Chapter 37 Introduction to R Markdown

Rmarkdown is an authoring framework for creating a variety of data-driven documents reproducibly with R. This e-book is itself a set of RMarkdown documents, assembled with the {bookdown} package. In many ways, Rmarkdown is a critical package for the R ecosystem, as it is a key enabler of reproducible reports in many formats. RMarkdown is a simple formatting syntax that allows you to mix text and code to document data analysis, and author MS Word, MS Powerpoint, HTML, PDF, web dashboards, web apps, and poster documents. Rmarkdown documents are fully reproducible and support more than a dozen output formats. If your data changes, or you decide to change a part of your analysis, you can reproduce the entire (new version) of the document with a single click of the Knit button (You can also use Cmd/Ctrl+Shift+K). This button is found at the top of the top left pane in RStudio.
When you click the Knit button a document will be generated that includes both text content as well as the output of any embedded R code chunks within the document. You can embed an R code chunk like this (which will run and produce output below. In this case, showing the contents of the covid_testing dataset.):
## Rows: 15,524
## Columns: 17
## $ subject_id <dbl> 1412, 533, 9134, 8518, 8967, 11048, 663, 2158, 3794, 4…
## $ fake_first_name <chr> "jhezane", "penny", "grunt", "melisandre", "rolley", "…
## $ fake_last_name <chr> "westerling", "targaryen", "rivers", "swyft", "karstar…
## $ gender <chr> "female", "female", "male", "female", "male", "female"…
## $ pan_day <dbl> 4, 7, 7, 8, 8, 8, 9, 9, 9, 9, 9, 9, 9, 9, 9, 10, 10, 1…
## $ test_id <chr> "covid", "covid", "covid", "covid", "covid", "covid", …
## $ clinic_name <chr> "inpatient ward a", "clinical lab", "clinical lab", "c…
## $ result <chr> "negative", "negative", "negative", "negative", "negat…
## $ demo_group <chr> "patient", "patient", "patient", "patient", "patient",…
## $ age <dbl> 0.0, 0.0, 0.8, 0.8, 0.8, 0.8, 0.8, 0.0, 0.0, 0.9, 0.9,…
## $ drive_thru_ind <dbl> 0, 1, 1, 1, 0, 0, 1, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, …
## $ ct_result <dbl> 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45…
## $ orderset <dbl> 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, …
## $ payor_group <chr> "government", "commercial", NA, NA, "government", "com…
## $ patient_class <chr> "inpatient", "not applicable", NA, NA, "emergency", "r…
## $ col_rec_tat <dbl> 1.4, 2.3, 7.3, 5.8, 1.2, 1.4, 2.6, 0.7, 1.0, 7.1, 2.5,…
## $ rec_ver_tat <dbl> 5.2, 5.8, 4.7, 5.0, 6.4, 7.0, 4.2, 6.3, 5.6, 7.0, 3.8,…The Knit button (at the top of the top left pane of RStudio) runs
render("file.Rmd", output = document-type)
for you in a
background (clean) session of R.
37.1 What Makes an Rmarkdown document?
An Rmarkdown document is a plain-text file with the *.Rmd file extension. It is composed of four types of content:
The YAML header (at the top), surrounded at top and bottom by 3 dashes (—)
Text narrative - the meat of your manuscript
Code chunks, surrounded at top and bottom by 3 back-ticks (```), and the
chunk headerin braces, like {r plot-figure}- Note that the first code chunk is always named
setupand is often used to load libraries and set up document options.
- Note that the first code chunk is always named
Inline code, which does calculations in your text to provide calculated values like means, medians, and p values.
This essentially provides an interface like a ‘lab notebook’ for data analysis. You can use code chunks to run the analysis, and text to document what you are doing in the analysis, how it worked, and interpret the results. When the analysis is ready, you can polish up your document to produce a final manuscript.
Code outputs, including tables and plots, are incorporated into the document.
You can choose to show or hide the code chunks in the final document with options in the chunk header, like:
{r, echo = FALSE} - runs code, but does not show it.
or
{r, echo = TRUE} - runs code and shows the code.
37.2 Trying out RMarkdown with a Mock Manuscript
Open up RStudio.
Start a new Project. Click - File - New Project… - Version Control > - Git >
Now paste the following into the Repository URL: box
https://github.com/higgi13425/rmd4medicine.git
add a directory name, like
rmd4medicineclick on
Create ProjectFind the Files tab in the lower right quadrant.
Click to open the
prepfolderClick to open the
mockstudy_analysis.Rmdfile.
This file should open in the top left quadrant of RStudio. If there are warnings at the top of the file that you need to install packages, click on the Install button.
37.3 Inserting Code Chunks
You can add code to your document to process your data and display results.
To insert a code chunk into your Rmarkdown document, click on the green
+c button at the top center of the top left pane in RStudio.
The dropdown menu will allow you to choose R code or several other computer languages, For now, click on R.
This inserts a gray code chunk, which starts and stops with 3 back-ticks. The starting back-ticks are followed by braces containing a lower-case r, designating what follows as R code.
You can name your individual code chunks with specific names, based on what they do. You can click to the right of the lower-case r, before the closing brace, and add a space, then a name for the code chunk. If the chunk name contains multiple words, connect these with hyphens, as in the code chunk below. Avoid spaces, periods, and underscores in chunk names.
## # A tibble: 6 × 17
## subject_id fake_first_name fake_last_name gender pan_day test_id clinic_name
## <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
## 1 1412 jhezane westerling female 4 covid inpatient wa…
## 2 533 penny targaryen female 7 covid clinical lab
## 3 9134 grunt rivers male 7 covid clinical lab
## 4 8518 melisandre swyft female 8 covid clinical lab
## 5 8967 rolley karstark male 8 covid emergency de…
## 6 11048 megga karstark female 8 covid oncology day…
## # ℹ 10 more variables: result <chr>, demo_group <chr>, age <dbl>,
## # drive_thru_ind <dbl>, ct_result <dbl>, orderset <dbl>, payor_group <chr>,
## # patient_class <chr>, col_rec_tat <dbl>, rec_ver_tat <dbl>37.3.1 Code Chunk Icons
You may have noticed 3 small icons at the top right of each code chunk. From left to right, these are a (settings) gear, a downward arrowhead with a green baseline (run all of the preceding chunks), and a rightward (run) arrow. Check these out and experiment with them.
| Icon | Uses |
|---|---|
| Settings Gear | Allows you to
|
| Run Chunks Above (down arrow) | Runs all of the preceding code chunks, including the setup chunk |
| Run Chunk (rightward arrow) | Runs the entire current chunk |
37.4 Including Plots
You can also embed plots using code chunks, for example:
covid %>%
ggplot() +
aes(x = pan_day, y = ct_result) +
geom_point() +
labs(title = "COVID Testing in First 100 Days of Pandemic",
x = "Pandemic Day, 2020",
y = "Cycle Threshold \n45 is a Negative Test")
Note that the echo = FALSE parameter was added to the top of the
plot code chunk to prevent printing of the R code that generated the
plot. This is an example of a chunk option.
37.5 Including Tables
You can also use code chunks to include tables in your document.
covid %>%
count(demo_group, gender) %>%
gt() %>%
tab_header(title = "Demographics of COVID Testing",
subtitle = "By Group and Gender") %>%
tab_source_note(source_note = "From CHOP, 2020") %>%
cols_label(demo_group = "Group",
gender = "Gender",
n = "Count")| Demographics of COVID Testing | ||
| By Group and Gender | ||
| Group | Gender | Count |
|---|---|---|
| client | female | 314 |
| client | male | 290 |
| misc adult | female | 1214 |
| misc adult | male | 1227 |
| other adult | female | 126 |
| other adult | male | 97 |
| patient | female | 6178 |
| patient | male | 6077 |
| unidentified | male | 1 |
| From CHOP, 2020 | ||
Note that this code chunk is using the {gt} package to format the table.
Other popular approaches to table formatting include the {flextable}
package and the knitr::kable() function.
37.6 Including Links and Images
37.6.1 Links
You can add hypertext links to your text (without a code chunk) with a description in square brackets followed immediately by the URL in (parentheses), like this:
[text description here](http://www.link.com)
As an example, the link to the Rmarkdown cheatsheet can be found at this link.
37.6.2 Images
You can add images to your text if they are in the same project as your
Rmarkdown document. You have to specify the path to the image file
correctly. It is often helpful to collect your images in an images
folder or a figures folder. If you have already generated your figures
with other R scripts, they can be placed into your manuscript document
from a figures folder.
You can add images to your text with an exclamation point, followed by a caption in square brackets followed immediately by the path to the image file in (parentheses), on a line of its own, separated from the text, like this:

You can also insert an image using a code chunk and the knitr function
include_image(), like this, which gives you more options to control
figure size, alignment, height, and width with code chunk options:
(note that echo = TRUE and eval=FALSE as code chunk options means that this code is shown, but not run)
An example shown below is an image of the “datasaurus dozen” used to
illustrate what summary measures can hide in data distributions, as seen
below in 12 data distributions with the same mean and standard
deviation, one of which happens to look like a T. Rex. You should always
visualize your data. There might be a dinosaur in there.
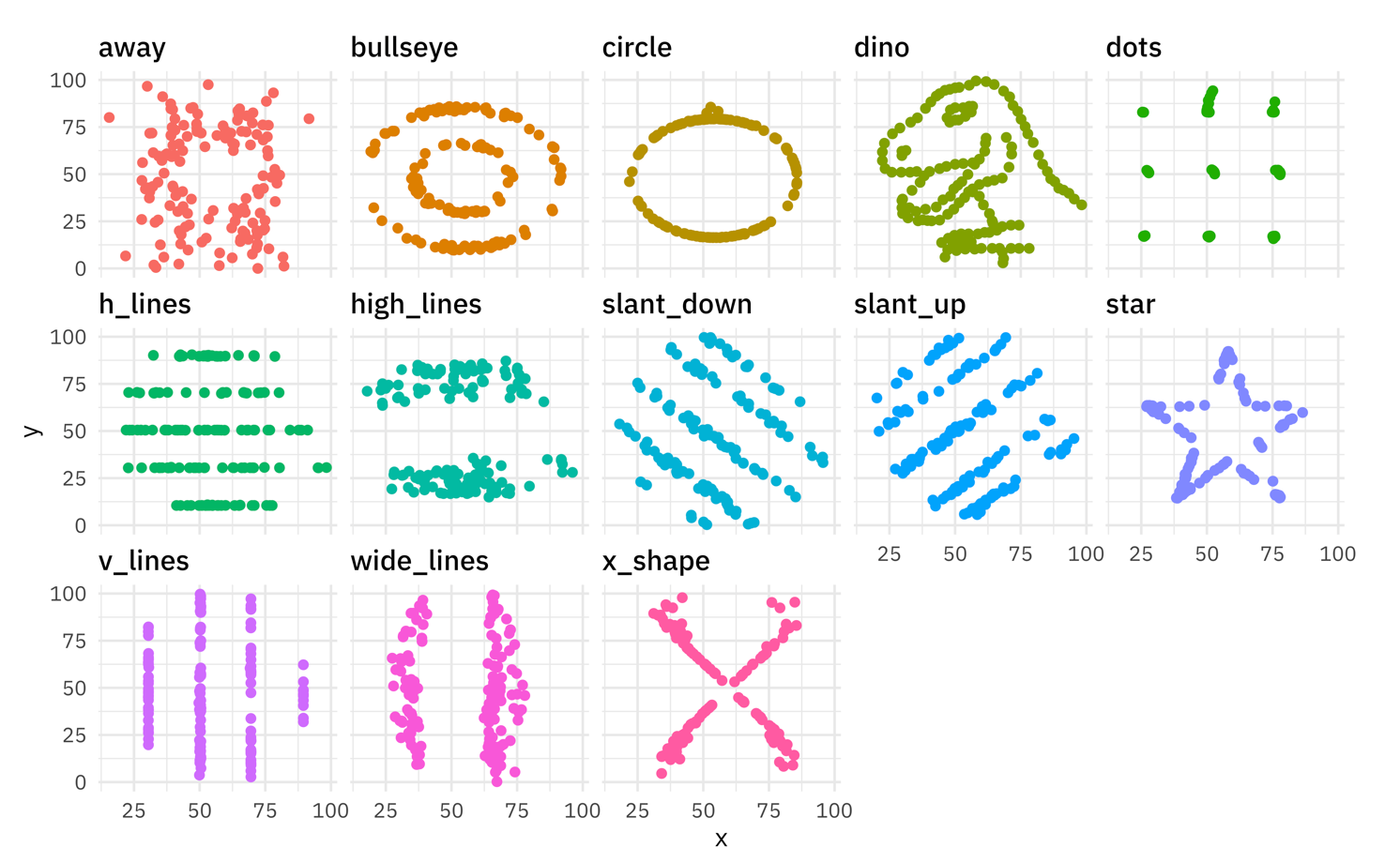
(note that echo = FALSE and eval=TRUE as code chunk options means that the code chunk used to include this image code is run, but not shown)
Source for Image: https://juliasilge.com/blog/datasaurus-multiclass/
37.7 Other languages in code chunks
You can use a number of different open-source languages in addition to R if needed to do your data analysis, including SQL, shell code with Unix Bash, C, C++ via Rcpp, Stan, and D3. Any of these options can be chosen from the Insert Code button dropdown (green +c button).
37.8 Code Chunk Options
When you are working through a data analysis, you usually want to
display the code that led to a result. For the final manuscript, you may
want to hide the code and just display the results. You can accomplish
this with echo=TRUE to display code, and echo=FALSE to hide the
code.
Code chunk options should be added to the top of each code chunk, in the
chunk header after the name of the code chunk, and separated from the
chunk name (and from each other) by commas.
The chunk header (material between the braces) must be written on one line. You must not break the line with a return, or it will not work.
| Option | Values | Output |
|---|---|---|
| eval | TRUE/FALSE | Whether or not the code is run. |
| echo | TRUE/FALSE | Show or hide the code |
| include | TRUE/FALSE | Whether or not the
resulting output of a
code chunk is displayed
in the document. FALSE
means that the code
will run, but will not
display results.
include = FALSE is
often used for the setup
chunk. |
| warning | TRUE/FALSE | Whether warnings generated from your code will be displayed in the document. |
| message | TRUE/FALSE | Whether messages generated from your code will be displayed in the document. |
| fig.align | default, left, right, center | Where on the page the
output figure should
align. Text options
should be in quotes,
likefig.align = "right" |
| fig.width | default = 7 | figure width in inches |
| fig .height | default = 7 | figure height in inches |
| error | TRUE/FALSE | If TRUE, will not stop
building the document if
there is an error in a
code chunk. |
| cache | TRUE/FALSE | If TRUE, will store
the results and not
re-run the chunk.
Helpful for long, slow
calculations. But watch
out for this if your
data change and your
results do not(!!). |
Note that there are many more chunk options which you can use if needed, and these can be found here.
37.9 How It All (Rmarkdown + {knitr} + Pandoc) Works

Rmarkdown is an R-flavored version of the markdown language. This is a
universal, open-source markup language for creating formatted documents
from plain text. Markdown documents end with the file extension *.md.
An open-source program named pandoc converts *.md documents to output
documents like MS Word, PDF, HTML, MS Powerpoint, etc.
When you click the Knit button or run the render() function, R Markdown feeds the .Rmd file to knitr, which executes all of the code chunks and creates a new markdown (.md) document which includes the code and its output.
The markdown file generated by {knitr} is then processed by pandoc
which is responsible for creating the finished format.
This may sound complicated, but R Markdown makes it extremely simple by encapsulating all of the above processing into a single render() function (or the Knit button).
37.10 Knitting and Editing (and re-Knitting() Your Rmd document
Find the Knit button at the top of the file, and click it to convert
this *.Rmd . It will automagically knit the document first to markdown
(to *.md and then from *.md to *.HTML.
Scroll through the document to see what has been created.
Go back to the Rmd document.
Now try the following:
in the YAML header, edit the author to your name
edit the date to the current (or a different) date
edit the output from
html_documenttoword_documentfind the
glimpsecode chunk. After the chunk nameglimpse, add a comma, then the optionecho=FALSEAdd this same option to a few of of the other code chunks (feel free to use copy-paste, but make sure you don’t end up with duplicate commas, as this will cause errors)
Now click on the Knit button again.
You should get a new version of the document, now in MS Word format, with the code chunks hidden, and a new author and date. The Word document is in a particular default format, but you can change this by specifying a template word file in the YAML header.
37.11 Try Out Other Chunk Options
Try adding different chunk options, including
include = FALSEeval = FALSE, echo = TRUEeval=TRUE, echo = FALSE
37.12 The setup chunk
The setup chunk is a special code chunk, which is usually the first
code chunk at the top of your RMarkdown document. Typically it includes
two types of code:
- libraries to be loaded
- data to be loaded in the background
and often looks something like this:
(note that for display purposes, I am using the chunk options
eval = FALSE, echo = TRUE, while in a real setup chunk, I would use
include = FALSE, which runs the code but does not display the code nor
the output. I also had to change the chunk name to setup2 because only
unique chunk names are allowed).
Scroll to the setup chunk at the top of your Rmd document to see what
a working setup chunk looks like.
37.13 Markdown syntax
Markdown is a popular markup language that allows you to add formatting
elements to text, including bold, italics, and code formatting.
We make text Bold by surrounding the words with double asterisks. We
make text Italic by surrounding the words with single underscores or
single asterisks. We make text Bold and Italic by surrounding the
words with triple underscores or triple asterisks. We can make text in
code-font by surrounding it with single back-ticks.
You can also format level 1 to level 5 headings. These are done by preceding the heading (on its own line) with 1-5 hashtags.
37.15 Line Breaks and Page Breaks
If you simply hit return in your *.Rmd document, you will see a line
break in your text. But this is only a semantic line break, as the
knitted document will smush these lines together.
You can create a deliberate line break by adding 2 or more spaces to
the end of a line of text. This will work in any output format.
The downside of this is that these line breaks are not visible, until you Knit the document.
Line breaks can be inserted (for HTML output) by
using
html
tags
The HTML tag for line breaks is <br>. But these are kind of
annoying to type,
and 2 spaces at the end of each line is pretty
easy, but less visible when you are looking at the text in the Rmd.
Note that you need a blank line before headers for them to be recognized as headers and formatted properly
37.16 Making Lists
37.16.1 Ordered Lists
You can create an ordered list by preceding items with numbers and period:
- First
- Second
- Third
37.18 Inline Code
You will often want to insert a result into your text, like a percent reduction in an endpoint, or a p value. Rather than copying and pasting from somewhere else (which are prone to error, or forgetting to update), you can do these calculations right in your text, using inline R code. For example, if you want to say that we evaluated NN COVID PCR tests for this study, you can calculate how many rows in your dataset with R, right in the text.
To do this, you bracket the code with single back-ticks, and start with
the letter r immediately after the first back-tick, so that Rmarkdown
knows that R code is coming. After the lower-case r, you can insert
your code expression, like nrow(covid) to give you the number of rows
(observations). Putting this together, this looks like
`r nrow(covid)`. When you insert this in the
middle of a sentence in your text (inline code), this lets you write
sentences in your manuscript like the following, which calculate the
actual number in the text (and automatically update when your data
changes):
We evaluated 15524 COVID PCR tests in this
study.
37.18.1 Try inserting some in-line R code
Try this yourself. Select the correct code to insert the proper results, as illustrated below (note that these would be surrounded by single back-ticks)
The mean cycle threshold in this study wasCorrect answers should produce this output when knit:
The mean cycle threshold in this study was 44.122 and the standard deviation was 3.98.
37.19 A Quick Quiz
- Which code chunk option hides the code?
- Which code chunk always comes first, and includes libraries and data import steps?
- What is the name of the code block (and the markup language it is
written in), set off with 3 hyphens(—) at the very top of your
Rmarkdown document, that tells
pandochow to format the final document? - What symbols do you use to make text bold in Rmarkdown?
- Which {knitr} function do you use to add images to your document with a code chunk?