Chapter 10 Special topic: Clinical Trial Data Analysis with R
Nhan Thi Ho, MD, PhD
10.1 Clinical trial design
10.1.1 Fundamentals of major randomized controlled trial design
| Choice of patients, centers | Choice of treatments | Choice of outcomes | Randomization | Blinding | Trial sample size |
|---|---|---|---|---|---|
| - Precise eligibility criteria - Multi-center trial is preferable - Appropriate geographic representation |
- Precise treatment regimens - Placebo control or active comparator - 3-arm-trial may sometimes be appropriate |
- Define the primary efficacy endpoint - Select components of composite primary endpoint - List secondary endpoints - Define safety endpoints |
- Allocation concealment - Proper statistical method for randomization - Ensure between group balance by stratification - Unequal randomization may be useful for new treatment |
- Double blind if practical - If not practical, blinded evaluation is essential - Blinding is important for soft endpoints |
- Sample size calculation using appropriate method - Anticipate realistic effect size - Compromise may be needed for a real world study - Determining needed number of events may be useful |
(Source: adapted from Pocock S.J et al. J Am Coll Cardiol. 2015;66(24):2757-66. (Pocock, McMurray, and Collier 2015))
10.1.2 Requirements
Clinical trial design should accomplish the following (Piantadosi 2005):
- Quantify and reduce errors due to chance
- Reduce or eliminate bias
- Yield clinically relevant estimates of effects and precision
- Be simple in design and analysis
- Provide a high degree of credibility, reproducibility, and external validity
- Influence future clinical practice
10.1.3 Common concepts
- One-way designs: Only one factor= treatment
- Two-way factorial design: various combinations of interventions
- Parallel design =typical design: patients are randomized to a treatment and remain on that treatment throughout the course of the trial.
- Randomization: is used to remove systematic error (bias) and to justify Type I error probabilities in experiments; an essential feature of clinical trials for removing selection bias.
- Selection bias: occurs when a physician decides treatment assignment and systematically selects a certain type of patient for a particular treatment.
- Blocking and stratification are used to control unwanted variation.
- Placebos or relevant controls.
- Treatment masking or blinding is an effective way to ensure objectivity of the person measuring the outcome variables. Double-masked: both investigators and patients are masked to the treatment. Single-masked: only patients or evaluators are masked (Friedman et al. 2015).
10.1.4 Nonrandomized Studies
10.1.4.1 Nonrandomized Concurrent Control Studies
- No randomization
- May match by some characteristics
- Bias in the allocation of participants to either the intervention or control group
- Possible: one center= one group
\(\rightarrow\) Analysis:
- Baseline comparison + testing
- Adjusting for covariates
10.1.4.2 Historical Controls and Databases
Nonrandomized and nonconcurrent.
- All new participants can receive the new intervention
- Historical control data can be obtained from two sources: literature, unpublished \(\rightarrow\) low quality, uncomparable
- Prone to bias
\(\rightarrow\) Analysis:
- Two sample comparison/ one sample comparing to constant
- Baseline comparison + testing
- Adjusting for covariates
10.1.5 Cross-Over Designs
Special case of a randomized control trial.
- Each participant to serve as his own control.
- In the simplest case (two period cross-over design): each participant will receive either intervention or control (A or B) in the first period and the alternative in the succeeding period. The order in which A and B are given to each participant is randomized \(\rightarrow\) approximately half of the participants receive the intervention in the sequence AB and the other half in the sequence BA. This is so that any trend from first period to second period can be eliminated in the estimate of group differences in response.
- There may be more than two periods, a wash-out period may be used between the periods.
10.1.5.1 Advantages
- Allows assessment of how each participant does on both A and B.
- Reduction in variability because the measured effect of the intervention is the difference in an individual participant’s response to intervention and control \(\rightarrow\) need smaller sample sizes to detect a specific difference in response (parallel or noncross-over designs need 2.4 times bigger in sample size)
10.1.5.2 Strict assumption
- The effects of the intervention during the first period must not carry over into the second period
10.1.5.3 Analysis: Significance tests
- Carry-over: Is the carry-over from A to B (group 1 2nd period) the same as from B to A (group 2 2nd period)? The null hypothesis being tested is that the carry-over effects are equal between groups 1 (A first) and group 2 (B first).
- Treatment: Note: We can only test the treatment effect if it can be assumed that the carryover effects are equal. Hence it is necessary to perform the test above first. If a significant difference is found in carry-over, then this subsequent test will be invalid. The null hypothesis being tested is that the treatment effects are equal.
- Period: test the period effect (i.e., is there a change over time irrespective of treatment) \(\rightarrow\) need to assume that there is no significant treatment effect. The null hypothesis being tested is that changes between treatments are equal irrespective of time.
10.1.6 Factorial Design
Test the effect of more than one treatment, permits an assessment of potential interactions among the treatments.
Analysis: multiple comparison.
10.1.7 Group Allocation Designs
A group of individuals, a clinic or a community are randomized to a particular intervention or control.
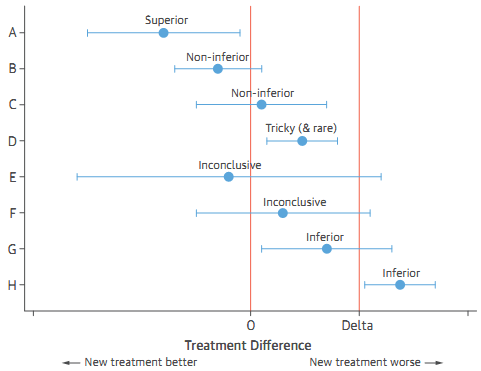
10.1.8 Studies of Equivalency and Noninferiority
Demonstrate that the new intervention is not worse, in terms of the primary response variable, than or at least as good as the standard by some predefined margin. (Usually, many clinical trials are designed to demonstrate that a new intervention is better than or superior to the control).
- The control or standard treatment must have been shown conclusively to be effective.
Possible results of noninferiority trials:
10.2 Sample size- power
The underlying theme of sample size calculation in all clinical trials is precision. Two circumstances:
- Precision for an estimator (e.g. 95% confidence interval).
- Statistical power for hypothesis testing, e.g., requiring 0.80 or 0.90 statistical power (1-\(\beta\)) for a hypothesis test when the significance level (\(\alpha\)) is 0.05 to detect the desired effect size (the clinically meaningful effect) (Chow et al. 2017).
10.2.1 Some R packages
Examples:
#install.packages("SampleSize4ClinicalTrials")
library("SampleSize4ClinicalTrials")
#Comparing blood glucose (mmol/l) in treatment vs. placebo, superior design
ssc_meancomp(design=2L, ratio=1, alpha=0.05, power=0.8, sd=2, theta=1, delta=0.5)## Treatment Control
## 1 198 19810.3 Randomization
Randomization tends to produce study groups comparable with respect to known as well as unknown risk factors, removes investigator bias in the allocation of participants, and guarantees that statistical tests will have valid false positive error rates.
- Simple: a patient is assigned a treatment without any regard for previous assignments (~fliping coin)
- Randomization in permuted blocks
- Stratified Randomization
- Adaptive randomization refers to any scheme in which the probability of treatment assignment changes according to assigned treatments of patients already in the trial.
10.3.1 R package
Examples:
#install.packages("randomizeR")
library(randomizeR)
# generate randomization sequences using the Random Allocation Rule for N = 50
myPar <- rarPar(50)
genSeq(myPar)##
## Object of class "rRarSeq"
##
## design = RAR
## seed = 99276
## N = 50
## groups = A B
##
## The sequence M:
##
## 1 A A B A B B A B B B ...10.4 Interim Analysis
10.4.1 Overview
“Interim analysis” or “early stopping” procedures are used to interpret the accumulating information during a clinical trial to monitor treatment effects as well as tracking safety issues. Reasons stopping a trial can be:
- Treatments are found to be convincingly different,
- Treatments are found to be convincingly not different,
- Side effects or toxicity are too severe to continue treatment, relative to the potential benefits,
- The data are of poor quality,
- Accrual is too slow to complete the study in a timely fashion,
- Definitive information is available from outside the study, making the trial unnecessary or unethical, this is also related to the next item…
- The scientific questions are no longer important because of other developments,
- Adherence to the treatment is unacceptably poor, preventing an answer to the basic question,
- Resources to perform the study are lost or no longer available, and/or
- The study integrity has been undermined by fraud or misconduct.
“The review, interpretation, and decision-making aspects of clinical trials based on interim data are necessary but prone to error. If the investigators learn of interim results, then it could affect objectivity during the remainder of the trial, or if statistical tests are performed repeatedly on the accumulating data, then the Type I error rate is increased”.
10.4.2 Methods used in interim analysis
“Although many statistical techniques are available to assist in monitoring, none of them should be used as the sole basis in the decision to stop or continue the trial”.
- Likelihood Methods
- Bayesian Methods
- Frequentist Methods: O’Brien-Fleming, Pocock, Haybittle-Peto
- Alpha Spending Function approach
- Futility Assessment with Conditional Power; Adaptive Designs
For details, please see Courses: Design and Analysis of Clinical Trials
10.4.3 Monitoring and Interim Reporting for Trials
10.4.3.1 Single-Center Trials
The report should include:
- Compliance with governmental and institutional oversight,
- Review of eligibility (low frequency of ineligible patients entering the trial),
- Treatment review (most patients are adhering to the treatment regimen),
- Summary of response,
- Summary of survival,
- Adverse events,
- Safety monitoring rules (possibly statistical criteria for evaluating safety endpoints), and
- Audit and other quality assurance reviews.
10.4.3.2 Multi-Center Trials
A DSMB typically examines the following issues:
- Are the treatment groups comparable at baseline?
- Are the accrual rates meeting initial projections and is the trial on its scheduled timeline?
- Are the data of sufficient quality?
- Are the treatment groups different with respect to safety and toxicity data?
- Are the treatment groups different with respect to efficacy data?
- Should the trial continue?
- Should the protocol be modified?
- Are other descriptive statistics, graphs, or analyses needed for the DSMB to make its decisions?
10.5 Estimating Clinical Effects
10.5.1 Dose-Finding (Phase I) Studies
What level of the drug is appropriate? \(\rightarrow\) Assess the distribution and elimination of drug in the human system.
- Pharmacokinetic (PK) models (compartmental models): to account for the absorption, distribution, metabolism, and excretion of a drug in the human system.
10.5.2 Safety and Efficacy (Phase II) Studies
- The main objectives is to estimate the frequency of adverse reactions and estimate the probability of treatment success: often are expressed in binary form, presence/absence of adverse reaction, success/failure of treatment, etc.,. Three common parameters of risk:
- Risk difference
- Relative risk
- Odds ratio
Statistical test of the null hypothesis H0: RR = 1 (or H0: RD = 0).
If safety and efficacy study is stratified according to some factor, such as clinical center, disease severity, gender, etc \(\rightarrow\) estimate the odds ratio while accounting for strata effects. Mantel-Haenszel Test for the Odds Ratio: assumes that the odds ratio is equal across all strata, although the rates, p1 and p2, may differ across strata \(\rightarrow\) calculates the odds ratio within each stratum and then combines the strata estimates into one estimate of the common odds ratio.
In some safety and efficacy studies, it is of interest to determine if an increase in the dose yields an increase (or decrease) in the response \(\rightarrow\) dose-response or trend analysis.
- Continuous response: Jonckheere-Terptsra (JT) trend test which is based on a sum of Mann-Whitney-Wilcoxon tests.
- Binary response: Cochran-Armitage (CA) trend test
- Survival Analysis For time-to-event outcomes: Kaplan-Meier survival curve (log rank test) (Pocock, McMurray, and Collier 2015).
10.6 Clinical Trial Analysis and reporting
10.6.1 Baseline Assessment
10.6.1.1 Description of Trial Participants
The typical Table 1 of the results publication. It provides documentation that can guide cautious extrapolation of trial findings to other populations with the same medical condition
10.6.1.2 Controlling for Imbalances in the Analysis
Imbalanced groups at baseline may be addressed by covariate-adjustment during analysis.
10.6.1.3 Subgrouping
Stratified analysis: help to identify the specific population most likely to benefit from, or be harmed by, the intervention; elucidate the mechanism of action of the intervention. Definition of such subgroups should rely only on baseline data, not data measured after initiation of intervention (except for factors such as age or gender which cannot be altered by the intervention).
10.6.2 Trial profile
Figure shows the flow of patients through the trial from the pre-randomization build-up to the post-randomization follow-up.
10.6.3 Tables/ plots of main outcomes
- The key table for any clinical trial displays the main outcomes by treatment group.
- The most common type of figure in trial is Kaplan-Meier plot of time-to-event outcomes, repeated measures over time (longitudinal data)
10.6.4 Estimates of treatment effects and their CIs
- Effect size (point estimate)
- Uncertainty (CIs)
10.6.4.1 p values and interpretation
- Should provide actual p value (not <)
- Useful to recognize the link between the p value and the 95% CI for the treatment difference
- Use 2-sided p values
- A small p value clarifies that an observed treatment difference appears greater than what could be attributed to chance, but this does not automatically mean that a real treatment effect is occurring (There may be biases in the study design and conduct).
- There is an important distinction between statistical significance and clinical relevance of a treatment effect \(\rightarrow\) The magnitude of treatment difference and its CI are a guide for clinical usefulness of treatment difference.
- For a small trial the magnitude of treatment needs to be very large to reach a statistically significant treatment effect.
10.6.5 Summary
The main steps in data analysis and reporting for clinical trials:
- What to include in result section
- Trial profile
- Baseline characteristics
- Main outcomes tables +/- figures
- Quantify association, uncertainty and evidence: estimate treatment effect (e.g. relative risk, odds ratio, absolute difference, number needed to treat, hazard ration for time to event outcomes, …), confidence interval and p-values.
10.6.6 Statisticial Controversy in Reporting of Clinical Trials
Problems and solutions:
| Problems | Solution |
|---|---|
| Multiplicity of data: how to make sense of all the options? | - Predefined Statistical Analysis Plan (SAP) - Prioritized primary endpoint - Balanced account of safety and efficacy - Careful interpretation of composite endpoints |
| Covariate adjustment: should key results be adjusted for baseline covariates? | - Adjust variables affecting prognosis - Predefined chosen variables and models - Consider covariate adjustment as primary analysis |
| Subgroup analysis: which subgroups should be explored? | - Predefined subgroups - Analyse using interaction tests not subgroup p-values - Interprete all subgroup findings cautiously |
| Individual benefits and risks: how to link trial findings to individualized patient care? | - Balance absolute benefits against absolute harms - Consider individual risk profile in determine their treatment benefit - Consider multivariable model instead of univariable subgroups |
| Intention to treat (ITT) analysis: how to deal with non-adherence during follow-up | - Prioritize ITT analysis - Continue follow up for patients withdrawing from treatment if possible - Minimize poor compliance and loss of follow-up - For non-inferiority trial, present both ITT and per protocol analyses |
| Interpreting surprising results: what to do with unexpected findings? | - Seek evidence to confirm (or not) as soon as possible - Be skeptical of large effects - Anticipate regression to the truth - Avoid alarmist reactions to unexpected safety signals |
10.7 Case study
The data for the examples below are from a multi-centered phase 1/2/3 COVID-19 vaccine trial in which the author Nhan Thi Ho participated as a core researcher/ data scientist (N. T. Ho et al. 2024). The actual interim and final analyses for this trial for publication and authority approval were done in SAS. A sample of trial data (with some modification for de-identification) are used in the following examples to demonstrate the use of R for clinical trial data analysis and report. The example data contain the following datasets:
- ran.ex: randomization list and analysis sets
- base.ex: baseline data
- ve.ex: time to event outcome (COVID-19) data
- sae.ex: serious adverse event data
- ae.ex: adverse event data
10.7.2 Baseline summary
Summary of baseline characteristics for ITT analysis set. The examples below are to illustrate the use of the package “arsenal” for descriptive summary tables.
d_controls <- tableby.control(
test = T,
total = F,
numeric.test = "kwt", cat.test = "chisq",
numeric.stats = c("meansd", "medianq1q3","meanCI","range","Nmiss2"),
cat.stats = c("countpct", "Nmiss2"),
stats.labels = list(
meansd = "Mean (SD)",
medianq1q3 = "Median (Q1, Q3)",
range = "Min - Max",
meanCI = "Mean (95%CI)",
Nmiss2 = "Missing"
)
)
dvar<- c("age","agecat","sex","riskcov")
mylabels <-as.list(c("Age (year)","Age category (year)","Sex","Risk category"))
names(mylabels)<-dvar
tabd <- tableby(as.formula(paste("treatment",paste(dvar,collapse="+"),sep="~")),
data = subset(base.ex, itt=="1"),
control = d_controls)
kable(summary(tabd,labelTranslations = mylabels, text=TRUE),
caption="Summary of baseline data")| Placebo (N=7345) | Vaccine (N=7753) | p value | |
|---|---|---|---|
| Age (year) | 0.004 | ||
| - Mean (SD) | 46.539 (13.406) | 45.964 (13.270) | |
| - Median (Q1, Q3) | 48.000 (37.000, 57.000) | 47.000 (36.000, 56.000) | |
| - Mean (95%CI) | 46.539 (46.232, 46.846) | 45.964 (45.668, 46.259) | |
| - Min - Max | 17.000 - 89.000 | 17.000 - 85.000 | |
| - Missing | 0 | 0 | |
| Age category (year) | 0.022 | ||
| - <=60 | 6199 (84.4%) | 6646 (85.7%) | |
| - >60 | 1146 (15.6%) | 1107 (14.3%) | |
| - Missing | 0 | 0 | |
| Sex | 0.684 | ||
| - Mean (SD) | 1.645 (11.650) | 1.634 (11.340) | |
| - Median (Q1, Q3) | 2.000 (1.000, 2.000) | 2.000 (1.000, 2.000) | |
| - Mean (95%CI) | 1.645 (1.378, 1.911) | 1.634 (1.382, 1.887) | |
| - Min - Max | 1.000 - 999.000 | 1.000 - 999.000 | |
| - Missing | 0 | 0 | |
| Risk category | 0.827 | ||
| - At_risk | 3705 (50.4%) | 3897 (50.3%) | |
| - Healthy | 3640 (49.6%) | 3856 (49.7%) | |
| - Missing | 0 | 0 |
10.7.3 Event summary
10.7.3.1 Summary with package “arsenal”
Summary number of event and follow-up time by group for analysis set mITT at Day 92. The function “tableby” of the package “arsenal” may also be used for overall summary and for subgroup stratification.
Overall:
o_controls <- tableby.control(
test = F,
total = F,
numeric.test = "kwt", cat.test = "chisq",
numeric.stats = "arsenal_sum",
cat.stats = c("countpct", "Nmiss2"),
stats.labels = list(arsenal_sum ="Sum"))
ovar<- c("event","ttevent","event3692","ttevent3692")
mylabels <- as.list(c("Event (any time)","Follow-up time (day)",
"Event between Day 36-92","Follow-up time between 36-92"))
names(mylabels)<-ovar
tabo <- tableby(as.formula(paste("treatment",paste(ovar,collapse="+"),sep="~")),
data = subset(ve.ex, mitt_d92=="1"),
control = o_controls)
kable(summary(tabo,labelTranslations = mylabels, text=TRUE))| Placebo (N=6825) | Vaccine (N=6849) | |
|---|---|---|
| Event (any time) | ||
| - Sum | 735.000 | 539.000 |
| Follow-up time (day) | ||
| - Sum | 618244.000 | 625475.000 |
| Event between Day 36-92 | ||
| - Sum | 432.000 | 220.000 |
| Follow-up time between 36-92 | ||
| - Sum | 601174.000 | 607577.000 |
Stratified by risk category:
tabo <- tableby(as.formula(paste("treatment",paste(ovar,collapse="+"),sep="~")),
data = subset(ve.ex, mitt_d92=="1"),
strata=riskcov,
control = o_controls)
kable(summary(tabo,labelTranslations = mylabels, text=TRUE))| riskcov | Placebo (N=6825) | Vaccine (N=6849) | |
|---|---|---|---|
| Healthy | Event (any time) | ||
| - Sum | 405.000 | 324.000 | |
| Follow-up time (day) | |||
| - Sum | 308028.000 | 309900.000 | |
| Event between Day 36-92 | |||
| - Sum | 222.000 | 133.000 | |
| Follow-up time between 36-92 | |||
| - Sum | 297987.000 | 299884.000 | |
| At_risk | Event (any time) | ||
| - Sum | 330.000 | 215.000 | |
| Follow-up time (day) | |||
| - Sum | 310216.000 | 315575.000 | |
| Event between Day 36-92 | |||
| - Sum | 210.000 | 87.000 | |
| Follow-up time between 36-92 | |||
| - Sum | 303187.000 | 307693.000 |
10.7.3.2 Manual calculation for later table combination
All data:
esum<-ve.ex %>%
filter(mitt_d92=="1") %>%
group_by(treatment) %>%
summarise_at(.vars = vars(event3692,ttevent3692),
.funs = sum) %>%
pivot_wider(
names_from = treatment,
values_from = c(event3692,ttevent3692)
)
esum## # A tibble: 1 × 4
## event3692_Placebo event3692_Vaccine ttevent3692_Placebo ttevent3692_Vaccine
## <dbl> <dbl> <dbl> <dbl>
## 1 432 220 601174 607577Stratify by subgroups and combine results:
esums<-ve.ex %>%
filter(mitt_d92=="1") %>%
group_by(treatment,riskcov) %>%
summarise_at(.vars = vars(event3692,ttevent3692),
.funs = sum) %>%
pivot_wider(
names_from = treatment,
values_from = c(event3692,ttevent3692)
)
esums## # A tibble: 2 × 5
## riskcov event3692_Placebo event3692_Vaccine ttevent3692_Placebo ttevent3692_Vaccine
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 Healthy 222 133 297987 299884
## 2 At_risk 210 87 303187 307693#Combined results
esuma<-bind_rows(esum, esums)
esuma<-esuma%>%
select(riskcov,contains("Placebo"),contains("Vaccine"))
esuma## # A tibble: 3 × 5
## riskcov event3692_Placebo ttevent3692_Placebo event3692_Vaccine ttevent3692_Vaccine
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 <NA> 432 601174 220 607577
## 2 Healthy 222 297987 133 299884
## 3 At_risk 210 303187 87 30769310.7.4 Vaccine efficacy
10.7.4.1 Cox model
10.7.4.1.1 All data
Select analysis set mITT at Day 92.
# Cox model all
cfit<-coxph(Surv(ttevent3692,event3692)~treatment,
data=subset(ve.ex, mitt_d92=="1"))
hrtab<-summary(cfit)$conf.int[,c(1,3:4)]
names(hrtab)<-c("HR","lower.95","upper.95")
# HR all
hrtab## HR lower.95 upper.95
## 0.498 0.424 0.586## efficacy lower.95 upper.95
## 0.502 0.414 0.576# Another way to display results
library(gtsummary)
tbl_regression(cfit, estimate_fun = function(x) {round(1 - exp(x) , 2)})| Characteristic | log(HR)1 | 95% CI1 | p-value |
|---|---|---|---|
| treatment | |||
| Placebo | — | — | |
| Vaccine | 0.5 | 0.58, 0.41 | <0.001 |
| 1 HR = Hazard Ratio, CI = Confidence Interval | |||
10.7.4.1.2 Stratified by risk category
For participants at risk of severe COVID-19:
datr<-subset(ve.ex,mitt_d92==1 & riskcov=="At_risk")
fitr<-coxph(Surv(ttevent3692,event3692)~treatment, data=datr)
tabr<-summary(fitr)$conf.int[,c(1,3:4)]
names(tabr)<-c("HR","lower.95","upper.95")
# HR riskcov=At_risk
tabr## HR lower.95 upper.95
## 0.404 0.314 0.518# VE riskcov=At_risk
ver<-1-tabr
ver<-c(ver[1],ver[3],ver[2])
names(ver)<-c("efficacy","lower.95","upper.95")
ver## efficacy lower.95 upper.95
## 0.596 0.482 0.686Healthy participants:
dath<-subset(ve.ex,mitt_d92==1 & riskcov=="Healthy")
fith<-coxph(Surv(ttevent3692,event3692)~treatment, data=dath)
tabh<-summary(fith)$conf.int[,c(1,3:4)]
names(tabh)<-c("HR","lower.95","upper.95")
# HR riskcov=Healthy
tabh## HR lower.95 upper.95
## 0.589 0.475 0.730# VE riskcov=Healthy
veh<-1-tabh
veh<-c(veh[1],veh[3],veh[2])
names(veh)<-c("efficacy","lower.95","upper.95")
veh## efficacy lower.95 upper.95
## 0.411 0.270 0.525Combine results:
#Combine efficacy results
ve.all<- round(rbind(ve, veh, ver),2)
ve.s<-paste0(ve.all[,1]," (", ve.all[,2],",",ve.all[,3],")")
#Combine event summary with efficacy results:
esumas<-cbind(esuma,ve.s)
esumas$riskcov<-c("All participants","- Healthy","- At risk of severe COVID-19")
kable(esumas,
caption = "Summary of vaccine efficacy",
col.names = c("Study population",
"Placebo <br> Total event",
"Placebo <br> Total follow-up (day)",
"Vaccine <br> Total event",
"Vaccine <br> Total follow-up (day)",
"Vaccine efficacy (95%CI)"))| Study population | Placebo Total event |
Placebo Total follow-up (day) |
Vaccine Total event |
Vaccine Total follow-up (day) |
Vaccine efficacy (95%CI) |
|---|---|---|---|---|---|
| All participants | 432 | 601174 | 220 | 607577 | 0.5 (0.41,0.58) |
| - Healthy | 222 | 297987 | 133 | 299884 | 0.41 (0.27,0.52) |
| - At risk of severe COVID-19 | 210 | 303187 | 87 | 307693 | 0.6 (0.48,0.69) |
10.7.4.2 Cumulative incidence plot
10.7.4.2.1 All data
fit<-survminer::surv_fit(Surv(ttevent3692,event3692)~treatment,
data=subset(ve.ex, mitt_d92=="1"))
cuminca<-ggsurvplot(fit, linetype="strata",
conf.int=FALSE,pval=TRUE,censor=FALSE,
pval.coord=c(43,0.15),
risk.table = TRUE,
risk.table.height = 0.25,
break.x.by =7,
xlim=c(36,92),
fun = "event",
legend.title ="",
tables.col = "strata",
legend.labs=c("Placebo","Vaccine"),
xlab="Follow-up day from Day 1",
ylab="COVID-19 cumulative incidence",
risk.table.y.text = FALSE)
cuminca$plot<-cuminca$plot+
scale_x_continuous(breaks = c(0,36,43,50,57,64,71,78,85,92))
cuminca$table<-cuminca$table+
scale_x_continuous(breaks = c(0,36,43,50,57,64,71,78,85,92))
cuminca 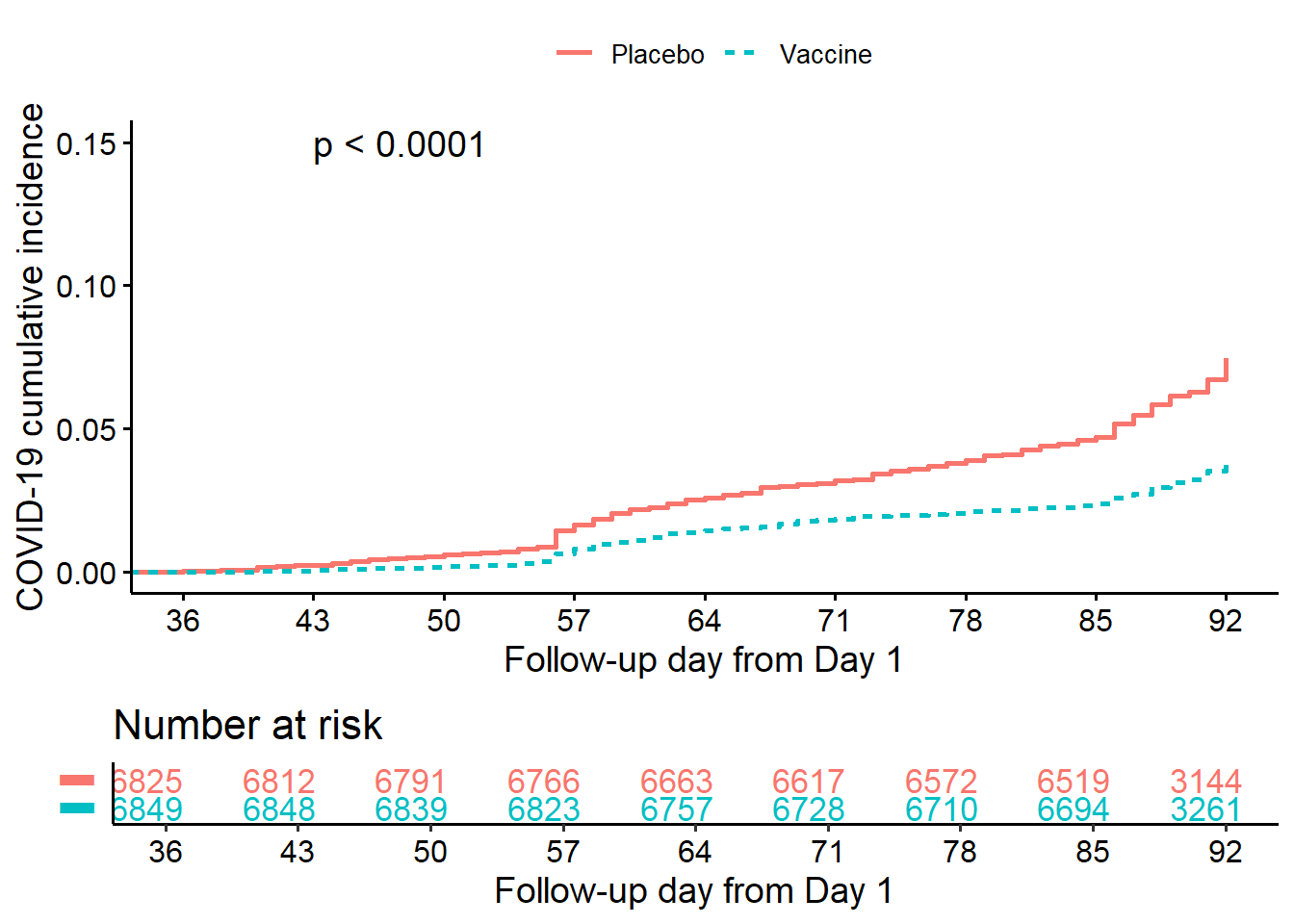
10.7.4.2.2 Stratified by risk category
fit<-survminer::surv_fit(Surv(ttevent3692,event3692)~treatment + riskcov,
data=subset(ve.ex, mitt_d92=="1"))
cumincr<-ggsurvplot(fit, linetype="strata",
conf.int=FALSE,pval=FALSE,censor=FALSE,
risk.table = TRUE,
risk.table.height = 0.3,
break.x.by =7,
xlim=c(36,92),
fun = "event",
legend.title ="",
legend.labs=c("Placebo.Healthy", "Placebo.At_risk",
"Vaccine.Healthy","Vaccine.At_risk"),
tables.col = "strata",
xlab="Follow-up day from Day 1",
ylab="COVID-19 cumulative incidence",
risk.table.y.text = FALSE)
cumincr$plot<-cumincr$plot+
scale_x_continuous(breaks = c(0,36,43,50,57,64,71,78,85,92))
cumincr$table<-cumincr$table+
scale_x_continuous(breaks = c(0,36,43,50,57,64,71,78,85,92))
cumincr 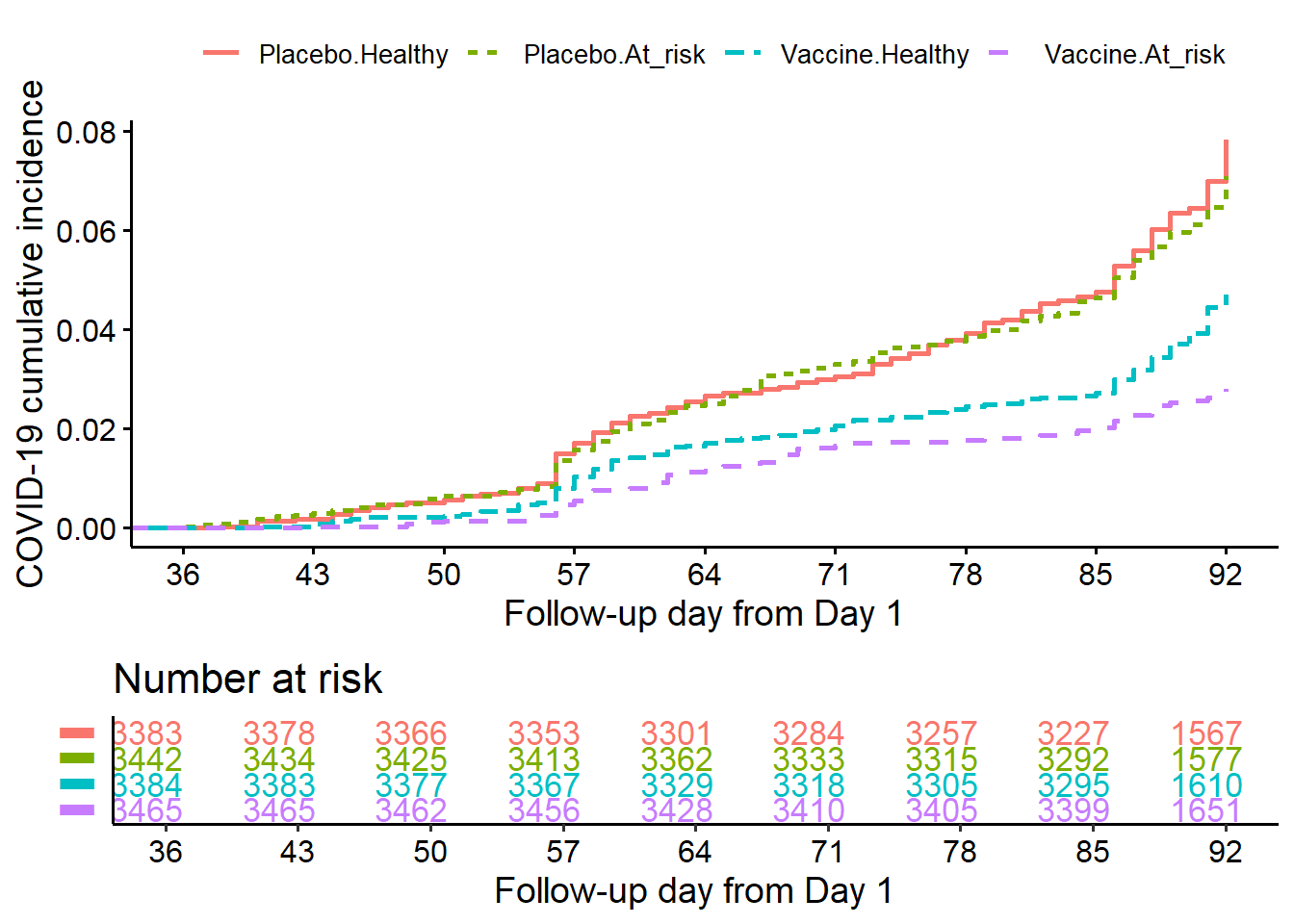
10.7.5 Safety data summary
The codes for safety analysis were adapted from the book R for Clinical Study Reports and Submission (Zhang et al. 2025).
10.7.5.2 Summary of AE by SOC and PT
Reformat AE data and summarize the number and percentage of participants by SOC:
t1 <- ae.ex %>%
group_by(treatment, soc) %>%
summarise(n = fmt_num(n_distinct(usubjid), digits = 0)) %>%
mutate(pt = soc, order = 0)%>%
mutate(n=as.numeric(n))
t1.pop<-merge(t1,pop,by = "treatment", all.x=T)
t1.np <- t1.pop %>%
mutate(
pct = fmt_num(n / tot * 100, digits = 1),
n = fmt_num(n, digits = 0),
pct = paste0("(", pct, ")")
)Reformat AE data and summarize the number and percentage of participants by SOC and PT:
t2 <- ae.ex %>%
group_by(treatment, soc, pt) %>%
summarise(n = fmt_num(n_distinct(usubjid), digits = 0)) %>%
mutate(order = 1)%>%
mutate(n=as.numeric(n))
t2.pop<-left_join(t2, pop, by="treatment")
t2.np <- t2.pop %>%
mutate(
pct = fmt_num(n / tot * 100, digits = 1),
n = fmt_num(n, digits = 0),
pct = paste0("(", pct, ")")
)
head(t2.np)## # A tibble: 6 × 7
## # Groups: treatment, soc [2]
## treatment soc pt n order tot pct
## <fct> <chr> <chr> <chr> <dbl> <int> <chr>
## 1 Vaccine Blood and lymphatic system disorders Anaemia " 1" 1 7753 ( 0.…
## 2 Vaccine Blood and lymphatic system disorders Lymphadenitis " 3" 1 7753 ( 0.…
## 3 Vaccine Blood and lymphatic system disorders Lymphadenopathy " 2" 1 7753 ( 0.…
## 4 Vaccine Cardiac disorders Acute myocardial infarction " 1" 1 7753 ( 0.…
## 5 Vaccine Cardiac disorders Angina pectoris " 1" 1 7753 ( 0.…
## 6 Vaccine Cardiac disorders Angina unstable " 1" 1 7753 ( 0.…Prepare reporting data for AE and make table:
np_ae<-bind_rows(t1.np, t2.np) %>%
pivot_wider(
id_cols = c(soc, order, pt),
names_from = treatment,
values_from = c(n,pct),
values_fill = fmt_num(0, digits = 0)
) %>%
arrange(soc, order, pt) %>%
mutate(Vaccine=paste0(n_Vaccine," ",pct_Vaccine),
Placebo=paste0(n_Placebo," ",pct_Placebo)) %>%
mutate(AE = case_when(
order==1 ~ paste0("- ",pt),
TRUE ~ pt)) %>%
select(AE,Placebo,Vaccine)
pop.r<-c("Participants in SAS",
pop$tot[pop$treatment %in% "Placebo"],
pop$tot[pop$treatment %in% "Vaccine"])
names(pop.r)<-names(np_ae)
np_ae_pop<-bind_rows(pop.r, np_ae)
# Show first 30 rows
kable(head(np_ae_pop,30),
caption = "Summary of adverse events by SOC & PT",
col.names = c("Adverse events SOC & PT",
"Placebo n (%)",
"Vaccine n (%)"))| Adverse events SOC & PT | Placebo n (%) | Vaccine n (%) |
|---|---|---|
| Participants in SAS | 7345 | 7753 |
| Blood and lymphatic system disorders | 5 ( 0.1) | 6 ( 0.1) |
| - Anaemia | 0 0 | 1 ( 0.0) |
| - Iron deficiency anaemia | 1 ( 0.0) | 0 0 |
| - Lymphadenitis | 2 ( 0.0) | 3 ( 0.0) |
| - Lymphadenopathy | 2 ( 0.0) | 2 ( 0.0) |
| Cardiac disorders | 411 ( 5.6) | 453 ( 5.8) |
| - Acute myocardial infarction | 2 ( 0.0) | 1 ( 0.0) |
| - Angina pectoris | 2 ( 0.0) | 1 ( 0.0) |
| - Angina unstable | 1 ( 0.0) | 1 ( 0.0) |
| - Arrhythmia | 2 ( 0.0) | 1 ( 0.0) |
| - Atrial fibrillation | 1 ( 0.0) | 0 0 |
| - Atrioventricular block complete | 0 0 | 1 ( 0.0) |
| - Bradycardia | 4 ( 0.1) | 6 ( 0.1) |
| - Chest discomfort | 11 ( 0.1) | 11 ( 0.1) |
| - Chest pain | 8 ( 0.1) | 14 ( 0.2) |
| - Dizziness | 19 ( 0.3) | 28 ( 0.4) |
| - Dyspnoea | 9 ( 0.1) | 4 ( 0.1) |
| - Heart valve incompetence | 0 0 | 1 ( 0.0) |
| - Mitral valve incompetence | 1 ( 0.0) | 0 0 |
| - Myocardial ischaemia | 3 ( 0.0) | 5 ( 0.1) |
| - Palpitations | 0 0 | 1 ( 0.0) |
| - Sinus arrhythmia | 0 0 | 1 ( 0.0) |
| - Syncope | 2 ( 0.0) | 1 ( 0.0) |
| - Tachycardia | 352 ( 4.8) | 382 ( 4.9) |
| Congenital, familial and genetic disorders | 1 ( 0.0) | 0 0 |
| - Atrial septal defect | 1 ( 0.0) | 0 0 |
| Ear and labyrinth disorders | 25 ( 0.3) | 43 ( 0.6) |
| - Ear infection | 1 ( 0.0) | 2 ( 0.0) |
| - Ear pain | 3 ( 0.0) | 4 ( 0.1) |
Above are some example codes for analysis and report of example clinical trial data. Ones may apply what have been learnt from previous chapters for further analysis and visualization.
10.8 Reference materials
10.8.2 Other sources
- Making sense of statistics in clinical trial report.
- Complete Analysis of a Crossover Trial
- Example Clinical Trial Paper 1: Lancet Infectious Diseases.
- Example Clinical Trial Paper 2: New England Journal of Medicine.
- Example Clinical Trial Paper 3: Lancet Global Health.
- Example Clinical Trial Paper 4: Nature Communications.