Chapter 17 Advanced Regression Topics
Get Started
This chapter continues the analysis of country-level life expectancy by taking into account other factors that might be related to health conditions in general. We will also address a few important issues related to evaluating multiple regression models. To follow along in R, you should load the countries2 data set, as well as the libraries for the DescTools and stargazer packages.
Incorporating Access to Health Care
One set of variables that we haven’t considered but that should be related to life expectancy are factors that measure access to health care. Generally speaking, countries with greater availability of health care resources (doctors, hospitals, research centers, etc) should have higher levels of life expectancy. In the table below, the regression model is expanded to include two additional variables, percent of the population living in urban areas and the number of doctors per 10,000 persons. These variables are expected to be positively related to life expectancy, as they are linked to the availability of health care services and facilities. Doctors per 10,000 persons is a direct measure of access to health care, and levels of access to health care are generally lower in rural areas than in urban areas.
These variables are added in the model below:
#Add % urban and doctors per 10k to the model
fit<-lm(data=countries2, lifexp~fert1520 +
mnschool+ log10(pop19_M)+urban+docs10k)
#Use information in 'fit' to create table of results
stargazer(fit, type="text",
dep.var.labels=c("Life Expectancy"), #Dependent variable label
covariate.labels = c("Fertility Rate", #Indep Variable Labels
"Mean Years of Education",
"Log10 Population", "% Urban",
"Doctors per 10,000"))
===================================================
Dependent variable:
---------------------------
Life Expectancy
---------------------------------------------------
Fertility Rate -3.237***
(0.341)
Mean Years of Education 0.413**
(0.163)
Log10 Population 0.082
(0.331)
% Urban 0.050***
(0.015)
Doctors per 10,000 0.049*
(0.027)
Constant 73.923***
(2.069)
---------------------------------------------------
Observations 179
R2 0.768
Adjusted R2 0.761
Residual Std. Error 3.606 (df = 173)
F Statistic 114.590*** (df = 5; 173)
===================================================
Note: *p<0.1; **p<0.05; ***p<0.01Here’s how I would interpret the results:
- The first thing to note is that four of the five variables are statistically significant (counting
docs10kas significant with a one-tailed test), and the overall model fit is improved in comparison to the fit for the three-variable model from Chapter 16. The only variable that has no discernible impact is population size. The model R2 is now .768, indicating that the model explains 76.8% of the variation in life expectancy, and the adjusted R2 is .761, an improvement over the previous three-variable model (adjusted R2 was .743). Additionally, the RMSE is now 3.606, also signaling less error compared to the previous model (3.768).
Interpretation of the individual variables should be something like the following:
Fertility rate is negatively and significantly related to life expectancy. For every one unit increase in the value of fertility rate, life expectancy is expected to decline by about 3.24 years, controlling for the influence of the other variables in the model.
Average years of education is positively related to life expectancy. For every unit increase in education level, life expectancy is expected to increase by about .413 years, controlling for the influence of the other variables in the model.
Percent of urban population is positively related to life expectancy. For every unit increase in the percent of the population living in urban areas, life expectancy is expected to increase by .05 years, controlling for the influence of other variables in the model.
Doctors per 10,000 population is positively related to life expectancy. For every unit increase in doctors per 10,000 population, life expectancy is expected to increase by about .05 years, controlling for the influence of the other variables in the model.
I am treating the docs10k coefficient as statistically significant even though the p-value is greater than .05. This is because the reported p-value (< .10) is a two-tailed p-value, and if we apply a one-tailed test (which makes sense in this case), the p-value is cut in half and is less than .05. The two-tailed p-value (not shown in the table) is .068, so the one-tailed value is .034. Still, in a case like this, where the level of significance is borderline, it is best to assume that the effect is pretty small. A weak effect like this is surprising for this variable, especially since there are strong substantive reasons to expect greater access to health care to be strongly related to life expectancy. Can you think of an explanation for this? We explore a couple of potential reasons for this weaker-than-expected relationship in the next two sections.
Multicollinearity
One potential explanation for the seemingly tepid role of docs10k in the life expectancy model is multicollinearity, a problem that can emerge when independent variables are highly correlated with each other. Recall from the discussions in Chapters 14 and 16 that when independent variables are strongly correlated with each other the simple bi-variate relationships are likely overstated and the independent variables lose strength when considered in conjunction with other overlapping explanations. Normally, this is not a major concern. In fact, one of the virtues of regression analysis is that the model sorts out overlapping explanations (this is why we use multiple regression). However, when the degree of overlap is very high, it can make it difficult for substantively important variables to demonstrate statistical significance, which can increase the probability of making a Type II error. Perfect collinearity violates a regression assumption (see next chapter), but generally high levels of collinearity can also cause problems, especially for interpreting significance levels.
Here’s how this problem comes about: the t-score for a regression coefficient (\(b\)) is calculated as \(t=\frac{b}{S_b}\), and the standard error of b (\(S_b\)) for any given variable is directly influenced by how that variable is correlated with other independent variables. The formula below provides a clue to how collinearity can affect the standard error of \(b_1\) in a model with two independent variables:
\[S_{b_1}=\sqrt{\frac{RMSE}{\sum(x_i-\bar{x}_1)^2(1-R^2_{1\cdot2})}}\]
The key to understanding how collinearity affects the standard error, which then affects the t-score, lies in part of the denominator of the formula for the standard error: \((1-R^2_{1\cdot2})\). As the correlation (and R2) between x1 and x2 increases, the denominator of the formula decreases in size, leading to larger standard errors and smaller t-scores. Because of this, high correlations among independent variables can make it difficult for variables to demonstrate statistical significance. This problem can be compounded when there are multiple strongly related independent variables. This problem is usually referred to as high multicollinearity, or high collinearity.
When you have good reasons to expect a variable to be strongly related to the dependent variable and it is not statistically significant in a multiple regression model, you should think about whether collinearity could be a problem. Is this a potential explanation for the borderline statistical significance for docs10k? It does make sense that docs10k is strongly related to other independent variables, especially since most of the variables reflect an underlying level of economic development. As a first step toward diagnosing this, let’s look at the correlation matrix for evidence of collinearity:
#Create "logpop" variable for exporting
countries2$lg10pop<-log10(countries2$pop19_M)
#Copy DV and IVs to new data set.
lifexp_corr<-na.omit(countries2[,c("lifexp", "fert1520", "mnschool",
"lg10pop", "urban", "docs10k")])
#Use variable in 'lifexp_corr' to produce correlation matrix
cor(lifexp_corr) lifexp fert1520 mnschool lg10pop urban docs10k
lifexp 1.00000 -0.839901 0.768438 -0.036340 0.602412 0.70310
fert1520 -0.83990 1.000000 -0.762732 0.070566 -0.518459 -0.67767
mnschool 0.76844 -0.762732 1.000000 -0.095843 0.578742 0.76661
lg10pop -0.03634 0.070566 -0.095843 1.000000 0.079091 -0.01679
urban 0.60241 -0.518459 0.578742 0.079091 1.000000 0.55433
docs10k 0.70310 -0.677672 0.766615 -0.016790 0.554334 1.00000Look across the row headed by docs10k, or down the docs10k column to see how strongly it is related the other variables. First, there is a strong bivariate correlation (r=.70) between doctors per 10,000 population and the dependent variable, life expectancy, so it is a bit surprising that its effect is so small in the regression model. Further, docs10k is also highly correlated with fertility rates (r= -.68), mean years of education (r=.77), and percent living in urban areas (r= .55). Based on these correlations, it seems plausible that collinearity could be causing a problem for the significance of slope for docs10k.
There are two important statistics that help you evaluate the extent of the problem with collinearity: The tolerance and VIF (variance inflation) statistic. Tolerance is calculated as:
\[ \text{Tolerance}_{x_1}=1-R^2_{x_1,x_2\cdot \cdot x_k}\]
You should recognize this from the denominator of the formula for the standard error of the regression slope; it is the part of the formula that inflates the standard error when the independent variables are highly correlated with each other. Note that as the proportion of variation in x1 that is accounted for by the other independent variables increases, the tolerance value decreases. The tolerance is the proportion of variance in one independent variable that is not explained by variation in the other independent variables.
The VIF statistic tells us how much the standard error of the slope is inflated due to inter-item correlation. The calculation of the VIF is based on the tolerance statistic:
\[\text{VIF}_{b_1}=\frac{1}{\text{Tolerance}_{b_1}}\] We can calculate tolerance for docs10k stats by regressing it on the other independent variables. In other words, treat docs10k as a dependent variable and see how much of its variation is accounted for by the other variables in the model. We can get this information below by running the regression model and then using the summary function to extract just the R2 value:
#Use 'docs10k' as the DV to Calculate its Tolerance
fit_tol<-lm(data=countries2, docs10k~fert1520 + mnschool
+ lg10pop + urban)
#Use information in 'fit_tol' to create a table of results
summary(fit_tol)$r.square[1] 0.62321Note that the R2=.623, so 62.3 percent of the variation in docs10k is accounted for by the other independent variables. Using this information, we get:
Tolerance: 1-.623 = .377
VIF (1/tolerance): 1/.377 = 2.65
So, is a VIF of 2.65 a lot? In my experience, no, but it is best to evaluate this number in the context of the VIF statistics for all variables in the model. Instead of calculating all of these ourselves, we can have R get the VIF statistics for us:
fert1520 mnschool log10(pop19_M) urban docs10k
2.5471 3.5018 1.0421 1.6294 2.6540 Does collinearity explain the marginally significant slope for docs10k? Yes and no. Sure, the slope for docs10k would probably have a smaller p-value if we excluded the variables that are highly correlated with it, but some loss of impact is almost always going to happen when using multiple regression; that’s the point of using it! Also, the issue of collinearity is not appreciably worse for docs10k than it is for fertility rate, and it is less severe than it is for mean years of education, so it’s not like this variable faces a higher hurdle than the others. Based on these results I would conclude that collinearity is a slight problem for docs10k, but that there is probably a better explanation for its level of statistical significance.
While there is no magic cutoff point for determining that collinearity is a problem that needs to be addressed, in my experience VIF values greater than 5 should get your attention, anything in the 7-10 range signals that there could be a problem worth looking into, and values greater than 10 should be taken seriously. You always have to consider issues like this in the context of the data and model you are using.
Even though collinearity is not a problem here, it is important to consider what to do if there is extreme collinearity in a regression model. Some people suggest dropping a variable, possibly the one with the highest VIF. I don’t generally like to do this, but it is something to consider, especially if you have several variables that are measuring different aspects of the same concept and dropping one of them would not hurt your ability to measure an important concept. For instance, suppose this model also included hospital beds per capita and nurses per capita. These variables, along with docs10k all measure different aspects of access to health care and should be strongly related to each other. If there were high VIF values for these variables, we could probably drop one or two of them to decrease collinearity and would still be able to consider access to health care as an explanation of differences in life expectancy.
Another alternative if there are multiple indicators of the same underlying concept in a model is to combine the variables into a single variable. Suppose again that this model included hospital beds per capita, nurses per capita, and docs10k as measures of access to health care. If there were a high level of collinearity among these variables, we could probably combine them into a single index of health care access. We did something like this in Chapter 4, where we created a single index of LGBTQ policy preferences based on outcomes from several separate LGBTQ policy questions. Indexing like this minimizes the collinearity problem without dropping any variables and also provides a broader measure of the underlying concept. There are many different ways to combine variables, but expanding on that here is a bit beyond the scope of this book.
Checking on Linearity
The relationship between docs10k and life expectancy presents an opportunity to explore another important potential problem that can come up in regression analysis. Theoretically, there should be a relatively strong relationship between these two variables, not one whose statistical significance in the multiple regression model depends upon whether you are using one- or two-tailed test. We have already explored collinearity as an explanation for the weak showing of docs10k, but that does not appear to be the primary explanation. While the bivariate correlation (r=.70) with the dependent variable is reported above, one thing we did not do earlier was examine a simple scatterplot for this relationship. Let’s do that now to see if it offers any clues.
#Scatterplot of "lifexp" by "docs10k"
plot(countries2$docs10k, countries2$lifexp, ylab="Life Expectancy",
xlab="Doctors per 10k Population")
#Add linear regression line
abline(lm(countries2$lifexp~countries2$docs10k))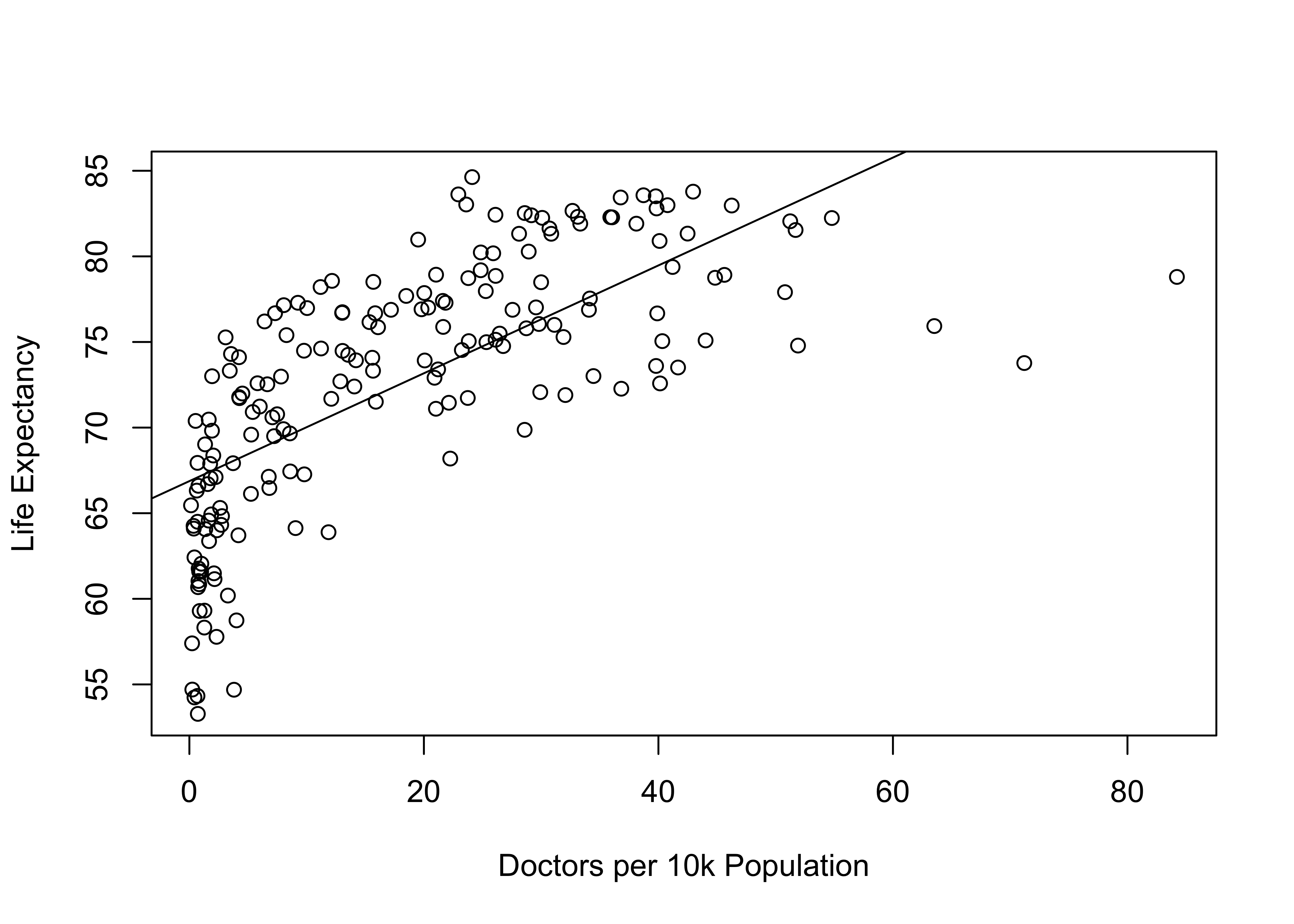
This is interesting. A rather strange looking pattern, right? Note that the linear prediction line does not seem to fit the data in the same way we have seen in most of the other scatterplots. Focusing on the pattern in the markers, this doesn’t look like a typical linear pattern. At low levels of the independent variable there is a lot of variation in life expectancy, but, on average, it is fairly low. Increases in doctors per 10k from the lowest values to about 15 leads to substantial increases in life expectancy but then there are diminishing returns from that point on. Looking at this plot, you can imagine that a curved line would fit the data points better than the existing straight line. One of the important assumptions of OLS regression is that all relationships are linear, that the expected change in the dependent variable for a unit change in the independent variable is constant across values of the independent variable. This is clearly not the case here. Sure, you can fit a straight line to the pattern in the data, but if the pattern in the data is not linear, then the line does not fit the data as well as it could, and we are violating an important assumption of OLS regression (see next Chapter).
Just as \(y=a + bx\) is the equation for a straight line, there are a number of possibilities for modeling a curved line. Based on the pattern in the scatterplot–one with diminishing returns–I would opt for the following model:
\[\hat{y}=a + b*log_{10}x\]
Here, we transform the independent variable by taking the logged values of its outcomes.
Log transformations are very common, especially when data show a curvilinear pattern or when a variable is heavily skewed. In this case, we are using a log(10) transformation. This means that all of the original values are transformed into their logged values, using a base of 10. As discussed in Chapter 5, a logged value is nothing more than the power to which you have to raise your log base (in this case, 10) in order to get the original raw score. Using logged values has two primary benefits: it minimizes the impact of extreme values, which can be important for highly skewed variables, and it enables us to model relationships as curvilinear rather than linear.
One way to assess whether the logged version of docs10k fits the data better than the raw version is to look at the bivariate relationships with the dependent variable using simple regression. The table of regression results shown below offers a side-by-side comparison of the two models:
#Life expectancy by "docs10k"
fit_raw<-lm(countries2$lifexp~countries2$docs10k,
na.action=na.exclude)
#Life expectancy by "log10(docs10k)"
fit_log<-lm(countries2$lifexp~log10(countries2$docs10k),
na.action=na.exclude)
#Use Stargazer to compare the results from two models
stargazer(fit_raw, fit_log, type="text",
dep.var.labels = c("Life Expectancy"),
covariate.labels = c("Doctors per 10k",
"Log10(Doctors per 10k)"))
===========================================================
Dependent variable:
----------------------------
Life Expectancy
(1) (2)
-----------------------------------------------------------
Doctors per 10k 0.315***
(0.024)
Log10(Doctors per 10k) 9.582***
(0.492)
Constant 66.874*** 63.404***
(0.581) (0.565)
-----------------------------------------------------------
Observations 186 186
R2 0.487 0.673
Adjusted R2 0.484 0.671
Residual Std. Error (df = 184) 5.294 4.227
F Statistic (df = 1; 184) 174.810*** 378.860***
===========================================================
Note: *p<0.1; **p<0.05; ***p<0.01All measures of fit, error, and relationship point to the logged (base 10) version of docs10k as the superior operationalization. Based on these outcomes, it is hard to make a case against using the log-transformed version of docs10k. You can also see this in the scatterplot in Figure 17.1, which includes the prediction lines from both the raw and logged models.
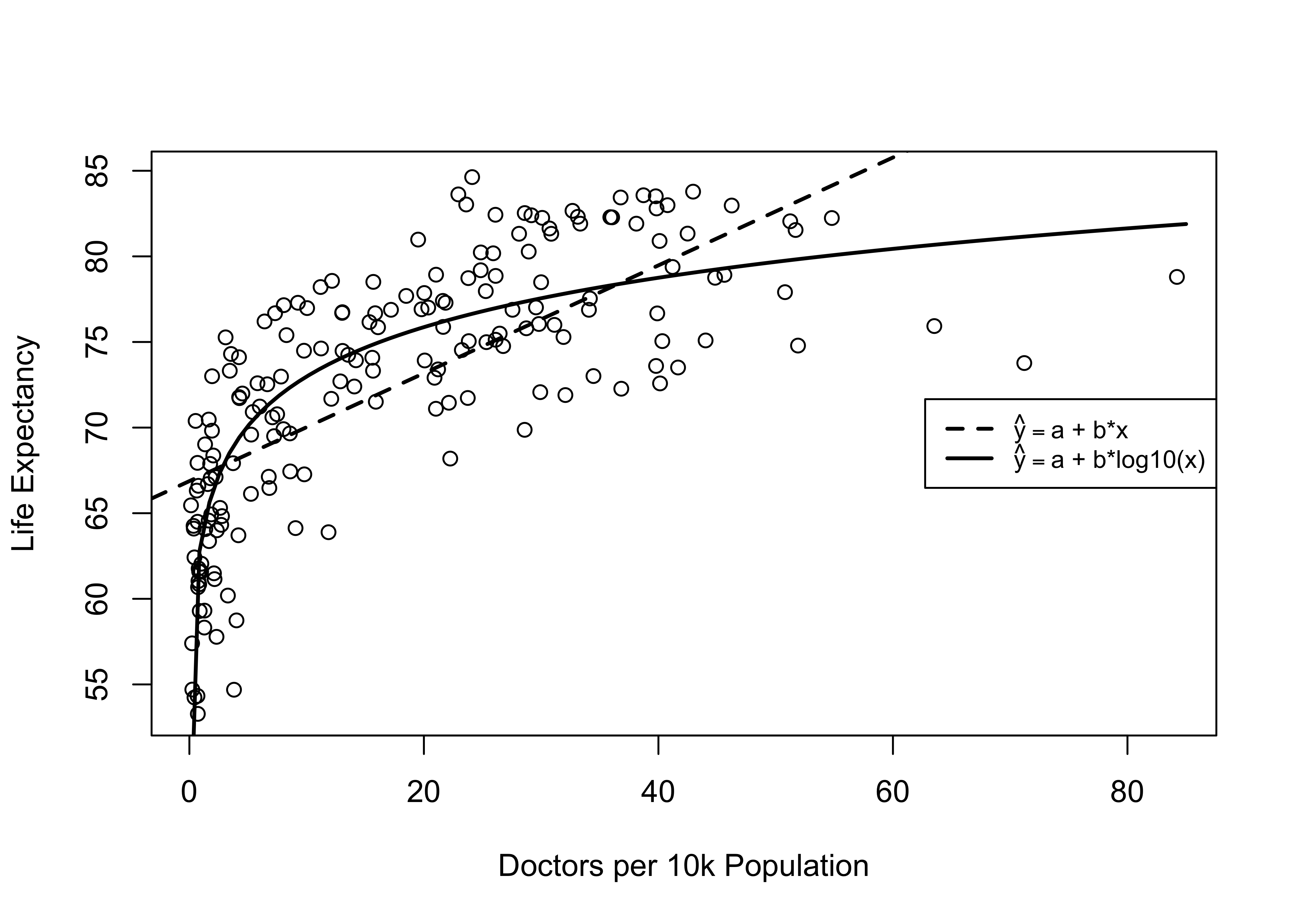
Figure 17.1: Alternative Models for the Relationship between Doctors per 10k and Life Expectancy
[Figure**************** 17.1 ****************about here]
Note that the curved, solid line fits the data points much better than the straight, dashed line, which is what we expect based on the relative performance of the two regression models above. The interpretation of the curvilinear relationship is a bit different than for the linear relationship. Rather than concluding that life expectancy increases as doctors per capita increases, we see in the scatterplot that life expectancy increases substantially as we move from nearly 0 to about 15 doctors per 10,000 people, but increases beyond that point have less and less impact on life expectancy.
This is illustrated further below in Figure 17.2 where the graph on the left shows the relationship between docs10k among countries below the median level (14.8) of doctors per 10,000 population, and the graph on the right shows the relationship among countries with more than the median level of doctors per 10,000 population. This is not quite as artful as Figure 17.1, with the plotted curved line, but it does demonstrate that the greatest gains in life expectancy from increased access to health care come at the low end of the scale, among countries with severely limited access. For the upper half of the distribution, there is almost no relationship between levels of docs10k and life expectancy.
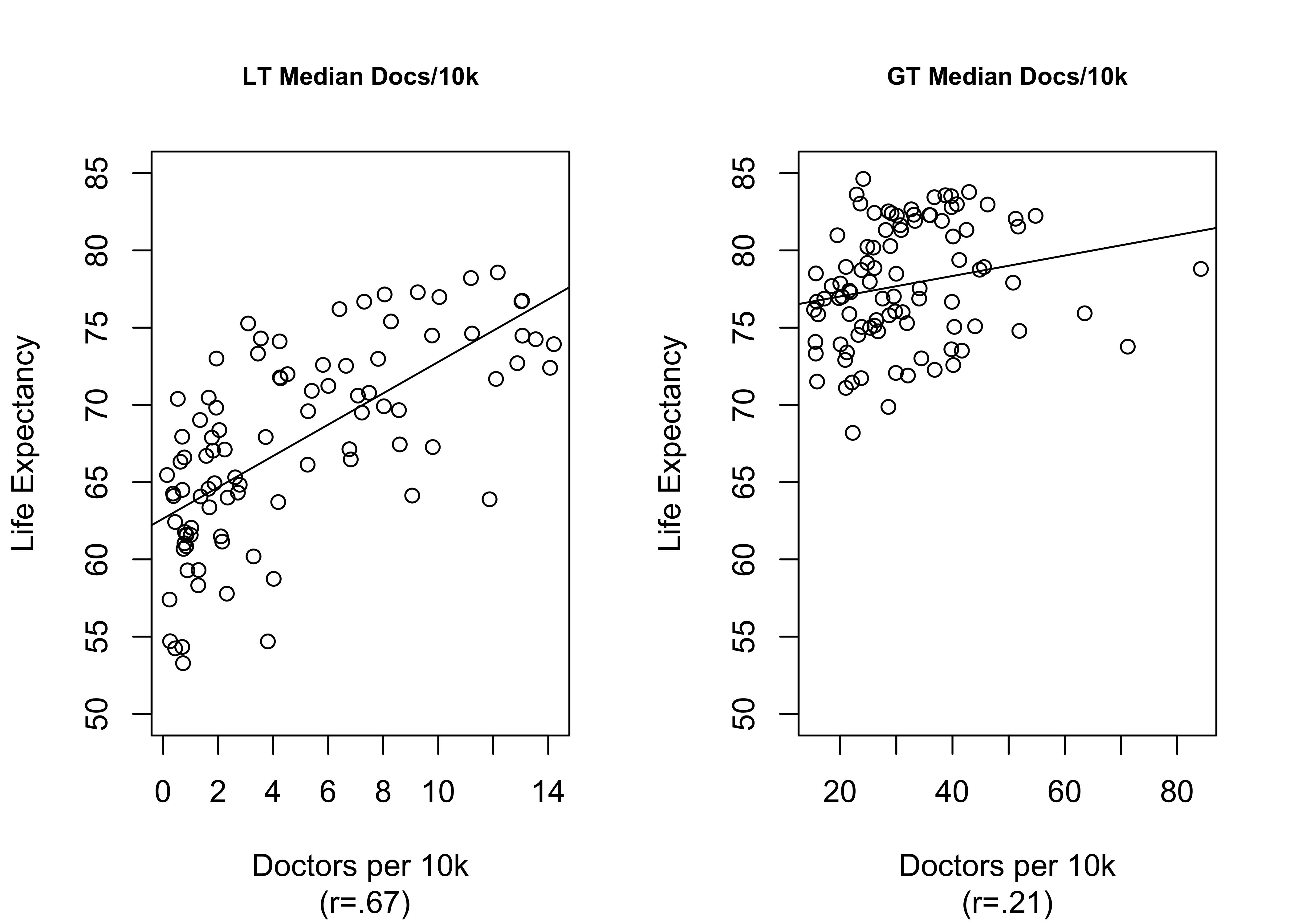
Figure 17.2: A Segmented View of a Curvilinear Relationship
[Figure**************** 17.2 ****************about here]
Finally, we can re-estimate the multiple regression model with the appropriate version of docs10k to see if this transformation has an impact on its statistical significance.
#Re-estimate five-variable model using "log10(docs10k)"
fit<-lm(data=countries2, lifexp~fert1520 + mnschool+
log10(pop19_M) + urban+
log10(docs10k), na.action = na.exclude)
#Using information in 'fit' to create a table of results
stargazer(fit, type="text", dep.var.labels=c("Life Expectancy"),
covariate.labels = c("Fertility Rate",
"Mean Years of Education",
"Log10(Population)",
"% Urban",
"log10(Doctors per 10k)"))
===================================================
Dependent variable:
---------------------------
Life Expectancy
---------------------------------------------------
Fertility Rate -2.808***
(0.399)
Mean Years of Education 0.356**
(0.163)
Log10(Population) 0.050
(0.329)
% Urban 0.045***
(0.015)
log10(Doctors per 10k) 2.424**
(0.973)
Constant 72.109***
(2.135)
---------------------------------------------------
Observations 179
R2 0.772
Adjusted R2 0.765
Residual Std. Error 3.577 (df = 173)
F Statistic 117.000*** (df = 5; 173)
===================================================
Note: *p<0.1; **p<0.05; ***p<0.01Accounting for the curvilinear relationship between docs10k and life expectancy made an important difference to the model. First, we now see a statistically significant impact from the logged version of the docs10k, whether using a one- or two-tailed test (the t-score changes from 1.83 in the original model to 2.49 in the model above). We need to be a little bit careful here when discussing the coefficient for the logged variable. Although there is a curvilinear relationship between docs10k and life expectancy, there is a linear relationship between log10(docs10k) and life expectancy. So, we can say that for every unit increase in the logged value of docs10k, life expectancy increases by 2.4 years, but we need to keep in mind that when we translate this into the impact of the original version of docs10k, the relationship is not linear. In addition to doctors per 10,000 population, the fertility rate, mean years of education, and percent urban population are also statistically significant, and there is less error in predicting life expectancy when using the logged version of docs10k: the RMSE dropped from 3.606 to 3.58, and the R2 increased from .768 to .772.
Stop and Think about Results
These findings underscore the importance of thinking hard about results that don’t make sense. There is a strong theoretical reason to expect life expectancy to be strongly related to doctors per capita, as greater access to health care should lead to better health outcomes. However, the initial regression results suggested that doctors per capita was tangentially related to life expectancy. Had we simply accepted this result, we would have come to an erroneous conclusion. Instead, by exploring explanations for the null finding, we were able to uncover a more nuanced and powerful role for the number of doctors per 10k population.
Of course, it is important to point out that one of the sources of “getting it wrong” in the first place was skipping a very important first step: examining the bivariate relationship closely, using not just the correlation coefficient but also the scatterplot. Had I started there, I would have understood the curvilinear nature of the relationship from the get go.
Which Variables have the Greatest Impact?
As a researcher, one thing you’re likely to be interested in is the relative impact of the independent variables; that is which variables have the greatest and least impact on the dependent variable? If we look at the regression slopes, we might conclude that the order of importance runs from highest-to-lowest in the following order: the fertility rate (b=-2.81) and log of doctors per 10k (b=2.42) are the most important and of similar magnitude, followed by education level in distant third (b=.36), followed by log of population (b=.05) and percent urban (b=.045), neither one of which seem to matter very much. But this rank ordering is likely flawed due to differences in the way the variables are measured. A tip off that something is wrong with this type of comparison is found in the fact that the slopes for urban population and the log of population size are of about the same magnitude, even though the slope for urban population is statistically significant (p<.01) while the slope for population size is not and has not been significant in any of the analyses shown thus far. How can a variable whose effect is not distinguishable from zero have an impact equal to one that has been statistically significant in all of the examples shown thus far? Seems unlikely.
Generally, the “raw” regression coefficients are not a good guide to the relative impact of the independent variables because the variables are measured on different scales. Recall that the regression slopes tell us how much Y is expected to change for a unit change in x. The problem we encounter when comparing these slopes is that the units of measure for the independent variables are mostly different from each other, making it very difficult to compare the “unit changes” in a fair manner.
To gain a sense of the impact of measurement scale on the size of the regression slope, consider the two plots below in Figure 17.3. Both plots show the relationship between the size of the urban population and country-level life expectancy. The plot on the left uses the percentage of a country’s population living in urban areas, and the plot on the right uses the proportion of a country’s population living in urban areas. These are the same variables, just using different scales. As you can see, other than the metric on the horizontal axis, the plots are identical. In both cases, there is a moderately strong positive relationship between the size of the urban population and life expectancy: as the urban population increases in size, life expectancy also increases. But look at the linear equations that summarize the results of the bi-variate regression models. The slope for the urban population variable in the second model is 100 times the size of the slope in the first model, not because the impact is 100 times greater but because of the difference in the scale of the two variables. This is the problem we encounter when comparing regression coefficients across independent variables measured on different scales.
[Figure**************** 17.3 ****************about here]
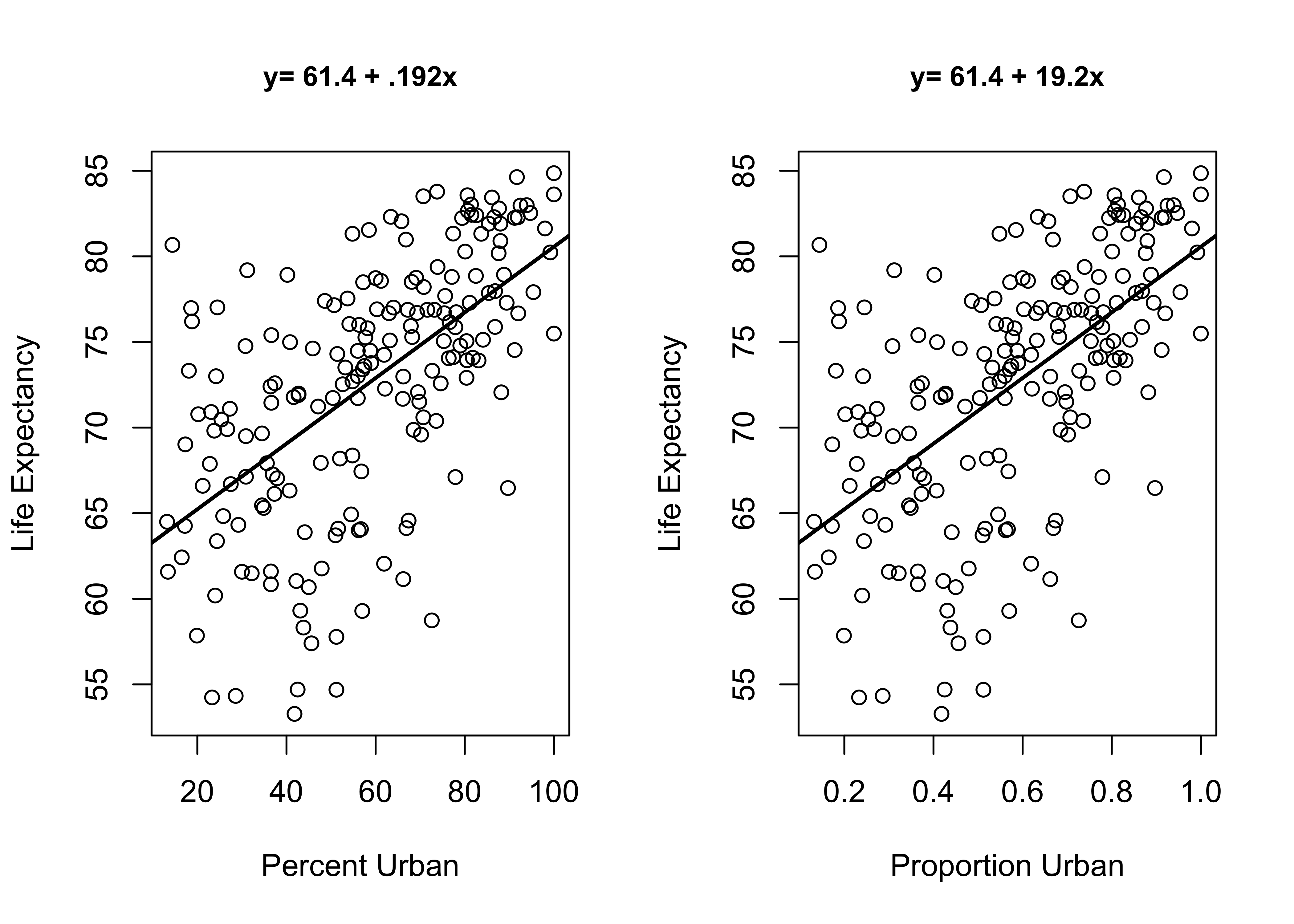
Figure 17.3: The Impact of Scale of the Independent Variable on the Size of Regression Coefficients
What we need to do is standardize the variables used in the regression model so they are all measured on the same scale. Variables with a common metric (same mean and standard deviation) will produce slopes that are directly comparable. One common way to standardize variables is to transform the raw scores into z-scores:
\[Z_i=\frac{x_i-\bar{x}}{S}\] All variables transformed in this manner will have \(\bar{x}=0\) and \(S = 1\). If we do this to all variables (including the dependent variable), we can get the standardized regression coefficients (sometimes confusingly referred to as “Beta Weights”). Fortunately, we don’t have to make these conversions ourselves since R provides the standardized coefficients. In order to get this information, we can use the StdCoef command from the DescTools package. The command is then simply StdCoef(fit), where “fit” is the object with the stored information from the linear model.
Estimate* Std. Error* df
(Intercept) 0.0000000 0.000000 173
fert1520 -0.4809160 0.068382 173
mnschool 0.1501100 0.068652 173
log10(pop19_M) 0.0056696 0.037140 173
urban 0.1390776 0.047084 173
log10(docs10k) 0.2061025 0.082729 173
attr(,"class")
[1] "coefTable" "matrix" The standardized coefficients are in the “Estimate” column. To evaluate relative importance, focus on and compare the absolute values. Based on the standardized coefficients, the ordering of variables from most-to-least in importance is a bit different than when we used the unstandardized coefficients: fertility rate (-.48) far outstrips all other variables, having more than twice the impact as doctors per 10,000 (.21), which is followed pretty closely level of education (.15) and urban population (.14), and population size comes in last (.006). These results give a somewhat different take on relative impact than if we relied on the raw coefficients.
We can also make more literal interpretations of these slopes. Since the dependent and independent variables are all transformed into z-scores, a “one unit change” in x is always a one standard deviation change in x. This means that we can interpret the standardized slopes as telling how many standard deviations y is expected to change for a one standard deviation change in x. For instance, the standardized slope for fertility (-.48) means that for every one standard deviation increase in the fertility rate we can expect to see a .48 standard deviation decline in life expectancy. Just to be clear, though, the primary utility of these standardized coefficients is to compare the relative impact of the independent variables.
Statistics vs. Substance
The model we’ve been looking at explains about 77% of the variation in country-level life expectancy using five independent variables. This is a good model fit, but it is possible to account for a lot more variance with even fewer variables. Take, for example, the alternative model below, which explains 99% of the variance using just two independent variables, male life expectancy and infant mortality.
#Alternative model
fit1<-lm(countries2$lifexp~countries2$malexp + countries2$infant_mort)
#Table with results from both models
stargazer(fit1, type="text",
dep.var.labels=c("Life Expectancy"),
covariate.labels = c("Male Life Expectancy",
"Infant Mortality"))
================================================
Dependent variable:
---------------------------
Life Expectancy
------------------------------------------------
Male Life Expectancy 0.810***
(0.018)
Infant Mortality -0.081***
(0.007)
Constant 17.423***
(1.413)
------------------------------------------------
Observations 184
R2 0.989
Adjusted R2 0.989
Residual Std. Error 0.787 (df = 181)
F Statistic 8,098.000*** (df = 2; 181)
================================================
Note: *p<0.1; **p<0.05; ***p<0.01We had been feeling pretty good about the model that explains 77% of the variance in life expectancy and then along comes this much simpler model that explains almost 100% of the variation. Not that it’s a contest, but this does sort of raise the question, “Which model is best?” The new model definitely has the edge in measures of error: the RMSE is just .787, compared to 3.58 in the five-variable model, and, as mentioned above, the R2=.99.
So, which model is best? Of course, this is a trick question. The second model provides a stronger statistical fit but at significant theoretical and substantive costs. Put another way, the second model explains more variance without providing a useful or interesting substantive explanation of the dependent variable. In effect, the second model is explaining life expectancy with other measures of life expectancy. Not very interesting. Think about it like this. Suppose you are working as an intern at the World Health Organization and your supervisor asks you to give them a presentation on the factors that influence life expectancy around the world. I don’t think they would be impressed if you came back to them and said you had two main findings:
- People live longer in countries where men live longer.
- People live longer in countries where fewer people die really young.
Even with your impressive R2 and RMSE, your supervisor would likely tell you to go back to your desk, put on your thinking cap, and come up with a model that provides a stronger substantive explanation, something more along the lines of the original model. With the results of the original model you can report back that the factors that are most strongly related to life expectancy across countries are fertility rates, access to health care, and education levels–things that policies and aid programs might be able to affect. The share of the population living in urban areas is also related to life expectancy, but this is not something that can be addressed as easily through policies and aid programs.
Next Steps
By now, you should be feeling comfortable with regression analysis and, hopefully, seeing how it can be applied to many different types of data problems. The last step in this journey of discovery is to examine some of the important assumptions that underlie OLS regression. Chapter 18 provides a brief overview of assumptions that need to be satisfied in order to be confident in the results of the life expectancy model presented above. As you will see, in some ways the model tested in this chapter does a good job of satisfying the assumptions, but in other ways the model still needs a bit of work. It is important to be familiar with these assumptions not just so you can think about them in the context of your own work, but also so you can use this information to evaluate the work of others.
Exercises
Concepts and Calculations
- The two correlation matrices below illustrate the pattern of relationships among variables for two different data sets, A and B.
| A | B | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Y | X1 | X2 | X3 | Y | X1 | X2 | X3 | |||
| Y | 1.0 | 0.55 | 0.32 | -0.75 | Y | 1.0 | 0.48 | 0.39 | -0.79 | |
| X1 | 0.55 | 1.0 | 0.25 | -0.43 | X1 | 0.48 | 1.0 | 0.30 | -0.85 | |
| X2 | 0.32 | 0.25 | 1.0 | 0.02 | X2 | 0.39 | 0.30 | 1.0 | -0.58 | |
| X3 | -0.75 | -0.43 | 0.02 | 1.0 | X3 | -0.79 | -0.85 | -0.58 | 1.0 |
- In which data set do you think multicollinearity is most likely to present a problem? Explain your answer.
- For which independent variable in the data set you chose is collinearity likely to present a problem? Why?
- What test would you advise using to get a better sense of the overall level of collinearity?
- In which of the following scatterplots does it appear that the relationship between X and Y is not linear? Describe the pattern in that scatterplot. What course of action do you recommend to correct this issue?
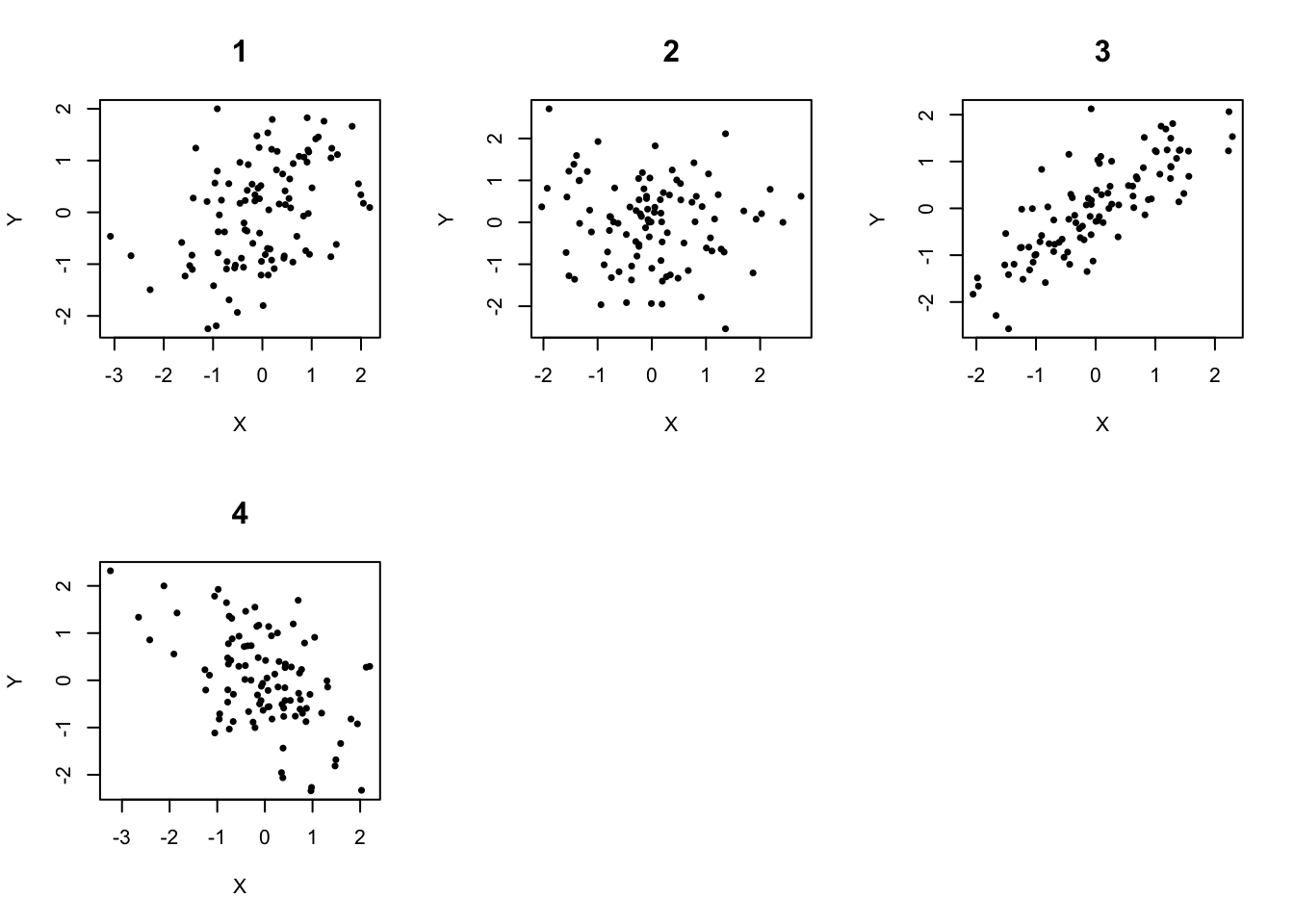
- The regression output copied below summarizes the impact and statistical significance of four independent variables (X1, X2, X3, and X4) on a dependent variable (Y).
Rank-order the independent variables from strongest to weakest in terms of their relative impact on the dependent variable. Explain the basis for your ranking.
Provide an interpretation of both the b value and the standardized coefficient for the variable that you have identified as having the greatest impact on the dependent variable.
| b | t-score | Standardized Coefficient | |
|---|---|---|---|
| Constant | -13.58 | 6.05** | |
| X1 | 0.79 | 2.45** | 0.35 |
| X2 | -3.55 | 1.98** | -0.17 |
| X3 | 3.78 | 2.78** | 0.31 |
| X4 | -0.15 | 3.52** | -0.43 |
| Note: **p<.05 |
R Problems
Building on the regression models from the exercises in the last chapter, use the states20 data set and run a multiple regression model with infant mortality (infant_mort) as the dependent variable and per capita income (PCincome2020), the teen birth rate (teenbirth), percent of low birth weight births (lowbirthwt), and southern region (south) as the independent variables.
- Produce a publication-ready table using
stargazer. Make sure you replace the R variable names with meaningful labels (see Chapter 16 for instructions on doing this usingstargazer).
Summarize the results of this model. Make sure to address matters related to the slopes of the independent variables and how well the group of variables does in accounting for state-to-state differences in infant mortality.
Generate the standardized regression coefficients and discuss the results. Any surprises, given the findings from the unstandardized coefficients? Make it clear that you understand why you need to run this test and what these results are telling you.
Run a test for evidence of multicollinearity. Discuss the results. Any surprises? Make it clear that you understand why you need to run this test and what these results are telling you.
- Use the code listed below to take a sample of 500 counties from
counties20largeand save the sample to a new object namedcounty500.
set.seed(1234)
#create a sample of 500 rows of data from the county20large data set
county500<-sample_n(county20large, 500) Choose one of the following to use as a dependent variable:
vcrim100k,gundeath100k,uninsured, andcvap_turnout20. Then choose four independent variables that you expect to be related to the dependent variable you selected. Justify your selection and be clear about your expectations. Consult the codebook for thecounties20largedata set to make sure you understand what these variables are.Summarize the results of this model. Make sure to address matters related to the slopes of the independent variables and how well the group of variables does in accounting for state-to-state differences in infant mortality.
Generate the standardized regression coefficients and discuss the results. Any surprises, given the findings from the unstandardized coefficients? Make it clear that you understand why you need to run this test and what these results are telling you.
Run a test for evidence of multicollinearity. Discuss the results. Any surprises? Make it clear that you understand why you need to run this test and what these results are telling you.