac" "gyrate"
## [9] "ncore" "para" "perim" "shape"Calculate all aggregation metrics on landscape level:
nlcd_landscape_aggr <- landscapemetrics::calculate_lsm(landscape = RasterMaps$nlcd,
level = "landscape",
type = "aggregation metric")
nlcd_landscape_aggr## # A tibble: 14 × 6
## layer level class id metric value
## <int> <chr> <int> <int> <chr> <dbl>
## 1 1 landscape NA NA ai 84.1
## 2 1 landscape NA NA cohesion 99.2
## 3 1 landscape NA NA contag 62.7
## 4 1 landscape NA NA division 0.804
## 5 1 landscape NA NA enn_cv 218.
## 6 1 landscape NA NA enn_mn 129.
## 7 1 landscape NA NA enn_sd 281.
## 8 1 landscape NA NA iji 43.0
## 9 1 landscape NA NA lsi 32.3
## 10 1 landscape NA NA mesh 2694.
## 11 1 landscape NA NA np 1648
## 12 1 landscape NA NA pd 12.0
## 13 1 landscape NA NA pladj 83.7
## 14 1 landscape NA NA split 5.09Calculate specific metrics:
nlcd_subset <- landscapemetrics::calculate_lsm(landscape = RasterMaps$nlcd,
what = c("lsm_c_pladj",
"lsm_l_pr",
"lsm_l_shdi"))
nlcd_subset## # A tibble: 10 × 6
## layer level class id metric value
## <int> <chr> <int> <int> <chr> <dbl>
## 1 1 class 11 NA pladj 77.9
## 2 1 class 12 NA pladj 52.5
## 3 1 class 31 NA pladj 48.8
## 4 1 class 42 NA pladj 89.9
## 5 1 class 52 NA pladj 58.6
## 6 1 class 71 NA pladj 81.6
## 7 1 class 90 NA pladj 41.8
## 8 1 class 95 NA pladj 32.8
## 9 1 landscape NA NA pr 8
## 10 1 landscape NA NA shdi 1.01The resulting tibble is easy to integrate into a workflow. For example, to get the ordered patch IDs of the 5% largest patches, the following code could be used.
The pipe operator %>% from the dplyr package passes the resulting object automatically to the next function as first argument.
Note: the last step (pulling the id variable only) could be done by adding this to the pipe: %>% dplyr::pull(id). Due to some package inconsistencies, this sometimes created an error. Here we extract the id variable in a separate step as a work-around.
id_largest <- nlcd_patch %>% # previously calculated patch metrics
dplyr::filter(metric == "area") %>% # only patch area
dplyr::arrange(-value) %>% # order by decreasing size
dplyr::filter(value > quantile(value, probs = 0.95)) ## g.t only patches larger than 95% quantile
id_largest <- id_largest$id ## g.t only patch id
id_largest## [1] 206 166 1265 1459 1549 1558 1421 427 1434 1385 226 3 386 1205 589
## [16] 1441 426 1059 1206 324 433 1195 315 1225 712 1266 1377 389 753 1528
## [31] 894 1510 286 563 240 1411 478 1364 435 1336 812 640 1376 1523 786
## [46] 467 485 1559 1244 284 1537 1574 718 814 499 864 955 1015 1430 443
## [61] 393 930 1250 548 851 885 1481 928 1554 43 164 281 1608 716 514
## [76] 802 1614 729 977 1488 37 357 1353Because the metric names are only abbreviated, there is also a way to include the full name in the results. For the wrapper, just set full_name = TRUE. For the rowbinded tibble, you can use the provided tibble called lsm_abbreviations_names that comes with the package and use e.g. dplyr::left_join() to combine it with your results.
Add full metrics name to result:
nlcd_subset_full_a <- landscapemetrics::calculate_lsm(RasterMaps$nlcd,
what = c("lsm_c_pladj",
"lsm_l_pr",
"lsm_l_shdi"),
full_name = TRUE)
nlcd_subset_full_a## # A tibble: 10 × 9
## layer level class id metric value name type function_name
## <int> <chr> <int> <int> <chr> <dbl> <chr> <chr> <chr>
## 1 1 class 11 NA pladj 77.9 percentage of l… aggr… lsm_c_pladj
## 2 1 class 12 NA pladj 52.5 percentage of l… aggr… lsm_c_pladj
## 3 1 class 31 NA pladj 48.8 percentage of l… aggr… lsm_c_pladj
## 4 1 class 42 NA pladj 89.9 percentage of l… aggr… lsm_c_pladj
## 5 1 class 52 NA pladj 58.6 percentage of l… aggr… lsm_c_pladj
## 6 1 class 71 NA pladj 81.6 percentage of l… aggr… lsm_c_pladj
## 7 1 class 90 NA pladj 41.8 percentage of l… aggr… lsm_c_pladj
## 8 1 class 95 NA pladj 32.8 percentage of l… aggr… lsm_c_pladj
## 9 1 landscape NA NA pr 8 patch richness dive… lsm_l_pr
## 10 1 landscape NA NA shdi 1.01 shannon's diver… dive… lsm_l_shdiAdd full metrics name to results calculated previously using left_join():
nlcd_subset_full_b <- dplyr::left_join(x = nlcd_subset,
y = lsm_abbreviations_names,
by = c("metric", "level"))
nlcd_subset_full_b## # A tibble: 10 × 9
## layer level class id metric value name type function_name
## <int> <chr> <int> <int> <chr> <dbl> <chr> <chr> <chr>
## 1 1 class 11 NA pladj 77.9 percentage of l… aggr… lsm_c_pladj
## 2 1 class 12 NA pladj 52.5 percentage of l… aggr… lsm_c_pladj
## 3 1 class 31 NA pladj 48.8 percentage of l… aggr… lsm_c_pladj
## 4 1 class 42 NA pladj 89.9 percentage of l… aggr… lsm_c_pladj
## 5 1 class 52 NA pladj 58.6 percentage of l… aggr… lsm_c_pladj
## 6 1 class 71 NA pladj 81.6 percentage of l… aggr… lsm_c_pladj
## 7 1 class 90 NA pladj 41.8 percentage of l… aggr… lsm_c_pladj
## 8 1 class 95 NA pladj 32.8 percentage of l… aggr… lsm_c_pladj
## 9 1 landscape NA NA pr 8 patch richness dive… lsm_l_pr
## 10 1 landscape NA NA shdi 1.01 shannon's diver… dive… lsm_l_shdib. Calculate patch-level landscape metrics for ‘Evergreen Forest’
To only get the results for class 42 (evergreen forest), you can just dplyr::filter() the tibble (or use any other subset method you prefer).
## # A tibble: 2,664 × 6
## layer level class id metric value
## <int> <chr> <int> <int> <chr> <dbl>
## 1 1 patch 42 164 area 8.1
## 2 1 patch 42 165 area 0.18
## 3 1 patch 42 166 area 3180.
## 4 1 patch 42 167 area 0.27
## 5 1 patch 42 168 area 3.51
## 6 1 patch 42 169 area 1.71
## 7 1 patch 42 170 area 0.18
## 8 1 patch 42 171 area 0.09
## 9 1 patch 42 172 area 0.09
## 10 1 patch 42 173 area 1.98
## # ℹ 2,654 more rowsAll functions make heavy use of connected components labeling to delineate patches (neighbouring cells of the same class). To get all patches of every class you can just use get_patches(). This returns a list of layers (one layer per input raster map, here only one) with a separate SpatRaster for each class within the layer.
## c.nnected components labeling of landscape
cc_nlcd <- landscapemetrics::get_patches(RasterMaps$nlcd, directions = 8)
# summarize the SpatRaster for class 42:
cc_nlcd$layer_1$class_42## class : SpatRaster
## dimensions : 426, 358, 1 (nrow, ncol, nlyr)
## resolution : 30, 30 (x, y)
## extent : 683282.5, 694022.5, 4992833, 5005613 (xmin, xmax, ymin, ymax)
## coord. ref. : NAD83 / UTM zone 11N (EPSG:26911)
## source(s) : memory
## name : lyr.1
## min value : 164
## max value : 385To get only a certain class, just specify the class argument and the neighbourhood rule can be chosen between 8-neighbour rule or 4-neighbour rule with the argument directions.
Note: although we only process a single class, the resulting object still is a list with the same structure as above. Thus, we access the SpatRaster for class 42 the same way:
## class : SpatRaster
## dimensions : 426, 358, 1 (nrow, ncol, nlyr)
## resolution : 30, 30 (x, y)
## extent : 683282.5, 694022.5, 4992833, 5005613 (xmin, xmax, ymin, ymax)
## coord. ref. : NAD83 / UTM zone 11N (EPSG:26911)
## source(s) : memory
## name : lyr.1
## min value : 164
## max value : 385To plot the patches you can use the show_patches() function. Here we show patches of class 42 (forest) and class 52 (shrubland). Note that the color indicates patch ID!
## $layer_1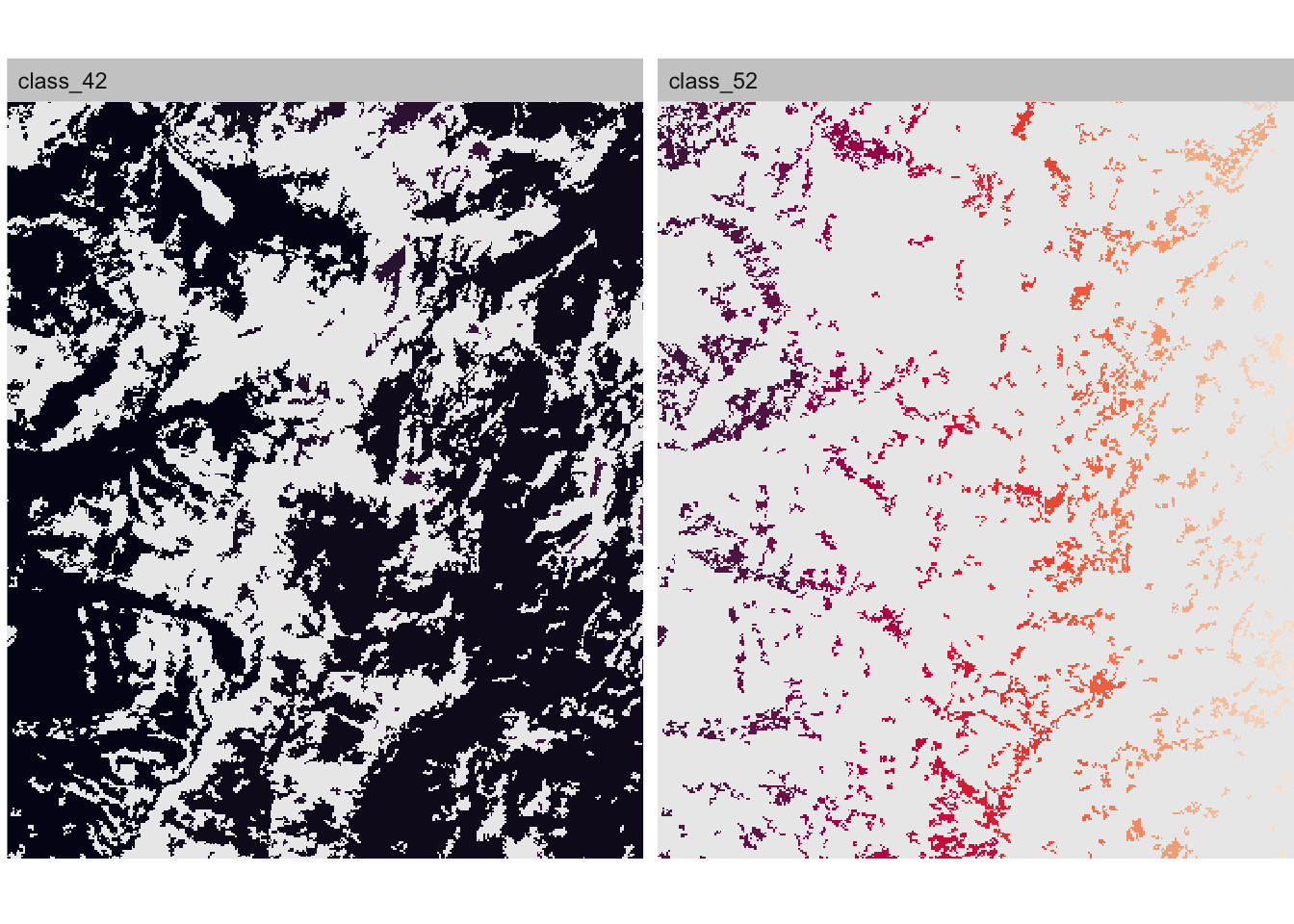
It is also possible to visualize only the core area of each patch using show_cores(). The core area is defined as all cells that are further away from the edge of each patch than a specified edge depth (e.g. 5 cells). Here we show core area with edge depth = 5 for class 42; try edge_depth = 1 for comparison:
## $layer_1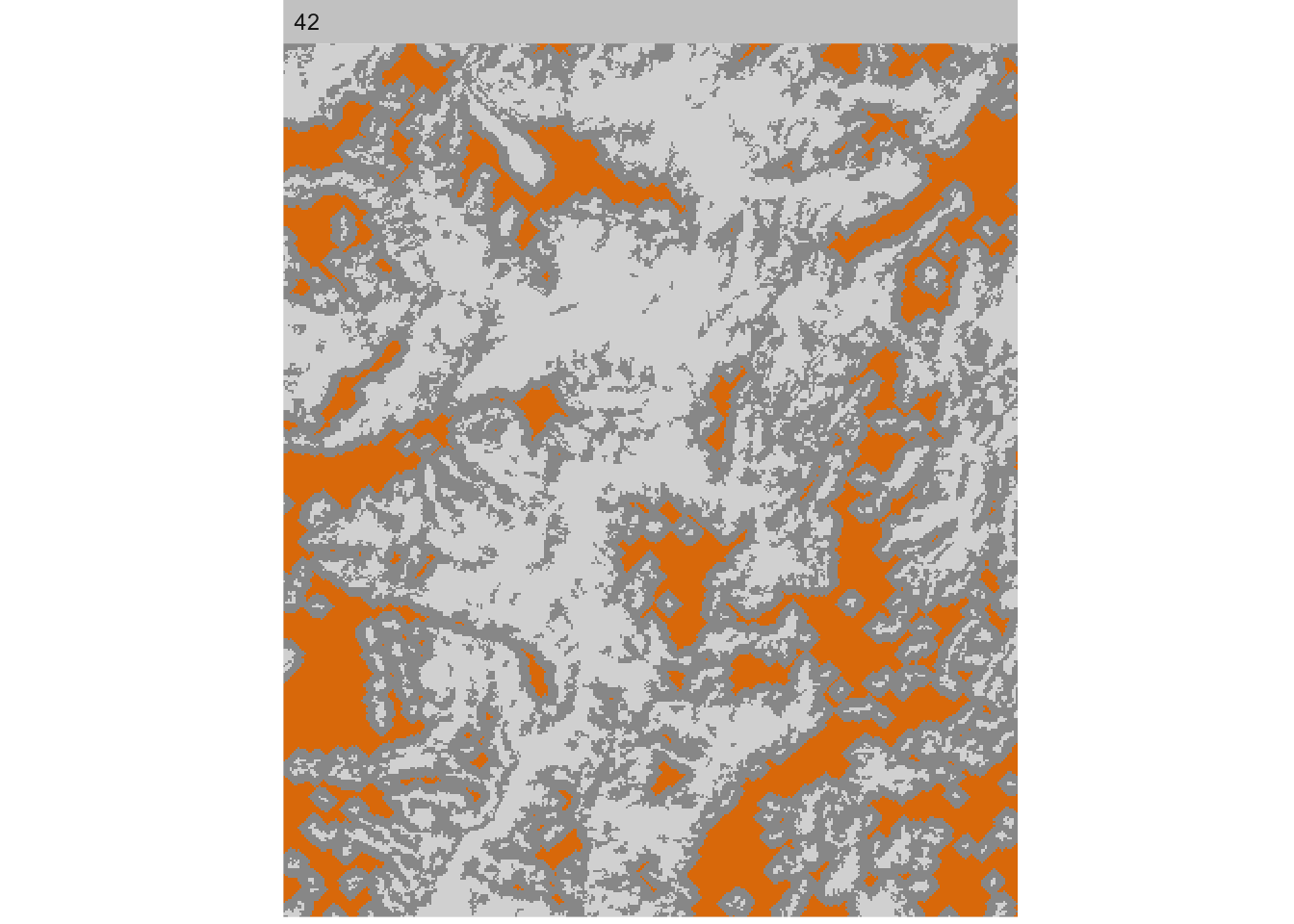
Note: this may create a warning “no non-missing arguments to min; returning Inf” for each patch that does not have any core area. Here we suppressed the warnings for the chunk with the chunk option warning=FALSE.
Lastly, you can plot the map and fill each patch with the corresponding metric value, e.g. patch size, using show_lsm(). Notice that there are two very large patches in class 42:
## $layer_1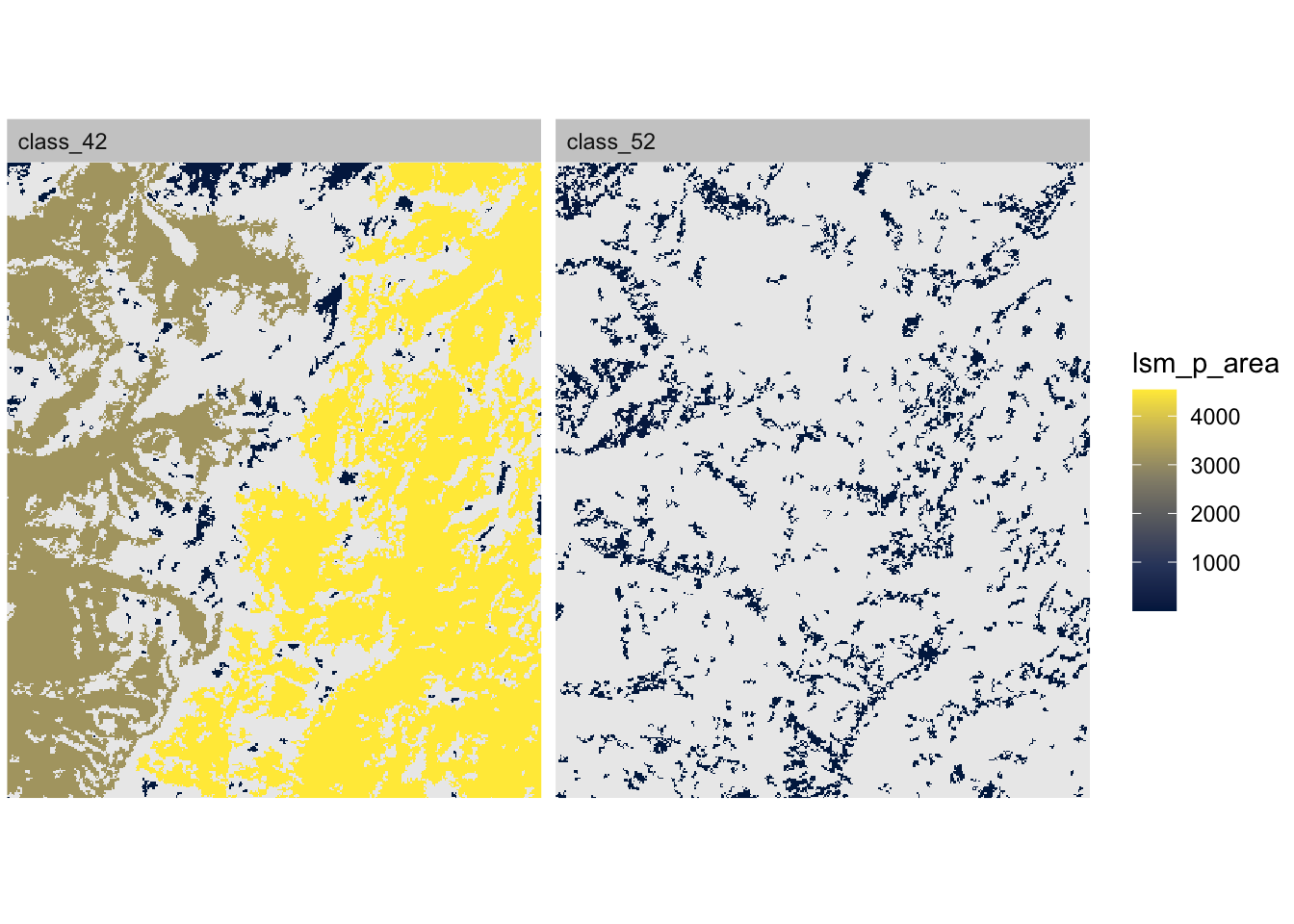
c. Extract forest patch size at sampling locations
Let’s add forest patch size to the Sites.sf data. To extract landscape metrics of the patch in which each sampling point is located, use extract_lsm(). Which metrics are extracted can be specified by the what argument (similar to calculate_lsm()). However, only patch-level metrics are available. Please be aware that the resulting tibble now has a new column, namely the ID of the sampling point (in the same order as the input points).
## e.tract patch area of all classes:
patch_size_sf <- extract_lsm(landscape = RasterMaps$nlcd, y = Sites.sf, what = "lsm_p_area")
## b.cause we are only interested in the forest patch size, we set all area of class != 42 to 0:
patch_size_sf_forest <- dplyr::mutate(patch_size_sf,
value = dplyr::case_when(class == 42 ~ value,
class != 42 ~ 0))
## a.d data to sf object:
Sites.sf$ForestPatchSize <- patch_size_sf_forest$value
Sites.sf$ForestPatchSize## [1] 3179.88 0.00 3179.88 0.00 6.66 4539.33 4539.33 4539.33 0.00
## [10] 4539.33 4539.33 23.76 23.76 0.00 0.00 4539.33 0.00 4539.33
## [19] 3.60 3179.88 0.00 4539.33 3.87 4539.33 0.00 0.00 23.76
## [28] 3179.88 0.00 4539.33 0.095. Sample landscape metrics within buffer around sampling locations
The package landscapemetrics has a built-in function sample_lsm to sample metrics in a buffer around sampling locations, which are provided with argument y. You can choose the shape of the buffer window (either a circle or a square) and, with the argument what, which metrics to sample (similar to calculate_lsm()).
The argument size specifies the buffer size in map units (e.g., meters): radius for circles, half of the side length for squares. Here, the value size = 500 results in a square window of 1000 m x 1000 m centered at the sampling location.
nlcd_sampled <- landscapemetrics::sample_lsm(landscape = RasterMaps$nlcd,
what = c("lsm_l_ta",
"lsm_c_np",
"lsm_c_pland",
"lsm_c_ai"),
shape = "square",
y = Sites.sf,
size = 500)## Warning: [crop] CRS do not match## # A tibble: 493 × 8
## layer level class id metric value plot_id percentage_inside
## <int> <chr> <int> <int> <chr> <dbl> <int> <dbl>
## 1 1 class 11 NA ai 88.6 1 98.0
## 2 1 class 31 NA ai NA 1 98.0
## 3 1 class 42 NA ai 93.3 1 98.0
## 4 1 class 52 NA ai 50 1 98.0
## 5 1 class 71 NA ai 65.9 1 98.0
## 6 1 class 11 NA np 2 1 98.0
## 7 1 class 31 NA np 1 1 98.0
## 8 1 class 42 NA np 2 1 98.0
## 9 1 class 52 NA np 4 1 98.0
## 10 1 class 71 NA np 8 1 98.0
## # ℹ 483 more rowsThe tibble now contains two additional columns. Firstly, the plot_id (in the same order as the input points) and secondly, the percentage_inside, i.e. what percentage of the buffer around the sampling location lies within the map. (In cases where the sampling location is on the edge of the landscape, the buffer around the sampling location could be only partly within the map). The value can also deviate from 100 % because the sampling locations are not necessarily in the cell center and the actually clipped cells lead to a slightly smaller or larger buffer area. A circular buffer shape increases this effect.
It is also possible to get the clippings of the buffer around sampling locations as a RasterLayer. For this, just set return_raster = TRUE.
# sample some metrics within buffer around sample location and returning sample
# plots as raster
nlcd_sampled_plots <- landscapemetrics::sample_lsm(landscape = RasterMaps$nlcd,
what = c("lsm_l_ta",
"lsm_c_np",
"lsm_c_pland",
"lsm_c_ai"),
shape = "square",
y = Sites.sf,
size = 500,
return_raster = TRUE)## Warning: [crop] CRS do not match## # A tibble: 493 × 9
## layer level class id metric value plot_id percentage_inside
## <int> <chr> <int> <int> <chr> <dbl> <int> <dbl>
## 1 1 class 11 NA ai 88.6 1 98.0
## 2 1 class 31 NA ai NA 1 98.0
## 3 1 class 42 NA ai 93.3 1 98.0
## 4 1 class 52 NA ai 50 1 98.0
## 5 1 class 71 NA ai 65.9 1 98.0
## 6 1 class 11 NA np 2 1 98.0
## 7 1 class 31 NA np 1 1 98.0
## 8 1 class 42 NA np 2 1 98.0
## 9 1 class 52 NA np 4 1 98.0
## 10 1 class 71 NA np 8 1 98.0
## # ℹ 483 more rows
## # ℹ 1 more variable: raster_sample_plots <list>The result will be a nested tibble containing the plot_id, the metrics and a RasterLayer with the clipped buffers (as a list). Attention: Because several metrics on class- and landscape-level the clipped buffers will be “repeated” several times.
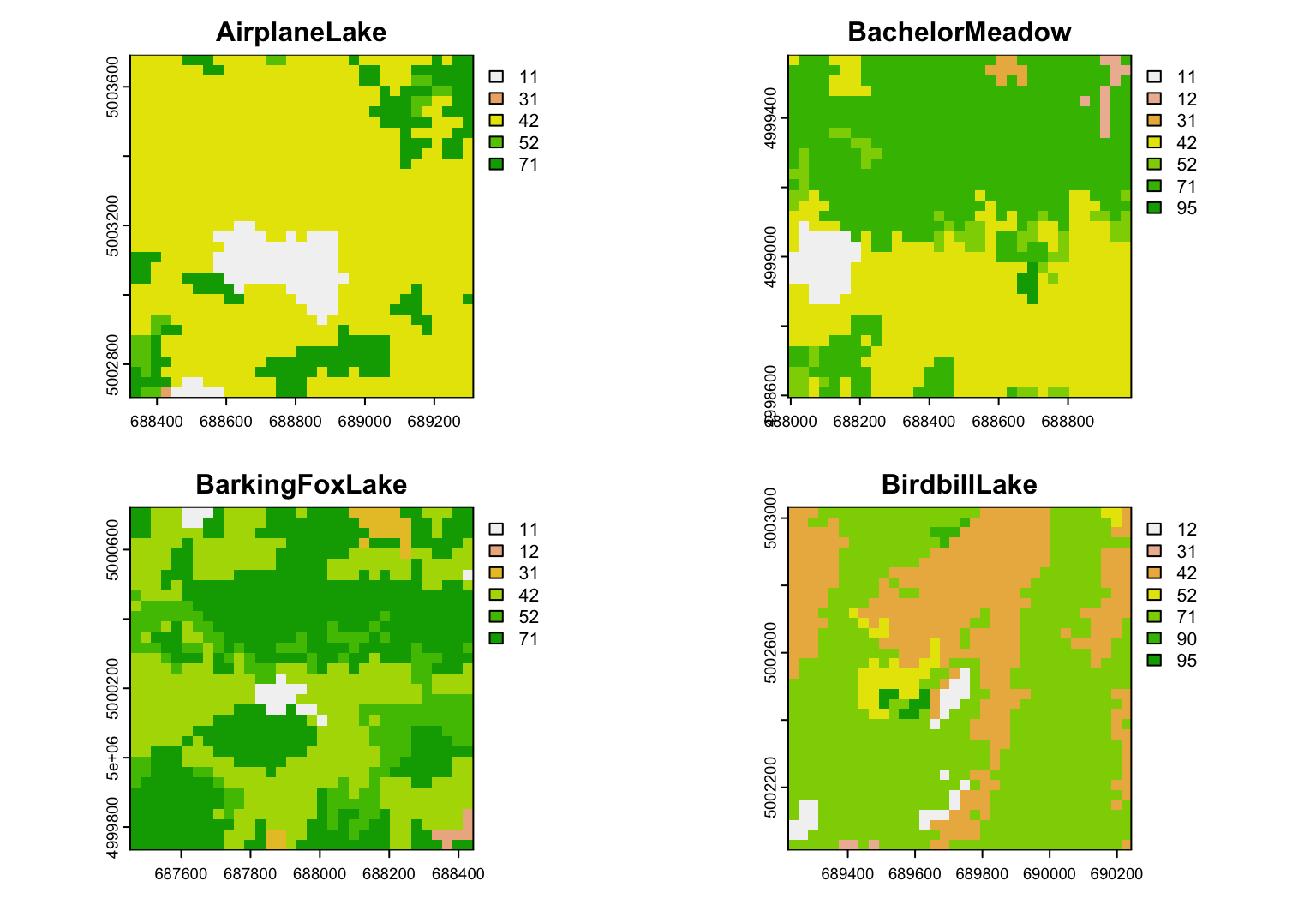
Here we show results for the first four sampling locations. We use a for loop to avoid repeating code.
unique_plots <- unique(nlcd_sampled_plots$raster_sample_plots)[1:4]
par(mfrow = c(2,2))
for(i in 1:4)
{
plot(unique_plots[[i]], type="classes",
main = paste(Sites.sf$SiteName[i]))
}
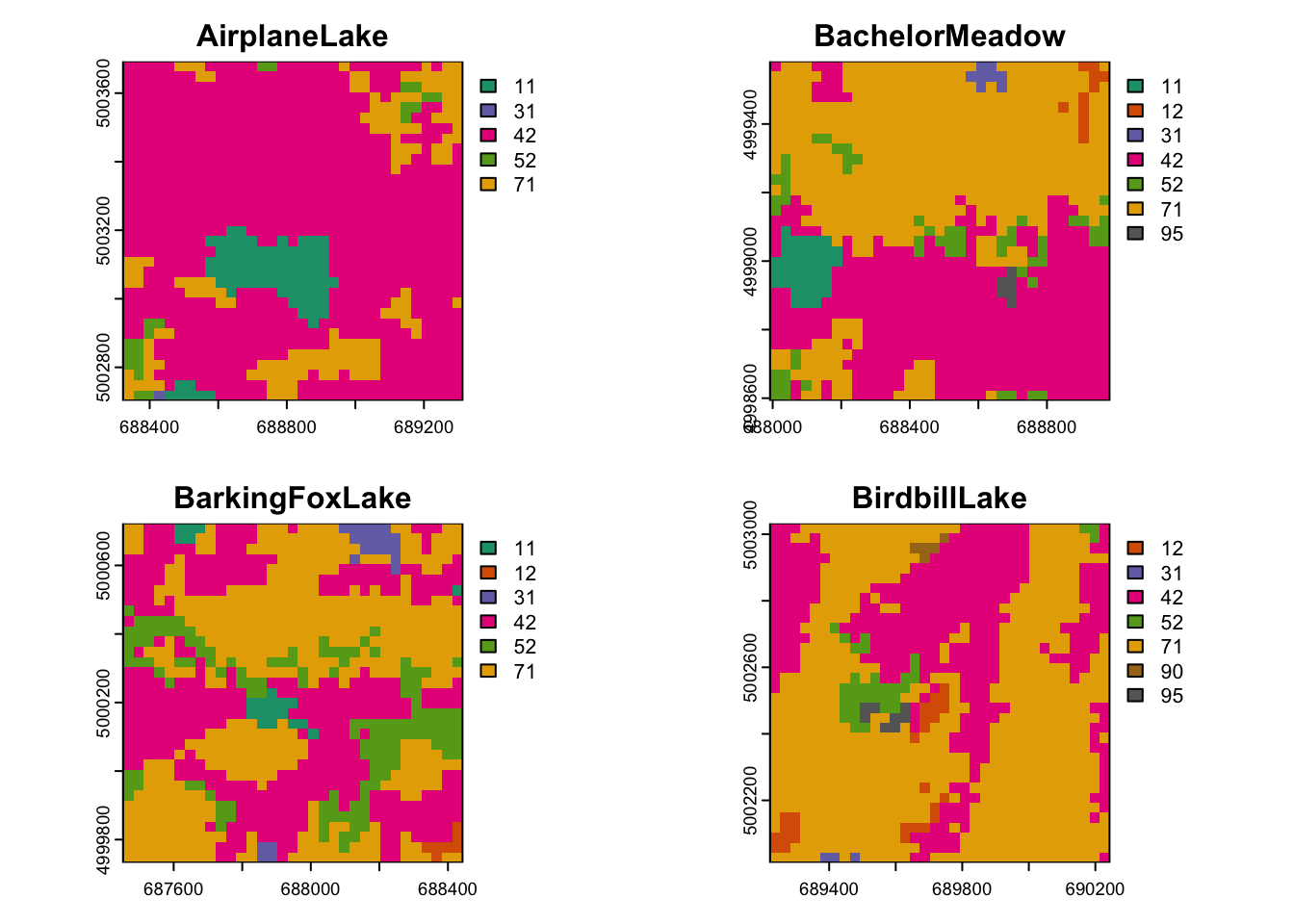
To use consistent colors for the land cover types, we need to tweak things a bit.
- Determine how many colors are needed (
nColor): number of cover types in rasternlcd. - Define a set of
nColorcolors, using a categorical color ramp from packageRColorBrewer:Colors - For each clipped buffer, we use the function
is.elementto determine for each land cover type if it is present in the buffer. The vector Present is of type logical (TRUE/FALSE). - Then we specify the colors to be used as the subset of Colors for which Present is TRUE. (A shorter alternative would be:
col=Colors[Present]).
nColors <- nrow(unique(RasterMaps$nlcd))
Colors <- RColorBrewer::brewer.pal(n = nColors, name = "Dark2")
par(mfrow = c(2,2))
for(i in 1:4)
{
Present <- is.element(unique(RasterMaps$nlcd)$nlcd, unique(unique_plots[[i]])$nlcd)
plot(unique_plots[[i]], type="classes",
col=Colors[Present==TRUE],
main = paste(Sites.sf$SiteName[i]))
}
b. Extract landscape metric of choice for a single cover type (as vector)
To extract a metrics you can just dplyr::filter() the resulting tibble and pull the value column.
Here we filter the results for class == 42 (forest) and metric pland (percentage of landscape) and pull the results as a vector:
percentage_forest_500_a <- dplyr::pull(dplyr::filter(nlcd_sampled,
class == 42,
metric == "pland"), value)
percentage_forest_500_a## [1] 77.50230 39.57759 38.01653 31.40138 40.40404 83.30450 69.16221 86.80927
## [9] 10.19284 72.81910 92.19467 34.04635 34.84848 27.09447 31.37255 56.23886
## [17] 59.77961 58.73440 37.64922 60.12111 46.16756 32.32323 23.35640 50.44563
## [25] 66.93405 30.57851 33.70064 26.53811 45.18717 38.05704 62.47772As an alternative, here’s the same workflow again, but using a pipe:
percentage_forest_500_b <- nlcd_sampled %>%
dplyr::filter(class == 42,
metric == "pland") %>%
dplyr::pull(value)
percentage_forest_500_b## [1] 77.50230 39.57759 38.01653 31.40138 40.40404 83.30450 69.16221 86.80927
## [9] 10.19284 72.81910 92.19467 34.04635 34.84848 27.09447 31.37255 56.23886
## [17] 59.77961 58.73440 37.64922 60.12111 46.16756 32.32323 23.35640 50.44563
## [25] 66.93405 30.57851 33.70064 26.53811 45.18717 38.05704 62.47772c. Extract landscape metric of choice for all cover types (as data frame)
To extract the landscape metric ‘prop.landscape’ for all cover types as a tibble, just filter dplyr::filter() the tibble again, but only use the metric as filter.
## f.lter for percentage of landscape
percentage_forest_500_df <- dplyr::filter(nlcd_sampled,
metric == "pland")
percentage_forest_500_df## # A tibble: 154 × 8
## layer level class id metric value plot_id percentage_inside
## <int> <chr> <int> <int> <chr> <dbl> <int> <dbl>
## 1 1 class 11 NA pland 7.25 1 98.0
## 2 1 class 31 NA pland 0.0918 1 98.0
## 3 1 class 42 NA pland 77.5 1 98.0
## 4 1 class 52 NA pland 1.93 1 98.0
## 5 1 class 71 NA pland 13.2 1 98.0
## 6 1 class 11 NA pland 3.49 2 98.0
## 7 1 class 12 NA pland 1.01 2 98.0
## 8 1 class 31 NA pland 0.735 2 98.0
## 9 1 class 42 NA pland 39.6 2 98.0
## 10 1 class 52 NA pland 5.33 2 98.0
## # ℹ 144 more rowsThe percent cover of all cover types should add up to ~ 100% (i.e., 1) for each site. We can check this with the function dplyr::summarize(). First, we need to group the data using the plot_id, then sum all percentages.
## g.oup by plot_id and sum all percentages
pland_sum_a <- dplyr::summarize(dplyr::group_by(percentage_forest_500_df,
by = plot_id),
sum_pland = sum(value))
pland_sum_a## # A tibble: 31 × 2
## by sum_pland
## <int> <dbl>
## 1 1 100
## 2 2 100
## 3 3 100
## 4 4 100
## 5 5 100
## 6 6 100
## 7 7 100
## 8 8 100
## 9 9 100
## 10 10 100
## # ℹ 21 more rowsSame workflow, but using a pipe:
pland_sum_b <- percentage_forest_500_df %>%
dplyr::group_by(plot_id) %>%
dplyr::summarize(sum_pland = sum(value))
pland_sum_b## # A tibble: 31 × 2
## plot_id sum_pland
## <int> <dbl>
## 1 1 100
## 2 2 100
## 3 3 100
## 4 4 100
## 5 5 100
## 6 6 100
## 7 7 100
## 8 8 100
## 9 9 100
## 10 10 100
## # ℹ 21 more rowsd. Extract all landscape metrics for a single cover type (as data frame)
Filterdplyr::filter() for class == 42 and add the sites names as coloumn to the resulting tibble.
## f.lter for class == 42 (forest)
forest_500_df <- dplyr::filter(nlcd_sampled,
class == 42)
## d.ta.frame with id and name of site
SiteName_df <- data.frame(id = 1:length(Sites.sf$SiteName), site_name = Sites.sf$SiteName)
## a.d site_name to metrics using plot_id and id of sampling sites
forest_500_df <- dplyr::left_join(forest_500_df, SiteName_df, by = c("plot_id" = "id"))
forest_500_df## # A tibble: 93 × 9
## layer level class id metric value plot_id percentage_inside site_name
## <int> <chr> <int> <int> <chr> <dbl> <int> <dbl> <chr>
## 1 1 class 42 NA ai 93.3 1 98.0 AirplaneLake
## 2 1 class 42 NA np 2 1 98.0 AirplaneLake
## 3 1 class 42 NA pland 77.5 1 98.0 AirplaneLake
## 4 1 class 42 NA ai 89.4 2 98.0 BachelorMeadow
## 5 1 class 42 NA np 7 2 98.0 BachelorMeadow
## 6 1 class 42 NA pland 39.6 2 98.0 BachelorMeadow
## 7 1 class 42 NA ai 79.4 3 98.0 BarkingFoxLake
## 8 1 class 42 NA np 10 3 98.0 BarkingFoxLake
## 9 1 class 42 NA pland 38.0 3 98.0 BarkingFoxLake
## 10 1 class 42 NA ai 81.2 4 104. BirdbillLake
## # ℹ 83 more rowsDone!
Check out this week’s bonus material to see:
- More details on how to create static and interactive maps of an
sfobject withtmap. - How to import and export ESRI shapefiles.
- How to plot a categorical raster map with a predefined color scheme, using
tmapandterra. package).