Chapter 22 Simple example of a t-test
mtcars %>%
filter(cyl ==4 | cyl ==6) %>%
mutate(cyl_f = as.factor(cyl)) %>%
t.test(mpg ~ cyl_f, data = .)
Welch Two Sample t-test
data: mpg by cyl_f
t = 4.7191, df = 12.956, p-value = 0.0004048
alternative hypothesis: true difference in means between group 4 and group 6 is not equal to 0
95 percent confidence interval:
3.751376 10.090182
sample estimates:
mean in group 4 mean in group 6
26.66364 19.74286 22.1 Common Problem
- Comparing two groups
- Mean or median vs. expected
- Two arms of study - independent
- Pre and post / spouse and partner / left vs right arm – paired groups
- Are the means significantly different?
- Or the medians (if not normally distributed)?
22.1.1 How Skewed is Too Skewed?
- Formal test of normality = Shapiro-Wilk test
- Use base data set called ToothGrowth
## ID age sex race diagnosis diagnosis.type
## 1 1 61 1 0 acute myeloid leukemia 1
## 2 2 62 1 1 non-Hodgkin lymphoma 0
## 3 3 63 0 1 non-Hodgkin lymphoma 0
## 4 4 33 0 1 Hodgkin lymphoma 0
## 5 5 54 0 1 acute lymphoblastic leukemia 0
## 6 6 55 1 1 myelofibrosis 1
## time.to.transplant prior.radiation prior.chemo prior.transplant recipient.cmv
## 1 5.16 0 2 0 1
## 2 79.05 1 3 0 0
## 3 35.58 0 4 0 1
## 4 33.02 1 4 0 1
## 5 11.40 0 5 0 1
## 6 2.43 0 0 0 1
## donor.cmv donor.sex TNC.dose CD34.dose CD3.dose CD8.dose TBI.dose C1/C2 aKIRs
## 1 0 0 18.31 2.29 3.21 0.95 200 0 1
## 2 0 1 4.26 2.04 NA NA 200 1 5
## 3 1 0 8.09 6.97 2.19 0.59 200 0 3
## 4 0 1 21.02 6.09 4.87 2.32 200 0 2
## 5 1 0 14.70 2.36 6.55 2.40 400 0 6
## 6 1 1 4.29 6.91 2.53 0.86 200 0 2
## cmv time.to.cmv agvhd time.to.agvhd cgvhd time.to.cgvhd
## 1 1 3.91 1 3.55 0 6.28
## 2 0 65.12 0 65.12 0 65.12
## 3 0 3.75 0 3.75 0 3.75
## 4 0 48.49 1 28.55 1 10.45
## 5 0 4.37 1 2.79 0 4.37
## 6 1 4.53 1 3.88 0 6.8722.1.2 Visualize the Distribution of data variables in ggplot
- Use geom_histogram or geom_density (pick one or the other)
- look at the distribution of CD3.dose or time.to.cmv
- Bonus points: facet by sex or race or donor.cmv
- Your turn to try it
library(tidyverse)
library(medicaldata)
data %>%
ggplot(mapping = aes(time.to.cmv)) +
geom_density() +
facet_wrap(~sex) +
theme_linedraw()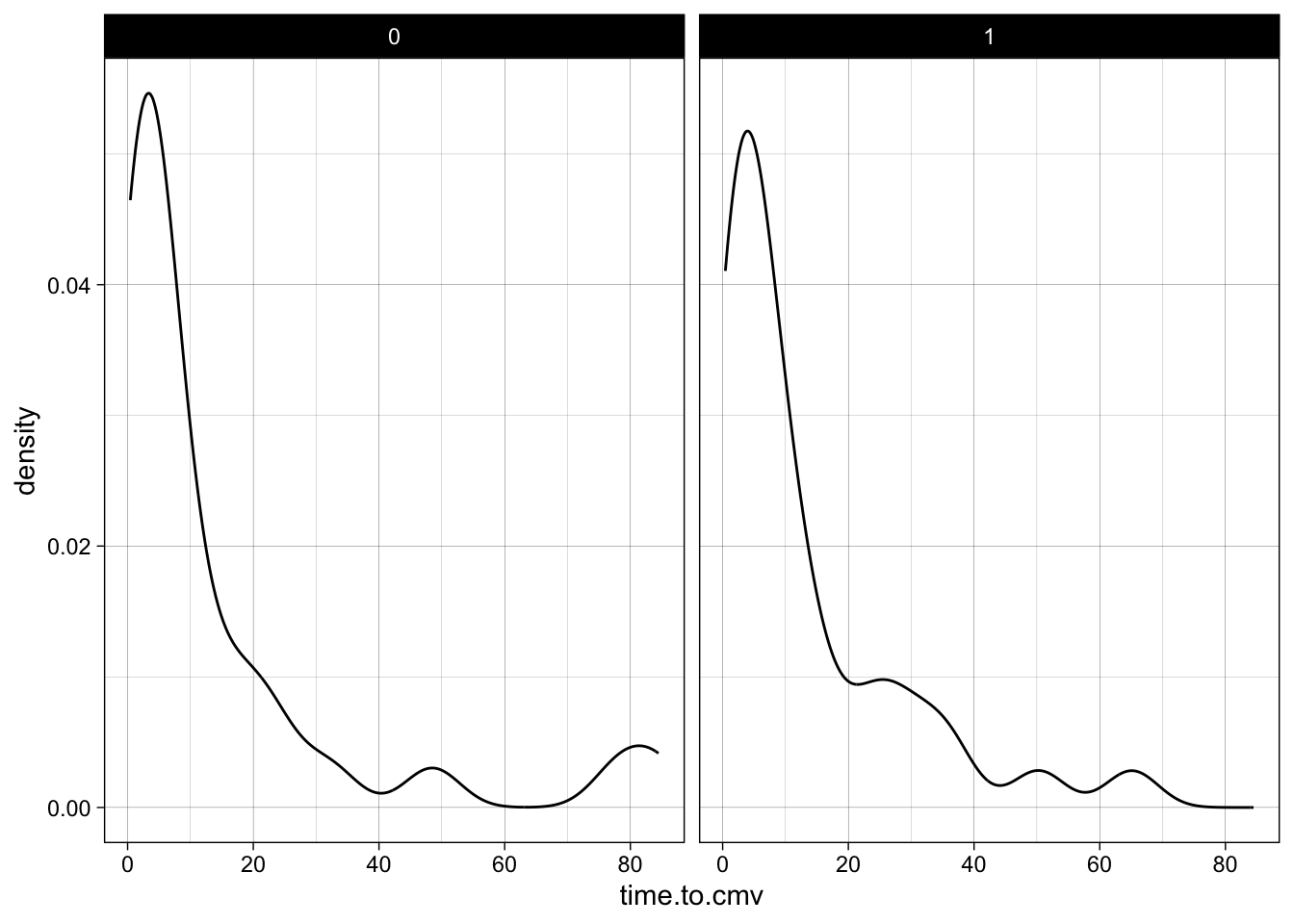
library(tidyverse)
library(medicaldata)
data %>%
ggplot(mapping = aes(time.to.cmv)) +
geom_histogram() +
facet_wrap(~race)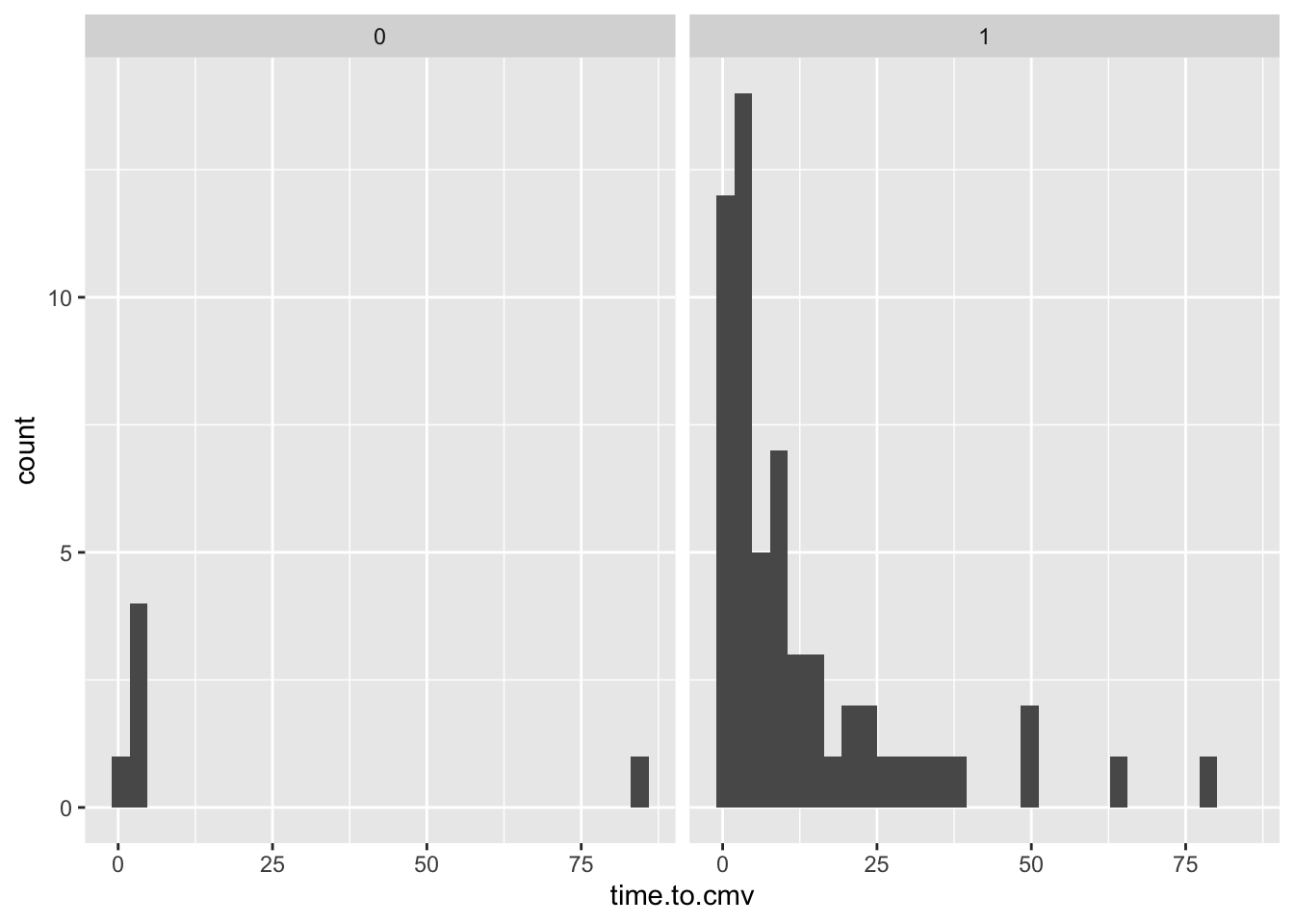
22.1.3 Visualize the Distribution of data$len in ggplot
- The OJ group is left skewed
- May be problematic for using means
- formally test with Shapiro-Wilk
##
## Shapiro-Wilk normality test
##
## data: .
## W = 0.68261, p-value = 1.762e-1022.1.4 Results of Shapiro-Wilk
- p-value = 0.1091
- p not < 0.05
- Acceptably close to normal
- OK to compare means rather than medians
- can use t test rather than wilcoxon test
- if p is < 0.05, use wilcoxon test
- also known as Mann-Whitney test
- a rank-based (non-parametric) test
22.1.5 Try it yourself
- use df <- msleep
## [1] 12.1 17.0 14.4 14.9 4.0 14.4- test the normality of total sleep hours in mammals
22.2 One Sample T test
- univariate test
- Ho: mean is 8 hours
- Ha: mean is not 8 hours
- can use t test because shapiro.test is NS
22.2.2 Interpreting the One Sample T test
##
## One Sample t-test
##
## data: df$sleep_total
## t = 4.9822, df = 82, p-value = 3.437e-06
## alternative hypothesis: true mean is not equal to 8
## 95 percent confidence interval:
## 9.461972 11.405497
## sample estimates:
## mean of x
## 10.43373- p is highly significant
- can reject the null, accept alternative
- sample mean 10.43, CI 9.46-11.41
22.2.3 What are the arguments of the t.test function?
- x = vector of continuous numerical data
- y= NULL - optional 2nd vector of continuous numerical data
- alternative = c(“two.sided”, “less”, “greater”),
- mu = 0
- paired = FALSE
- var.equal = FALSE
- conf.level = 0.95
- documentation
22.3 Insert flipbook for ttest here
Below is a flipbook.
It illustrates a bit of how to do a t-test.
click on it and you can use the arrow keys to proceed forward and back through the slides, as you add lines of code and more results occur.
Let’s start with a flipbook slide show. When the title slide appears, you can step through each line of the code to see what it does. The right/left and/or up/down arrows will let you move forward and backward in the code.
You can use the arrow keys to go through it one step at a time (forward or backward, depending on which arrow key you use), to see what each line of code actually does.
Give it a try below. See if you can figure out what each line of code is doing.
22.4 Fine, but what about 2 groups?
- consider df$vore
## prostate$AA n percent
## 0 261 0.8259494
## 1 55 0.1740506- hypothesis - herbivores need more time to get food, sleep less than carnivores
- how to test this?
- normal, so can use t test for 2 groups
22.4.1 Setting up 2 group t test
- formula interface: outcome ~ groupvar
library(tidyverse)
library(medicaldata)
df %>%
filter(vore %in% c("herbi", "carni")) %>%
t.test(formula = sleep_total ~ vore, data = .)- Try it yourself
- What do the results mean?
22.4.2 Results of the 2 group t test
##
## Welch Two Sample t-test
##
## data: sleep_total by vore
## t = 0.63232, df = 39.31, p-value = 0.5308
## alternative hypothesis: true difference in means between group carni and group herbi is not equal to 0
## 95 percent confidence interval:
## -1.911365 3.650509
## sample estimates:
## mean in group carni mean in group herbi
## 10.378947 9.50937522.4.3 Interpreting the 2 group t test
- Welch t-test (not Student)
- Welch does NOT assume equal variances in each group
- p value NS
- accept null hypothesis
- Ho: means of groups roughly equal
- Ha: means are different
- 95% CI crosses 0
- Carnivores sleep a little more, but not a lot
22.4.4 2 group t test with wide data
- You want to compare column A with column B (data are not tidy)
- Do mammals spend more time awake than asleep?
22.4.5 Results of 2 group t test with wide data
##
## Welch Two Sample t-test
##
## data: df$sleep_total and df$awake
## t = -4.5353, df = 164, p-value = 1.106e-05
## alternative hypothesis: true difference in means is not equal to 0
## 95 percent confidence interval:
## -4.498066 -1.769404
## sample estimates:
## mean of x mean of y
## 10.43373 13.5674722.5 3 Assumptions of Student’s t test
- Sample is normally distributed (test with Shapiro)
- Variances are homogeneous (homoskedasticity) (test with Levene)
- Observations are independent
- not paired like left vs. right colon
- not paired like spouse and partner
- not paired like measurements pre and post Rx
22.5.1 Testing Assumptions of Student’s t test
- Normality - test with Shapiro
- If not normal, Wilcoxon > t test
- Equal Variances - test with Levene
- If not equal, Welch t > Student’s t
- Observations are independent
- Think about data collection
- are some observations correlated with some others?
- If correlated, use paired t test
22.6 Getting results out of t.test
- Use the tidy function from the broom package
- Do carnivores have bigger brains than insectivores?
22.7 Reporting the results from t.test using inline code
- use backticks before and after, start with r
- i.e. My result is [backtick]r code here[backtick].
- The mean brain weight for carnivores was 0.0792556
- The mean brain weight for herbivores was 0.02155
- The difference was 0.0577056
- The t statistic for this Two Sample t-test was 1.1995501
- The p value was 0.2534631
- The confidence interval was from -0.05 to 0.16