Chapter 4 Handling data in R
Motivating scenarios: We just got our data, how do we get it into R and explore?
Learning goals: By the end of this chapter you should be able to get data into R and explore it. Specifically, students will be able to:
filter,mutate,arrange, andselectdata with thedplyrpackage.
- Pipe %>% these operations together.
4.1 Intro
FIGURE 4.1: Watch this video about manipulaitng data with dplyr (2 min and 09 sec). Note this is part of an intro to R series I am developing for CBS – comments appreciated
Through the rest of this chapter we will buy in fully to the tidyverse, so make sure it’s installed and load it at the top of your script by typing and entering library(tidyverse). In the next portions of this section, we focus on how to load and process data with tidyverse tools in R.
But first, we describe the primary structure that holds data in the tidyverse, a tibble. A tibble is a data structure in which each column is a vector and (ideally) entries in a row are united by relating to the same observation (e.g. individual at a time). Each variable associated with an observation is a column (i.e. a vector), and all so while all entries in a column must be of the same class, each column in a tibble can have its own class. Tibbles do not ensure that our data are tidy but they do make this easier.
If you have spent time in base R you are likely familiar with matrices, arrays, and data frames - don’t even worry about these. A tibble is much like a data frame, but has numerous features that make them easier to deal with. If you care, see chapter 10 of R for Data Science (Grolemund and Wickham 2018) for more info. Anyways, in this course we will focus on vectors and tibbles, ignoring arrays and matrices, and avoiding lists for as long as possible.
4.2 OPTIONAL Entering data into R
We usually import data into R with the read_csv() function, but we can also manually enter datasets into R. This is useful for small datasets for practice and quick analyses.
There are a bunch of ways we can create tibbles manually. I present the simplest and most common way this is done.
toad_data <- tibble( # This makes the data
individual = c("a", "b", "c"),
species = c("Bufo spinosus", "Bufo bufo", "Bufo bufo"),
sound = c("chirp", "croak","ribbit"),
weight = c(2, 2.6, 3),
height = c(2,3,2)
)
toad_data # This shows the data## # A tibble: 3 × 5
## individual species sound weight height
## <chr> <chr> <chr> <dbl> <dbl>
## 1 a Bufo spinosus chirp 2 2
## 2 b Bufo bufo croak 2.6 3
## 3 c Bufo bufo ribbit 3 2Feel free to read about more ways to make tibbles if you desire. Many of these options are fun and useful, but a distraction from our major mission
4.3 Dealing with data in R

Looking at the toad.data tibble above, we can get a sense of the utility of a tibble. We can see, not only the first few values of the data set, but also the class of each variable (chr for character, fct for factor, dbl for double – a continuous class of data).
Recall that we can get a sense of a dataset with the glimpse() function in dplyr, and scroll through it with View().
While glimpse() and View() are among the very handy tidyverse functions, the real utility of tidyverse is that it gives us a unified way to deal with data. Usually when we get data we want to handle/clean it, summarize it, visualize the results and develop a statistical model from it. This is where tidyverse really shines!
We first focus on handling data with the dplyr package. Today we’ll talk about using it to filter, arrange, mutate, and summarize our data, and to select columns of interest.
I say this all the time but: learning dplyr + ggplot was one of the highest payoff things I've done in my career.
— Joshua G. Schraiber🌹 (@jgschraiber) January 22, 2019
4.3.1 mutate your data
FIGURE 4.2: Watch this video about how to add columns with the mutate function (6 min and 07 sec). Note this is part of an intro to R series I am developing for CBS – comments appreciated

In our toad_data, we have height and weight. Let’s add a column for BMI (weight divided by height) with the mutate() function.
toad_data <- mutate(toad_data, BMI = height / weight)
toad_data## # A tibble: 3 × 6
## individual species sound weight height BMI
## <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 a Bufo spinosus chirp 2 2 1
## 2 b Bufo bufo croak 2.6 3 1.15
## 3 c Bufo bufo ribbit 3 2 0.6674.3.2 Nice helper verbs
FIGURE 4.3: Watch this video about using the filter(), arrange(),select() and rename() functions to handle data in R (10 min and 46 sec). Note this is part of an intro to R series I am developing for CBS – comments appreciated
4.3.2.1 arrange rows
You might want to sort BMI from lowest to highest, or vice-versa. The arrange() function is here for you!
arrange(toad_data, BMI) # arrange from lowest to highest## # A tibble: 3 × 6
## individual species sound weight height BMI
## <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 c Bufo bufo ribbit 3 2 0.667
## 2 a Bufo spinosus chirp 2 2 1
## 3 b Bufo bufo croak 2.6 3 1.15arrange(toad_data, desc(BMI)) # arrange from highest to lowest## # A tibble: 3 × 6
## individual species sound weight height BMI
## <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 b Bufo bufo croak 2.6 3 1.15
## 2 a Bufo spinosus chirp 2 2 1
## 3 c Bufo bufo ribbit 3 2 0.6674.3.2.2 filter your data
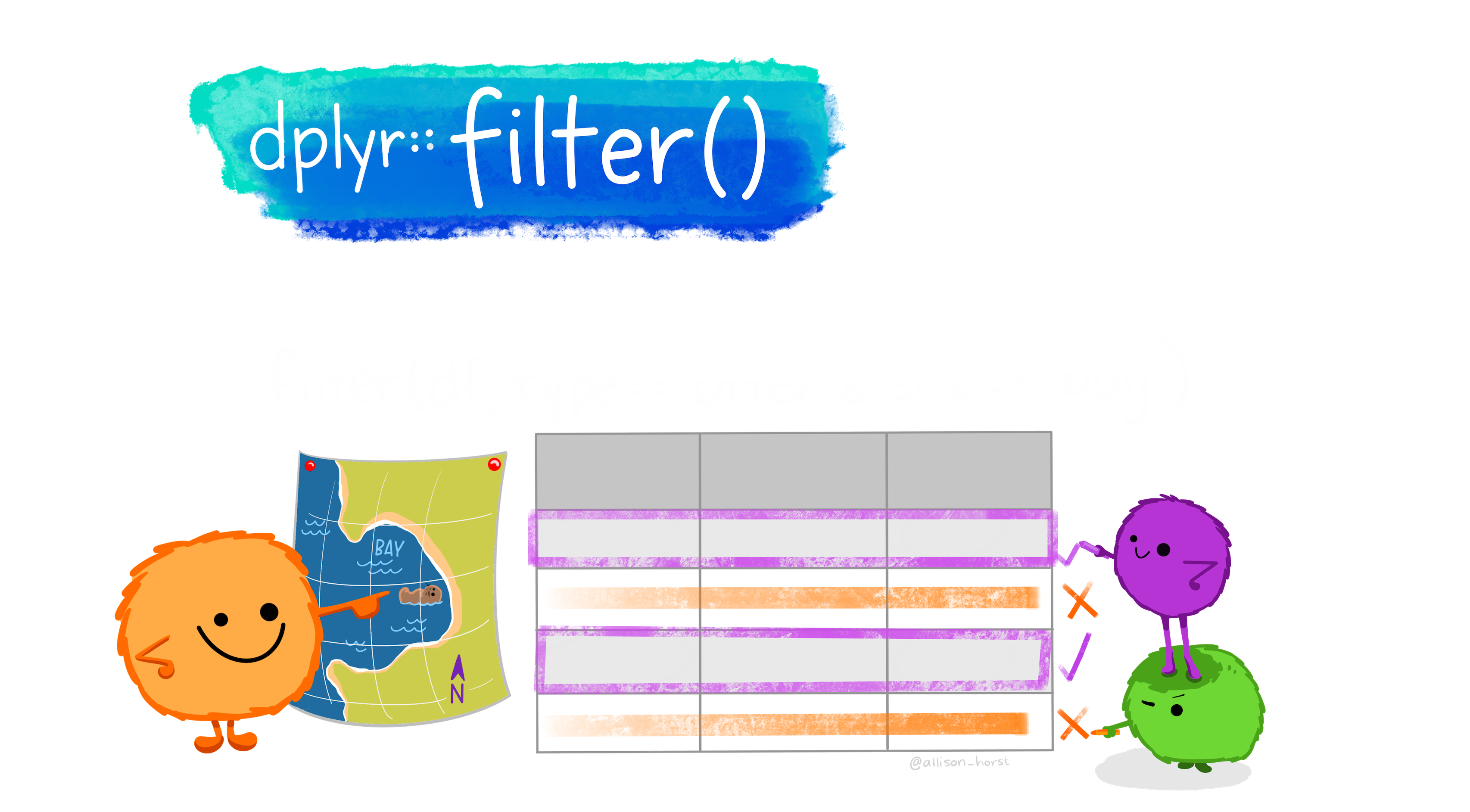
Say we only wanted to deal with individuals of the species, Bufo bufo. We can filter() our data to only have them!
dplyr::filter(toad_data, species == "Bufo bufo" )## # A tibble: 2 × 6
## individual species sound weight height BMI
## <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 b Bufo bufo croak 2.6 3 1.15
## 2 c Bufo bufo ribbit 3 2 0.6674.3.2.3 select your columns
Let’s say we didn’t care about the height or weight, and we just wanted individual, species, sound, and BMI. We can select() those columns as follows:
dplyr::select(toad_data, individual, species, sound, BMI)## # A tibble: 3 × 4
## individual species sound BMI
## <chr> <chr> <chr> <dbl>
## 1 a Bufo spinosus chirp 1
## 2 b Bufo bufo croak 1.15
## 3 c Bufo bufo ribbit 0.667A negative sign in select means remove, so select(toad_data, -height, - weight) will give the same result as the code above.
dplyr::select(toad_data, -height, -weight)## # A tibble: 3 × 4
## individual species sound BMI
## <chr> <chr> <chr> <dbl>
## 1 a Bufo spinosus chirp 1
## 2 b Bufo bufo croak 1.15
## 3 c Bufo bufo ribbit 0.6674.3.2.4 pull() vectors from tibbles
Some things we do in R require vectors to be pulled out of tibbles. We can achieve this with the pull() function. For example, to get the BMI as a vector, type
pull(toad_data, var = BMI)## [1] 1.0000 1.1538 0.66674.3.2.5 The pipe %>% operator
In the previous section we learned how to do a bunch of things to data. For example, in our toad dataset, below, we
- Use the
mutate()function made a new column forBMIby dividing weight by height.
- Sort the data with the
arrange()function.
We also saw how we could select() columns, and filter() for rows based on logical statements.
We did each of these things one at a time, often reassigning variables a bunch. Now, we see a better way, we combine operations with the pipe %>% operator.
Say you want to string together a few things – like you want make a new tibble, called sorted_bufo_bufo by:
- Only retaining Bufo bufo samples
- Calculating
BMI
- Sorting by BMI, and
- Getting rid of the column with the species name.
The pipe operator, %>%, makes this pretty clean by allowing us to pass results from one operation to another.
%>% basically tells R to take the data and keep going!
sorted_bufo_bufo <- toad_data %>% # initial data
dplyr::filter(species == "Bufo bufo") %>% # only Bufo bufo
dplyr::mutate(BMI = height / weight) %>% # calculate BMI
dplyr::arrange(BMI) %>% # sort by BMI
dplyr::select(-species) # remove species
sorted_bufo_bufo| individual | sound | weight | height | BMI |
|---|---|---|---|---|
| c | ribbit | 3.0 | 2 | 0.6667 |
| b | croak | 2.6 | 3 | 1.1538 |
4.3.3 Summarize your data
FIGURE 4.4: Watch this video about using the `f summarizing data in R (9 min and 43 sec). Note this is part of an intro to R series I am developing for CBS – comments appreciated
We often want high-level summaries of our data to e.g. stimate parameters. We use the summarise() function to reduce a data set to summaries that you ask it for. So, we can use tell R to summarize the toad_data data as follows.
toad_data %>%
dplyr::summarise(mean_weight = mean(weight),
mean_height = mean(height),
mean_BMI = mean(BMI))## # A tibble: 1 × 3
## mean_weight mean_height mean_BMI
## <dbl> <dbl> <dbl>
## 1 2.53 2.33 0.940Say we wanted separate means for each species in our toad data set. We can combine the group_by() function from with the summarise() function introduced above.
toad_data %>%
group_by(species) %>%
dplyr::summarise(mean_BMI = mean(BMI))## # A tibble: 2 × 2
## species mean_BMI
## <chr> <dbl>
## 1 Bufo bufo 0.910
## 2 Bufo spinosus 14.4 Think about reproducibility
Science is iterative, social / collaborative, and builds on previous work. You should know that
- Your first analysis is almost never your last.
- Your will need to be able to explain EXACTLY what you did to others.
- People might want to take you’ve done, understand it, and do it again on this or another data set, perhaps changing it a bit.
Think about this when you work in R. I suggest asking yourself the following questions:
(1) Could I explain what my code is doing to a colleague? What if I hadn’t looked at it in a month? Could someone who’s pretty good with R look at my code and understand what I was trying to do?
(2) Could my code work well (within minimal changes) on another data set? How about on another computer?
If your work is fully reproducible, the answers should all be yes.
One of the great things about scripting-based analyses that you can do in R (or similar programs) is that it facilitates us answering “yes” to most of these questions — take advantage of this. Always save your R script. Use enough commenting (#) to make sure it makes sense. Make sure it works from start to finish without anything in your R environment. etc…
4.5 Functions covered in Handling data in R
All require the tidyverse package
tibble(): Entering data as a tibble. Give each column a name and assign its values with an equals sign, =. Separate columns with a comma ,.glimpse(): Show the name of every column in your data frame, as well as their class and first few values.view(): Look at your entire tibble as a scrollable spreadsheet.mutate(): Add a new column, usually as some function of existing columns.arrange(): Sort rows from low to high (or from high to low with arrange(desc())) values of a specified column.filter(): Limit your dataset to those with certain values in specifed columns.select(): Select columns of interest (or remove ones you do not care about with select(-)) from your tibble. ```
4.5.1 dplyr cheat sheet
There is no need to memorize anything, check out this handy cheat sheet!
](images/dplyr_cheat1.png)
FIGURE 4.5: download the dplyr cheat sheet
](images/dplyr_cheat2.png)
FIGURE 4.6: download the dplyr cheat sheet