Capítulo 9 Bioconductor
O projeto Bioconductor desenvolve, apoia e dissemina software de código aberto gratuito que facilite a análise rigorosa e reprodutível de dados de ensaios biológicos atuais e emergentes (Huber et al. 2015).
Conta com mais de 2000 pacotes. A versão atual (3.17) requer a versão 4.3.0 do R.
Para instalar pacotes do Bioconductor precisa do pacote BiocManager:
if (!require(“BiocManager”, quietly = TRUE)) install.packages(“BiocManager”) BiocManager::install(version = “3.16”)
Depois os pacotes podem ser instalados assim:
BiocManager::install(c(“GenomicFeatures”, “AnnotationDbi”))
Para procurar pacotes:
## [1] "easyDifferentialGeneCoexpression" "ExpressionAtlas"
## [3] "ExpressionNormalizationWorkflow" "geneExpressionFromGEO"
## [5] "GeneExpressionSignature" "lungExpression"
## [7] "maqcExpression4plex" "NormExpression"
## [9] "RVerbalExpressions" "statsExpressions"O site é: https://bioconductor.org/
9.1 Pacote msa
Vamos instalar o pacote msa (Multiple Sequence Alignment)(Bodenhofer et al. 2015): An R Package for Multiple Sequence Alignment Enrico Bonatesta, Christoph Kainrath, and Ulrich Bodenhofer Institute of Bioinformatics, Johannes Kepler University Linz, Austria
BiocManager::install(“msa”)
Vamos carregar algumas sequências usando o pacote ape (ver capítulo 7):
library(ape)
id_seq<-c("NM_022942.1","NM_009738.3","NM_001313861.2")
seq_bche<-read.GenBank(id_seq)
seq_bche## 3 DNA sequences in binary format stored in a list.
##
## Mean sequence length: 3435
## Shortest sequence: 1878
## Longest sequence: 6209
##
## Labels:
## NM_022942.1
## NM_009738.3
## NM_001313861.2
##
## Base composition:
## a c g t
## 0.310 0.201 0.197 0.292
## (Total: 10.3 kb)Temos que salvar em formato fasta, para depois usar esse arquivo no msa.
O formato FASTA é um formato baseado em texto para representar tanto sequências de nucleotideos quanto sequências de aminoácidos. O formato também permite sequências de nomes e comentários precedendo as sequências.
Agora vamos fazer o alinhamento usando o pacote msa, onde informamos o tipo de sequência em type (pode ser dna,rna ou protein), em method escolhemos o método de alinhamento, sendo as opções: “ClustalW”, “ClustalOmega” e “Muscle”. Iremos utilizar o ClustalW.
## use default substitution matrix## CLUSTAL 2.1
##
## Call:
## msa("seq_bche.fasta", type = "dna", method = "ClustalW")
##
## MsaDNAMultipleAlignment with 3 rows and 6219 columns
## aln
## [1] ------------------------------------...------------------------------------
## [2] CTTCTCTCCTGAAGCCTGCCTGCAATGGTGGAGGAA...GAATCATATGCACAAAATAAAAACTTCCTTGTATAA
## [3] ------------------------------------...------------------------------------
## Con ------------------------------------...------------------------------------Para ver o alinhamento vamos usar outro pacote, o ggmsa (Zhou et al. 2022): instalar o pacote ggmsa BiocManager::install(“ggmsa”)
Precisamos fazer algumas transformações no objeto alin antes de usar no ggmsa:
library(ggmsa)
# converter para o formato do ape
ali_bche<-msaConvert(alin, type="ape::DNAbin")
# salvar no formato fasta
write.FASTA(ali_bche, file = "ali_bche.fas")
# ler o arquivo fasta
ali_bche2<-read.FASTA("ali_bche.fas")Agora vamos gerar a figura com o alinhamento:
## Warning: No shared levels found between `names(values)` of the
## manual scale and the data's fill values.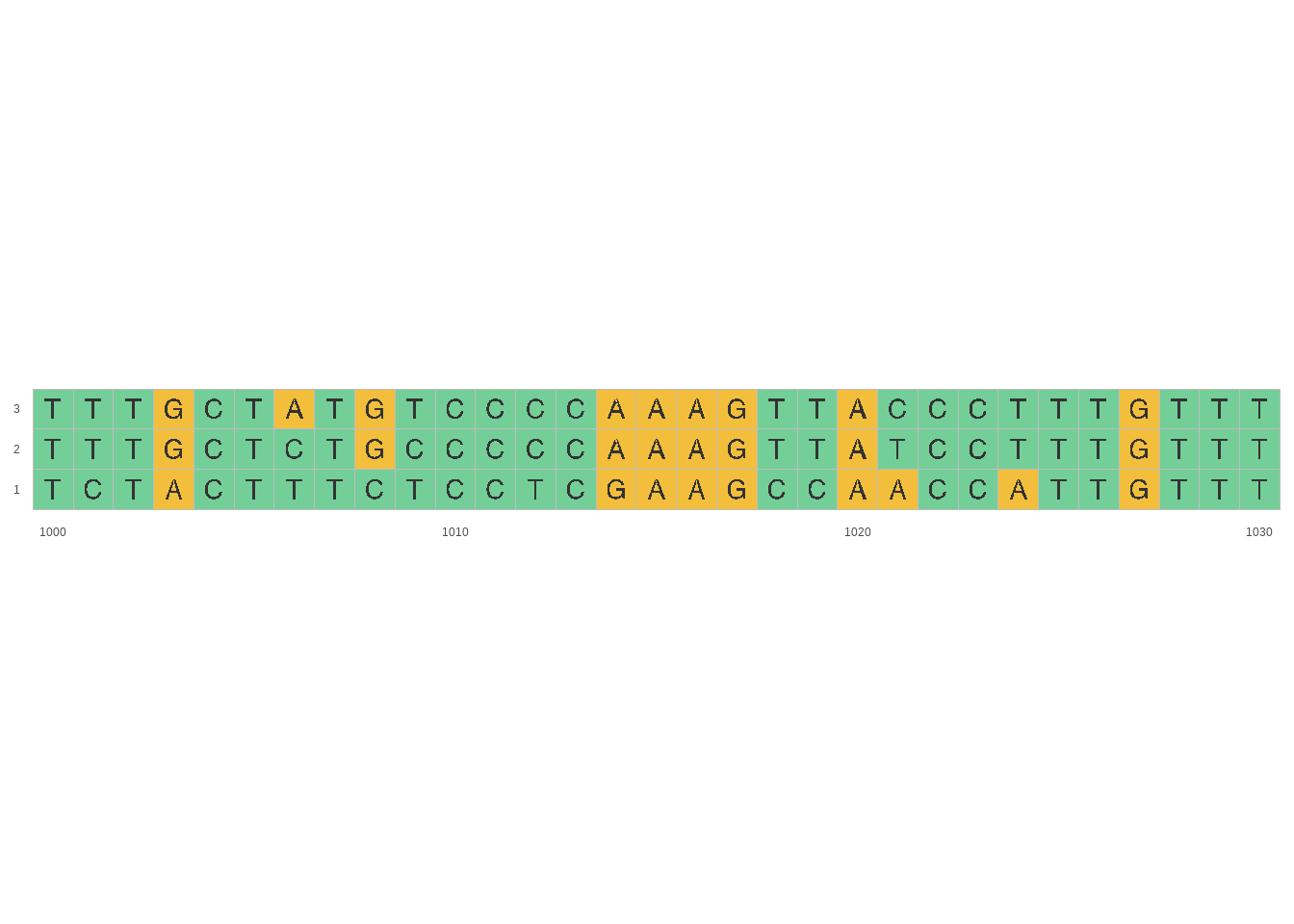
Acrescentando uma camada:
## Warning: No shared levels found between `names(values)` of the
## manual scale and the data's fill values.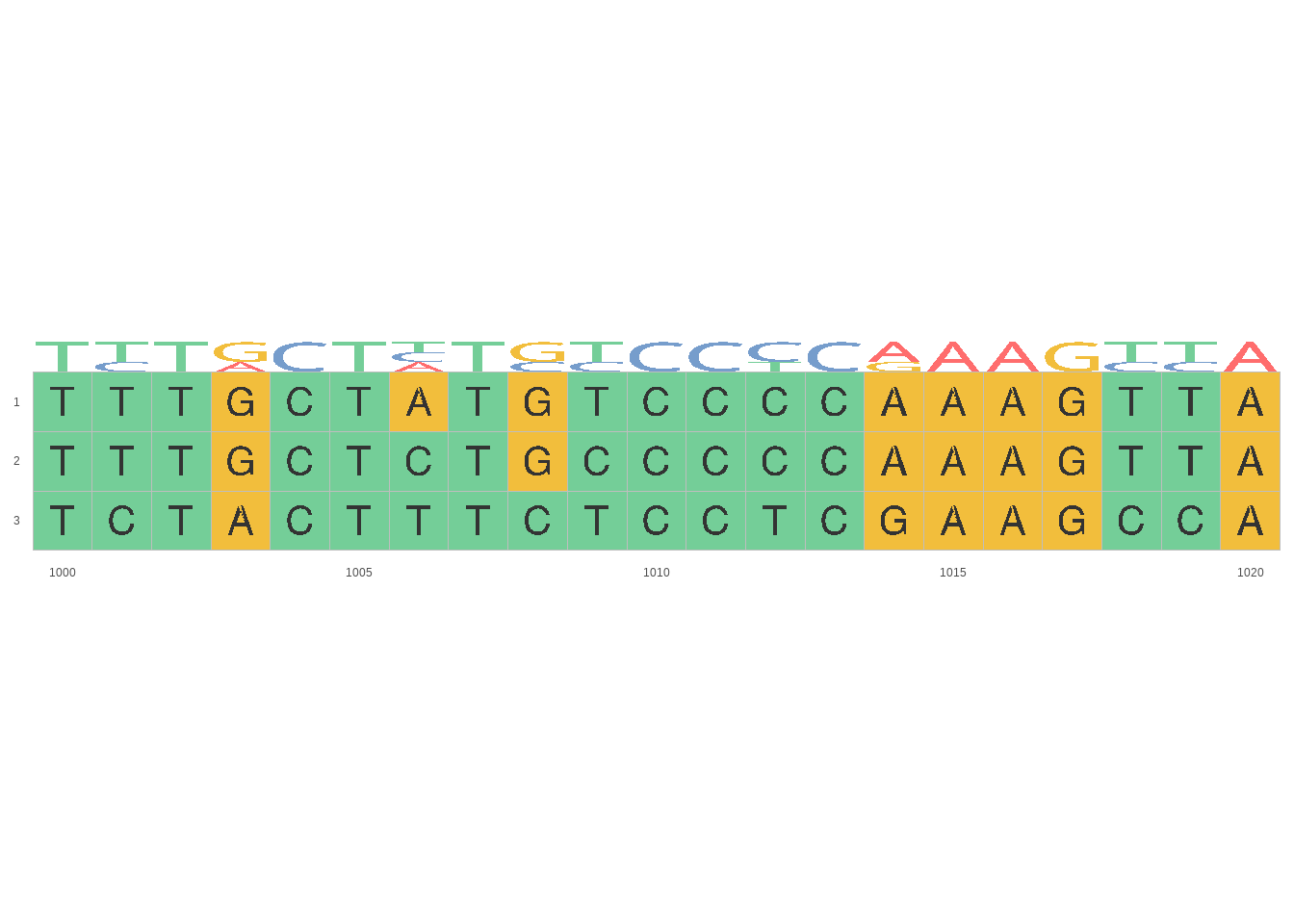
Acrescentando a sequência consenso:
ggmsa(ali_bche2, 1000, 1020, char_width = 0.8, seq_name = T) + geom_seqlogo(color = "Chemistry_NT") + geom_msaBar()## Coordinate system already present. Adding new
## coordinate system, which will replace the existing
## one.## Warning: No shared levels found between `names(values)` of the
## manual scale and the data's fill values.
## No shared levels found between `names(values)` of the
## manual scale and the data's fill values.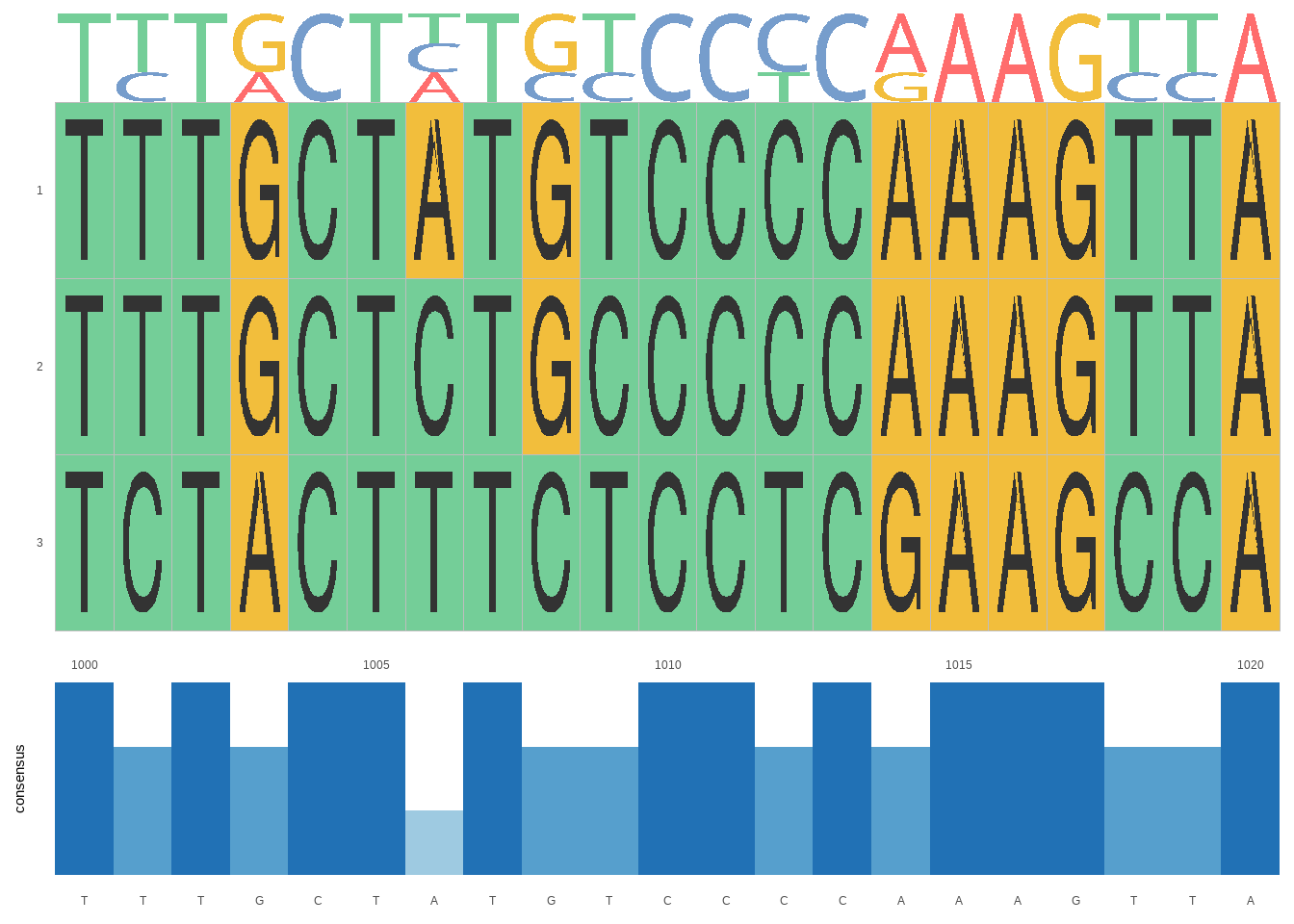
Esquema de cores
ggmsa predefine vários esquemas de cores para renderização. Podemos usar available_msa() para listar os esquemas de cores disponíveis.
Observe que aminoácidos (proteína) e nucleotídeos (DNA/RNA) têm nomes diferentes.
DNA:
Chemistry_NT Shapely_NT Taylor_NT Zappo_NT
Proteínas:
Clustal Chemistry_AA Shapely_AA Zappo_AA Taylor_AA LETTER CN6 Hydrophobicity
Abaixo estão as camadas de anotação suportadas pelo ggmsa:
Annotation modules Type Description
geom_seqlogo() geometric layer automatically generated sequence logos for a MSA
geom_GC() annotation module shows GC content with bubble chart
geom_seed() annotation module highlights seed region on miRNA sequences
geom_msaBar() annotation module shows sequences conservation by a bar chart
geom_helix() annotation module depicts RNA secondary structure as arc diagrams(need extra data)
9.2 Pacote multiMiR
Para insatalar: BiocManager::install(“multiMiR”)
Os microRNAs (miRNAs) regulam a expressão promovendo a degradação ou reprimindo a tradução de transcritos alvo. Os sítios alvo de miRNA foram catalogados em bancos de dados com base em validação experimental e previsão computacional usando uma variedade de algoritmos.
O pacote R multiMiR (http://multimir.org), é uma coleção abrangente de interações miRNA-alvo previstas e validadas e suas associações com doenças e medicamentos. O multiMiR inclui vários novos recursos não disponíveis em pacotes R existentes (Ru et al., n.d.).
As informações de cada banco de dados externo são armazenadas em uma tabela no banco de dados multiMiR. Para ver uma lista das tabelas, podemos usar a função multimir_dbTables().
## Welcome to multiMiR.
##
## multiMiR database URL has been set to the
## default value: http://multimir.org/
##
## Database Version: 2.3.0 Updated: 2020-04-15## [1] "diana_microt" "elmmo" "map_counts" "map_metadata" "microcosm"
## [6] "mir2disease" "miranda" "mirdb" "mirecords" "mirna"
## [11] "mirtarbase" "pharmaco_mir" "phenomir" "pictar" "pita"
## [16] "tarbase" "target" "targetscan"Podemos ver os detalhes de cada banco:
## map_name source_name source_version source_date
## 1 diana_microt DIANA-microT 5 Sept, 2013
## 2 elmmo EIMMo 5 Jan, 2011
## 3 microcosm MicroCosm 5 Sept, 2009
## 4 mir2disease miR2Disease Mar 14, 2011
## 5 miranda miRanda Aug, 2010
## 6 mirdb miRDB 6 June, 2019
## 7 mirecords miRecords 4 Apr 27, 2013
## 8 mirtarbase miRTarBase 7.0 Sept, 2017
## 9 pharmaco_mir Pharmaco-miR (Verified Sets)
## 10 phenomir PhenomiR 2 Feb 15, 2011
## 11 pictar PicTar 2 Dec 21, 2012
## 12 pita PITA 6 Aug 31, 2008
## 13 tarbase TarBase 8 2018
## 14 targetscan TargetScan 7.2 March, 2018
## source_url
## 1 http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index
## 2 http://www.mirz.unibas.ch/miRNAtargetPredictionBulk.php
## 3 http://www.ebi.ac.uk/enright-srv/microcosm/cgi-bin/targets/v5/download.pl
## 4 http://www.mir2disease.org
## 5 http://www.microrna.org/microrna/getDownloads.do
## 6 http://mirdb.org
## 7 http://mirecords.biolead.org/download.php
## 8 http://mirtarbase.mbc.nctu.edu.tw/php/download.php
## 9 http://www.pharmaco-mir.org/home/download_VERSE_db
## 10 http://mips.helmholtz-muenchen.de/phenomir/
## 11 http://dorina.mdc-berlin.de
## 12 http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html
## 13 http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=tarbasev8%2Findex
## 14 http://www.targetscan.org/cgi-bin/targetscan/data_download.cgi?db=vert_61Entre os 14 bancos de dados externos, oito contêm interações miRNA-alvo previstas, três têm interações miRNA-alvo validadas experimentalmente e os três restantes contêm associações miRNA-droga/doença.
Para verificar essas categorias e bancos de dados de dentro do R, temos um conjunto de quatro funções auxiliares:
## [1] "diana_microt" "elmmo" "microcosm" "miranda" "mirdb"
## [6] "pictar" "pita" "targetscan"## [1] "mirecords" "mirtarbase" "tarbase"## [1] "mir2disease" "pharmaco_mir" "phenomir"## [1] "predicted"Para ver quantos registros existem nesses 14 bancos de dados externos, nos referimos à função multimir_dbCount.
## map_name human_count mouse_count rat_count total_count
## 1 diana_microt 7664602 3747171 0 11411773
## 2 elmmo 3959112 1449133 547191 5955436
## 3 microcosm 762987 534735 353378 1651100
## 4 mir2disease 2875 0 0 2875
## 5 miranda 5429955 2379881 247368 8057204
## 6 mirdb 1990425 1091263 199250 3280938
## 7 mirecords 2425 449 171 3045
## 8 mirtarbase 544588 50673 652 595913
## 9 pharmaco_mir 308 5 0 313
## 10 phenomir 15138 491 0 15629
## 11 pictar 404066 302236 0 706302
## 12 pita 7710936 5163153 0 12874089
## 13 tarbase 433048 209831 1307 644186
## 14 targetscan 13906497 10442093 0 24348590A função list_multimir() listar todos os miRNAs únicos, genes alvo, drogas e doenças no banco de dados multiMiR. Uma opção para limitar o número de registros retornados foi adicionada para ajudar nos testes e na exploração.
miRNAs<- list_multimir("mirna", limit = 10)
genes<- list_multimir("gene", limit = 10)
drugs<- list_multimir("drug", limit = 10)
diseases<- list_multimir("disease", limit = 10)
miRNAs## mature_mirna_uid org mature_mirna_acc mature_mirna_id
## 1 7829 ath MIMAT0000184 ath-miR163
## 2 7833 ath MIMAT0000201 ath-miR170-3p
## 3 7831 ath MIMAT0000206 ath-miR173-5p
## 4 7828 ath MIMAT0001001 ath-miR400
## 5 7837 ath MIMAT0001004 ath-miR403-3p
## 6 7830 ath MIMAT0001011 ath-miR408-3p
## 7 7838 ath MIMAT0003931 ath-miR472-3p
## 8 7846 ath MIMAT0003933 ath-miR774a
## 9 7839 ath MIMAT0003934 ath-miR775
## 10 7841 ath MIMAT0003937 ath-miR778## target_uid org target_symbol target_entrez target_ensembl
## 1 112854 ath AT1G02860
## 2 112855 ath AT1G06180
## 3 112856 ath AT1G06580
## 4 112857 ath AT1G15125
## 5 112868 ath At1g31280
## 6 112869 ath At1g50055
## 7 112870 ath At1g51480
## 8 112871 ath At1g53290
## 9 112872 ath At1g62720
## 10 112873 ath At1g62930## drug
## 1 3,3'-diindolylmethane
## 2 5-fluoroucil
## 3 abt-737
## 4 alitretinoin
## 5 arabinocytosine
## 6 arsenic trioxide
## 7 azd6244
## 8 bleomycin
## 9 butyrate
## 10 calcitriol## disease
## 1 ACTH-INDEPENDENT MACRONODULAR ADRENAL HYPERPLASIA; AIMAH
## 2 ACUTE LYMPHOBLASTIC LEUKEMIA (ALL)
## 3 ACUTE MYELOGENEOUS LEUKEMIA (AML)
## 4 ACUTE MYELOID LEUKEMIA (AML)
## 5 ACUTE PROMYELOCYTIC LEUKEMIA (APL)
## 6 ADENOMA
## 7 ADENOMAS, MULTIPLE COLORECTAL
## 8 ADRENOCORTICAL CARCINOMA
## 9 ALCOHOLIC LIVER DISEASE (ALD)
## 10 ALZHEIMER'S DISEASE
## 11 ALZHEIMER DISEASE, SUSCEPTIBILITY TO
## 12 AMYOTROPHIC LATERAL SCLEROSIS, SUSCEPTIBILITY TO
## 13 ANAPLASTIC THYROID CARCINOMA (ATC)
## 14 ANXIETY DISORDER
## 15 ASTHMA
## 16 AUTISM, SUSCEPTIBILITY TO
## 17 BECKER MUSCULAR DYSTROPHY
## 18 BLADDER CANCER
## 19 BREAST CANCER
## 20 BURKITT LYMPHOMAget_multimir() é a principal função no pacote para recuperar interações miRNA-alvo previstas e validadas e suas associações de doenças e medicamentos do banco de dados multiMiR.
Podemos pesquisar por miRNA:
## Searching mirecords ...
## Searching mirtarbase ...
## Searching tarbase ...##
## validated
## 1378## database mature_mirna_acc mature_mirna_id target_symbol target_entrez
## 1 mirecords MIMAT0000443 hsa-miR-125a-5p ERBB2 2064
## 2 mirecords MIMAT0000443 hsa-miR-125a-5p ERBB3 2065
## 3 mirecords MIMAT0000443 hsa-miR-125a-5p LIN28
## 4 mirecords MIMAT0000443 hsa-miR-125a-5p LIN28
## 5 mirecords MIMAT0000443 hsa-miR-125a-5p ARID3B 10620
## 6 mirecords MIMAT0000443 hsa-miR-125a-5p TP53 7157
## target_ensembl experiment
## 1 ENSG00000141736 Western blot//activity assay//Luciferase activity assay
## 2 ENSG00000065361 Western blot//activity assay//Luciferase activity assay
## 3 ENSG00000131914
## 4 ENSG00000131914
## 5 ENSG00000179361 Western blot
## 6 ENSG00000141510 Western blot
## support_type pubmed_id type
## 1 17110380 validated
## 2 17110380 validated
## 3 16227573 validated
## 4 16227573 validated
## 5 19881956 validated
## 6 19818772 validatedPodemos pesquisar por medicamento:
## Searching mir2disease ...
## Searching pharmaco_mir ...
## Searching phenomir ...## database mature_mirna_acc mature_mirna_id target_symbol target_entrez
## 1 pharmaco_mir MIMAT0000772 hsa-miR-345-5p ABCC1 4363
## 2 pharmaco_mir MIMAT0000720 hsa-miR-376c-3p ALK7
## 3 pharmaco_mir MIMAT0000423 hsa-miR-125b-5p BAK1 578
## 4 pharmaco_mir hsa-miR-34 BCL2 596
## 5 pharmaco_mir MIMAT0000318 hsa-miR-200b-3p BCL2 596
## 6 pharmaco_mir MIMAT0000617 hsa-miR-200c-3p BCL2 596
## target_ensembl disease_drug paper_pubmedID type
## 1 ENSG00000103222 cisplatin 20099276 disease.drug
## 2 cisplatin 21224400 disease.drug
## 3 ENSG00000030110 cisplatin 21823019 disease.drug
## 4 ENSG00000171791 cisplatin 18803879 disease.drug
## 5 ENSG00000171791 cisplatin 21993663 disease.drug
## 6 ENSG00000171791 cisplatin 21993663 disease.drugPodemos pesquisar por gene:
example3 <- get_multimir(org = "hsa",
target = "BCHE",
table = "predicted",
summary = TRUE,
predicted.cutoff = 35,
predicted.cutoff.type = "p",
predicted.site = "all")## Searching diana_microt ...
## Searching elmmo ...
## Searching microcosm ...
## Searching miranda ...
## Searching mirdb ...
## Searching pictar ...
## Searching pita ...
## Searching targetscan ...##
## predicted
## 450## database mature_mirna_acc mature_mirna_id target_symbol target_entrez
## 1 diana_microt MIMAT0015021 hsa-miR-3148 BCHE 590
## 2 diana_microt MIMAT0015021 hsa-miR-3148 BCHE 590
## 3 diana_microt MIMAT0022489 hsa-miR-5696 BCHE 590
## 4 diana_microt MIMAT0022489 hsa-miR-5696 BCHE 590
## 5 diana_microt MIMAT0019060 hsa-miR-4522 BCHE 590
## 6 diana_microt MIMAT0019060 hsa-miR-4522 BCHE 590
## target_ensembl score type
## 1 ENSG00000114200 0.998 predicted
## 2 ENSG00000114200 0.998 predicted
## 3 ENSG00000114200 0.974 predicted
## 4 ENSG00000114200 0.974 predicted
## 5 ENSG00000114200 0.971 predicted
## 6 ENSG00000114200 0.971 predicted## mature_mirna_acc mature_mirna_id target_symbol target_entrez target_ensembl
## 1 MIMAT0004518 hsa-miR-16-2-3p BCHE 590 ENSG00000114200
## 2 MIMAT0000275 hsa-miR-218-5p BCHE 590 ENSG00000114200
## 3 MIMAT0003294 hsa-miR-625-5p BCHE 590 ENSG00000114200
## 4 MIMAT0004615 hsa-miR-195-3p BCHE 590 ENSG00000114200
## 5 MIMAT0004796 hsa-miR-576-3p BCHE 590 ENSG00000114200
## 6 MIMAT0005937 hsa-miR-1279 BCHE 590 ENSG00000114200
## diana_microt elmmo microcosm miranda mirdb pita targetscan predicted.sum
## 1 2 2 1 2 1 0 2 6
## 2 0 2 1 1 0 1 1 5
## 3 2 0 1 1 0 2 1 5
## 4 2 2 0 2 1 0 2 5
## 5 2 0 1 1 0 1 1 5
## 6 2 2 0 1 1 0 1 5
## all.sum
## 1 6
## 2 5
## 3 5
## 4 5
## 5 5
## 6 5Você pode ter uma lista de genes envolvidos em um processo biológico comum. É interessante verificar se alguns, ou todos, desses genes são direcionados pelo(s) mesmo(s) miRNA(s).
example4 <- get_multimir(org = 'hsa',
target = c('AKT2', 'CERS6', 'S1PR3', 'SULF2'),
table = 'predicted',
summary = TRUE,
predicted.cutoff.type = 'n',
predicted.cutoff = 500000)## Number predicted cutoff (predicted.cutoff) 500000 is larger than the total number of records in table pictar. All records will be queried.## Number predicted cutoff (predicted.cutoff) 500000 is larger than the total number of records in table targetscan. All records will be queried.## Searching diana_microt ...
## Searching elmmo ...
## Searching microcosm ...
## Searching miranda ...
## Searching mirdb ...
## Searching pictar ...
## Searching pita ...
## Searching targetscan ...example4.counts <- addmargins(table(example4@summary[, 2:3]))
example4.counts <- example4.counts[-nrow(example4.counts), ]
example4.counts <- example4.counts[order(example4.counts[, 5], decreasing = TRUE), ]
head(example4.counts)## target_symbol
## mature_mirna_id AKT2 CERS6 S1PR3 SULF2 Sum
## hsa-miR-129-5p 0 1 2 1 4
## hsa-miR-330-3p 0 1 2 1 4
## hsa-miR-144-3p 0 1 2 0 3
## hsa-miR-3180-5p 0 1 2 0 3
## hsa-miR-325-3p 1 1 0 1 3
## hsa-miR-34a-5p 0 1 2 0 3