Chapter 6 Linear regression with R
Nhan Thi Ho, MD, PhD
6.1 Introduction to linear regression
Linear regression evaluates the relationship between a dependent variable (outcome) and one or more independent variables (predictors). From a sample data, linear regression is used to estimate population parameters characterizing these relationships and their uncertainties. Linear regression helps us estimate the effect of a predictor while controlling for other variables and helps predict outcomes for future observations.
6.2 Simple linear regression
6.2.1 Simple linear model with R
Simple linear regression quantifies the relationship between one outcome and one predictor. Below is an generated example data:
## x y
## 1 3.620757 0.8449674
## 2 3.035641 0.8304951
## 3 3.773154 0.9622945
## 4 4.272489 0.9096735
## 5 3.370975 0.8072324
## 6 2.837146 0.8259437In which:
- y is outcome or response or dependent variable (DV)
- x is predictor or regressor or independent variable (IV)
Simple linear regression assumes the linear relationship between x and y. The formula for simple linear regression model may be written as: \[y = \beta_0 + \beta_1 x + \epsilon\] In which:
- \(\beta_0\) is the intercept or the predicted \(y\) when \(x=0\)
- \(\beta_1\) is the slope or the predicted change in \(y\) for a one-unit increase in \(x\)
- \(\epsilon\) is the error term or the deviation of the predicted \(y\) value from the observed \(y\).
Simple linear model may be fitted in R as below (R Core Team 2024b):
The lm() returns complex objects which may be examined by:
coef(): model coefficientssummary(): coefficients, standard errors, p-values, model F-test, etc.predict(): model predictionsresiduals(): various kinds of residualsconfint(): confidence intervals for coefficientsplot(): diagnostic plots
The result output may be summarized as below:
##
## Call:
## lm(formula = y ~ x, data = df)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.25771 -0.07126 0.00101 0.05635 0.32485
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.525215 0.019149 27.43 <2e-16 ***
## x 0.111954 0.006056 18.48 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.1034 on 298 degrees of freedom
## Multiple R-squared: 0.5342, Adjusted R-squared: 0.5326
## F-statistic: 341.7 on 1 and 298 DF, p-value: < 2.2e-16The summary() outputs show:
- Model formula
- Summary of residual distribution
- Estimated coefficients, standard errors, t-values, and p-values (for testing a hypothesis that a coefficient equals to zero).
- A quick visual shortcut to p-values by
* - Residual standard error measuring the average amount that observed responses deviate from the fitted regression line.
- Degrees of freedom (\(df\)) represent the number of independent observations (\(N\)) in the data that are available to estimate a parameter. For linear regression, \(p =\) the number of predictors \(+ 1\) and \(df = N - p\) .
- R-square (\(R^2\)), a measure to test if the model is a good fit, shows the amount of variance of y explained by x. \[R^2 = Explained \ variation \ of \ the \ model \ / \ Total \ variation \ of \ the \ model\] . If the model fit the data well, \(R^2\) is near 1 and if the model fit the data poorly, \(R^2\) is near 0. \(Adjusted \ R^2\) shows the same as \(R^2\) but adjusted by the number of cases and number of variables. When the number of variables is small and the number of outcomes is very large then \(Adjusted \ R^2\) is closer to \(R^2\).
- \(F\)-statistic: indicates whether the overall model is statistically significant (at least one predictor is related to the outcome)
The summary output above show that x is positively and significantly associated with y. One unit increase in x is associated with 0.1119536 increase of y.
Confidence intervals may be computed with confint():
## 2.5 % 97.5 %
## (Intercept) 0.4875300 0.5628993
## x 0.1000348 0.1238724The intercept, the slope and residuals are visualized as below:
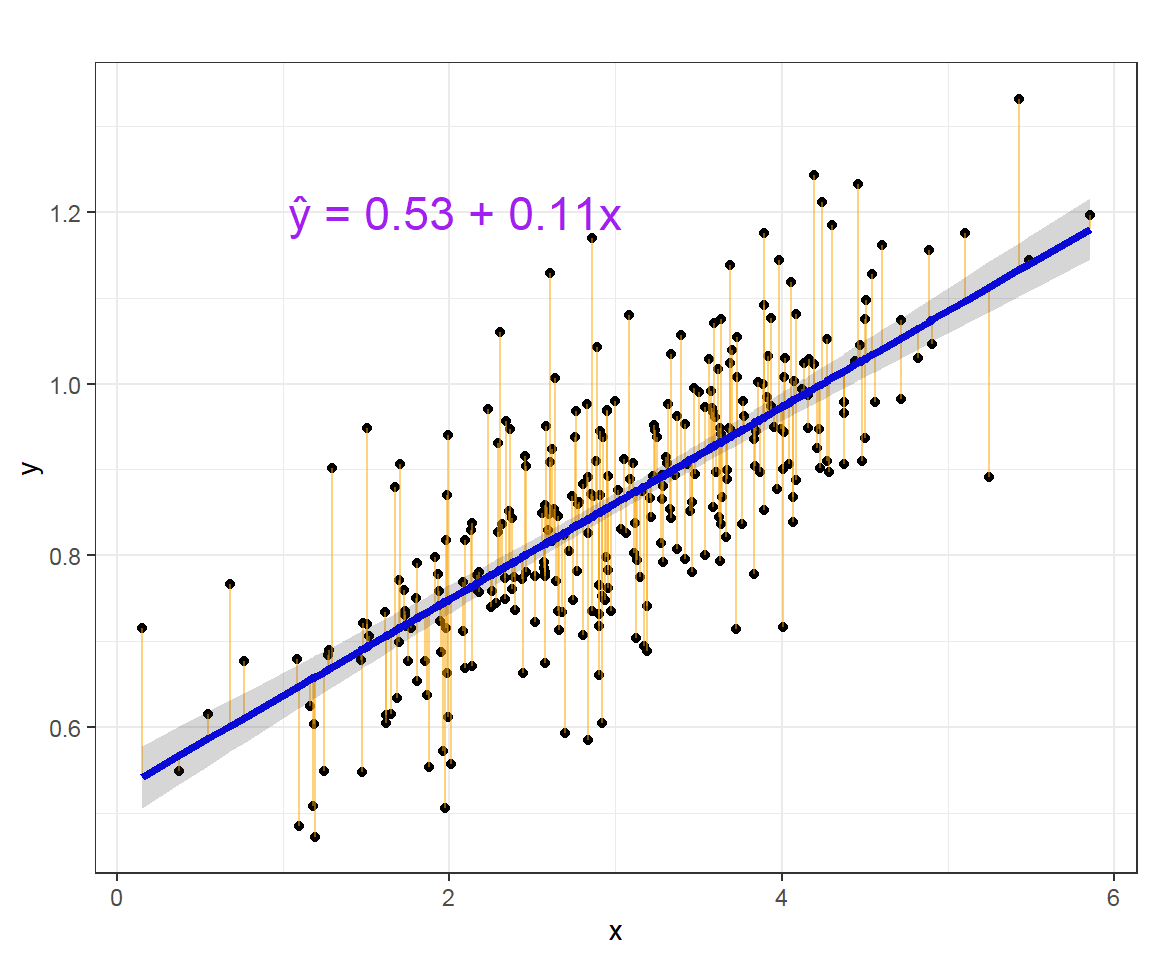
The straight blue line in the image above represents the predicted values. The model fit may be assessed by plotting the predicted values to evaluate if they look reasonable against the observed data.The shaded area represent the confidence interval of predicted values.
The orange vertical line from the straight line to the observed data value is the residual. For a good model, the sum of the residuals should be approximately zero or as low as possible and the residual plot should look random. If there is pattern in the residual plot, the simple linear model may not fit the data well and there may be a need to transform the variable (e.g. adding quadratic, or cubic term,…). The residuals may plotted in R as below:
6.2.2 Linear model assumptions
Below are assumptions for inferences made from a regression model.
- L: Linear relationship between the mean outcome (Y) and the predictor variable (X)
- I: The errors are independent
- N: The outcomes are normally distributed at each level of predictor variable (X)
- E: The variance of the outcomes is equal for all levels of predictor variable (X)
If the assumptions are violated, the model estimates and inferences may not be valid.
6.2.3 Checking linear model assumptions
6.2.3.1 Linear assumption
If linear assumption is met, the residuals should be random and there should be no systematic trend or pattern in residual plot.
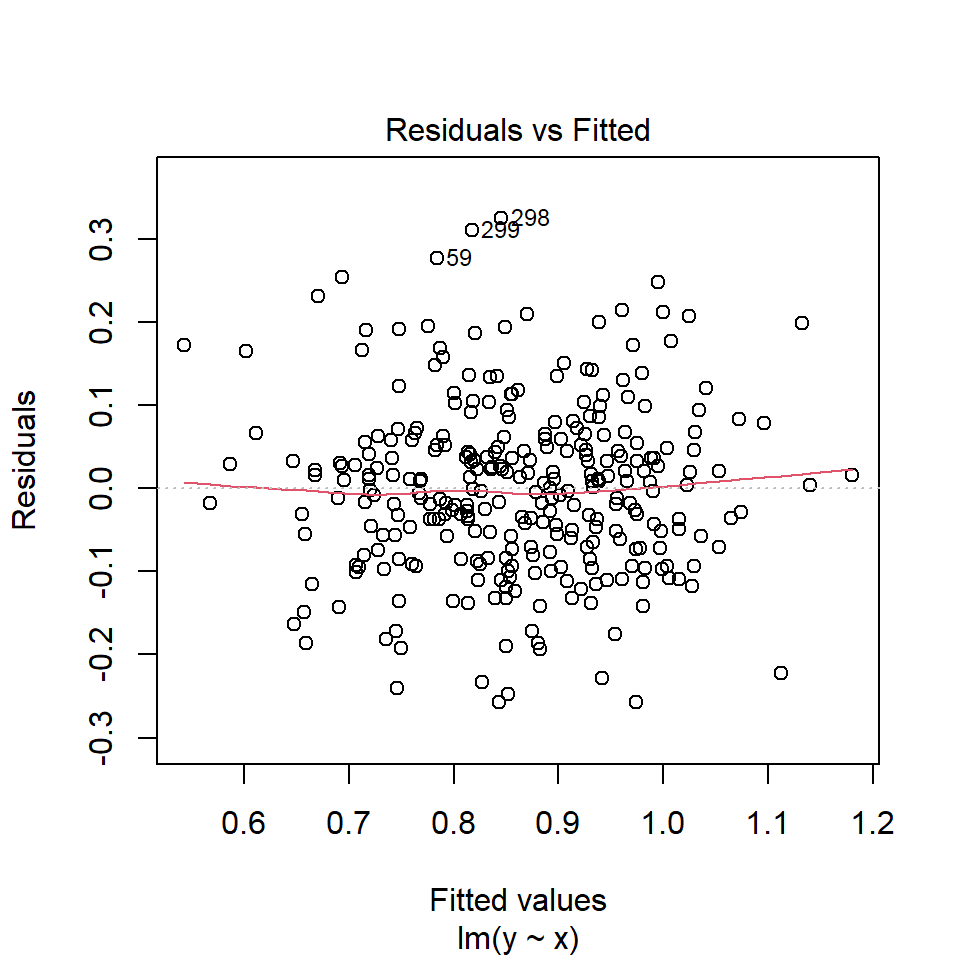
Diagnostic plots may be done by plot() on the lm object. The first plot is the residuals vs fitted values plot.
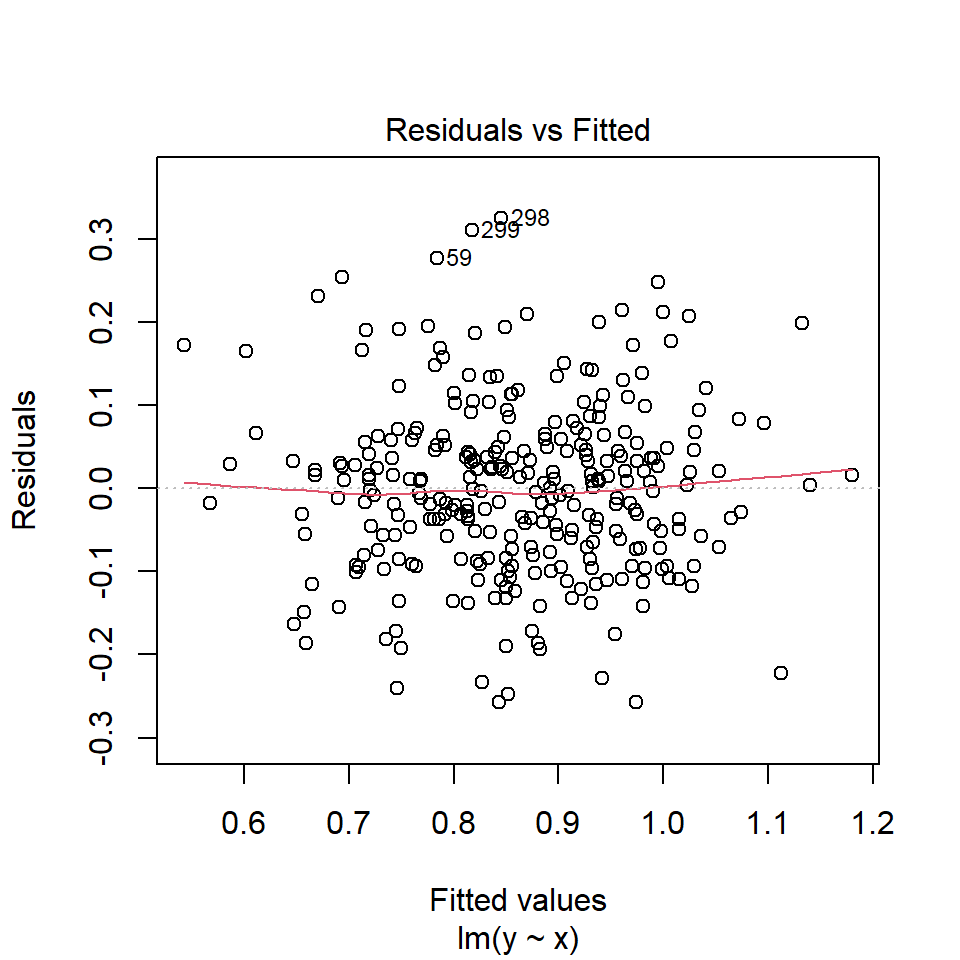
In our example, there is no trend in residual plot.
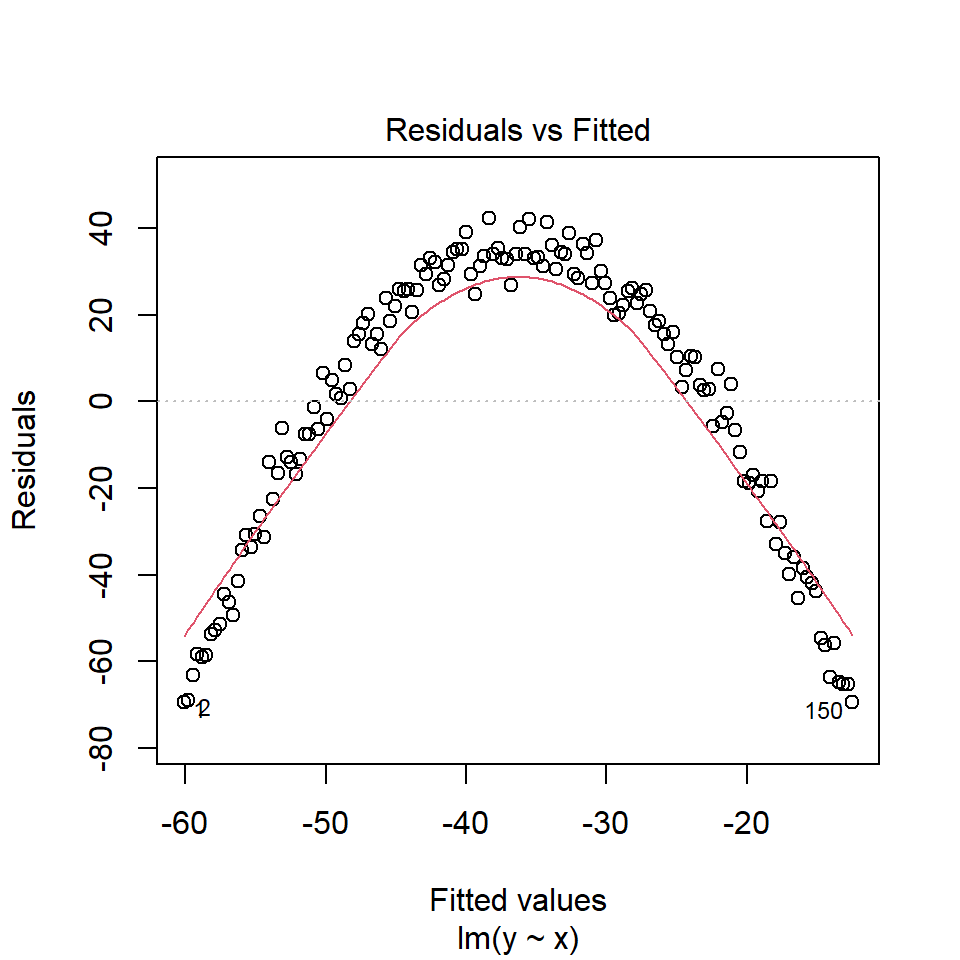
If there is a trend in the residuals, it suggests a nonlinear relationship between predictor and outcome variables. In this case, one may consider adding non-linear term to the model (e.g. cubic, quadratic, splines,…) or using non-linear transformations (e.g. logarithm) to either predictor or outcome variables.
Below is an example plot showing a clear trend of residuals:
6.2.3.2 Equal variance assumption (Homoscedasticity)
If this assumption is violated, standard errors will be inaccurate resulting in inaccurate test statistics, confidence intervals and p-values. For this assumption, the diagnostic plot of residuals vs. fitted values should show random scatter around the zero line with equal spread across the range of fitted values as in the residual plot of our example.
Unequal variance or heteroscedasticity is often observed in the form of increasing variance of the residuals with increasing fitted values:
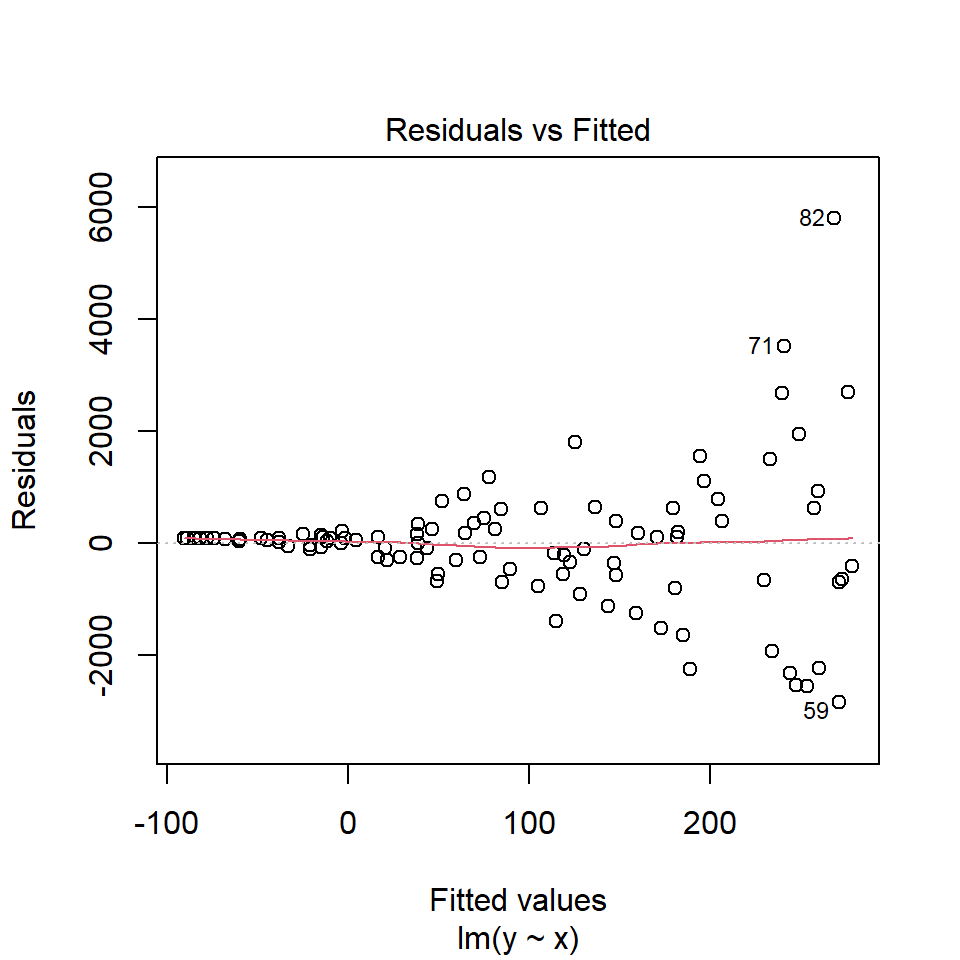
If equal variance assumption is violated, one may consider:
- Transformation of variables (e.g., log transformation of the outcome variable)
- Using robust standard errors (heteroscedastic-consistent or “sandwich”) (may be done with R package “sandwich”)
- Using a mixed model for heteroscedasticity modeling (may be done with R package “nlme”)
6.2.3.3 Independence assumption
If this assumption is violated, it may affect the accuracy of confidence intervals and p-values.
Common causes of data dependency are:
- Sampling in clusters
- Repeated measurements of the same subjects
The approaches to handle this assumption violation are:
- Model data dependency with mixed/multilevel models, fixed effect models, … (may be done with R package “lme4” )
- Using cluster robust standard errors
6.2.3.4 Normal distribution assumption
If the errors are not normally distributed, the accuracy of confidence interval and p-values may be affected, especially in case of small sample size (the impact is less with large sample size).
Histogram and q-q plot of residuals may be used as diagnostic plots:
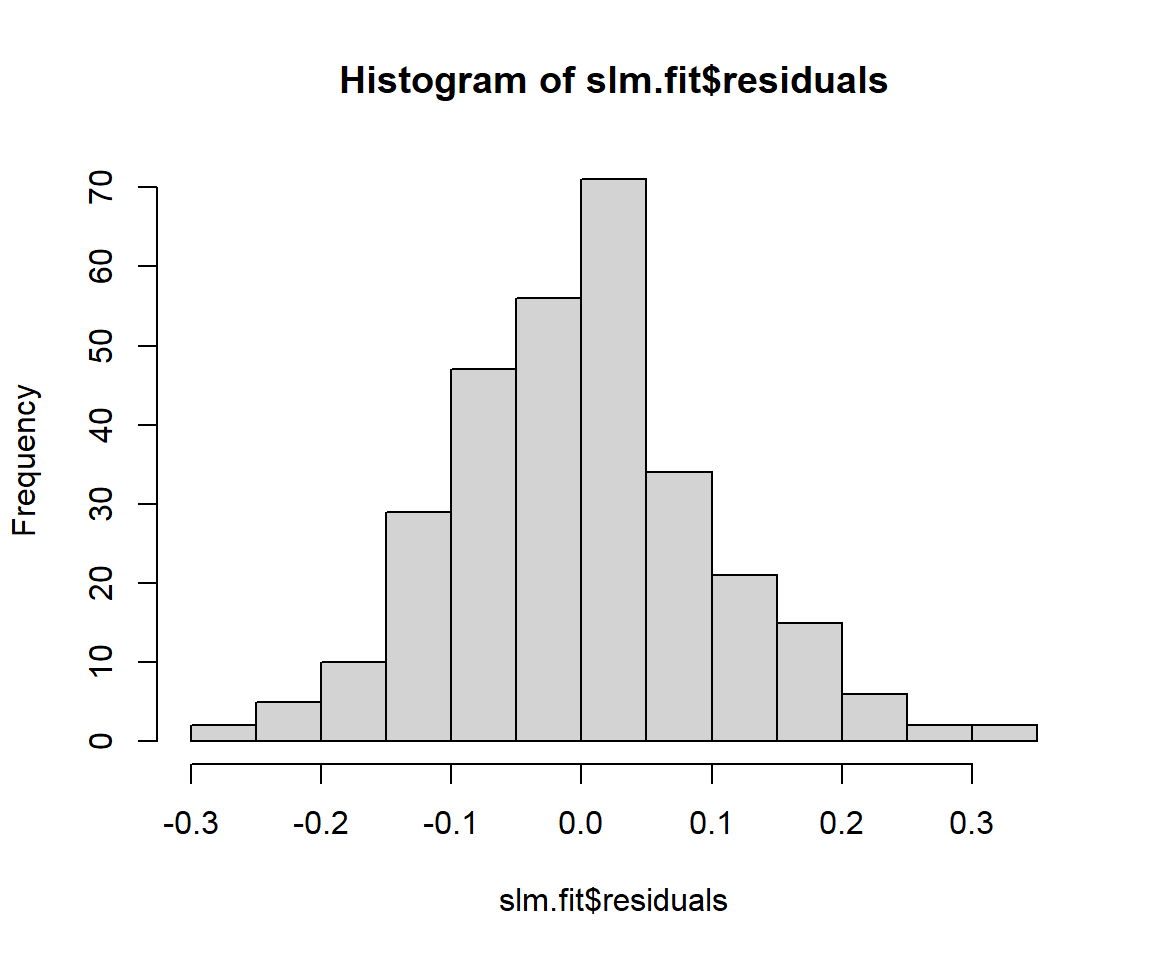
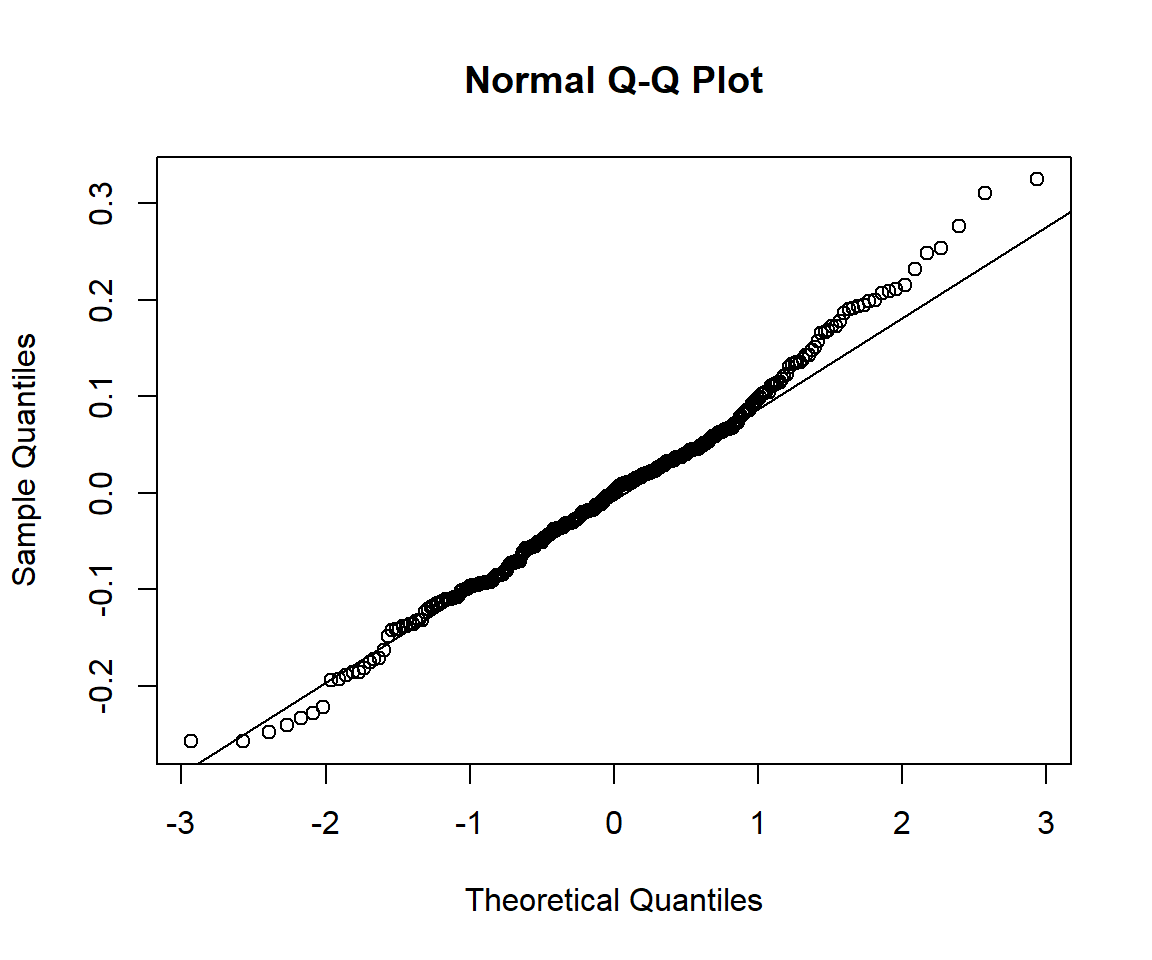
If the errors are not normally distributed, the q-q plot will show deviation from the diagonal line. The approaches to handle this issue may be:
- Transformation (e.g. logarithm,…) of the outcome variable
- Using using generalized linear model (may be done with
glm()function)
6.2.4 Checking influential observations
Influential observations change model estimates substantially when included vs when excluded from the data. Model coefficients change when a particular observation is excluded may be evaluated by influence diagnostic measures:
- dfbeta measures how a specific coefficient changes when deleting an observation
- Cook’s distance measures how much all coefficients change when deleting an observation
Influence diagnostic measures may be done with influence.measures() on an lm object, which results in several influence measures including dfbetas for each coefficient and Cook’s distance.
If there are any unusually large influence measures, it will be flagged by an * in the infl column.
6.2.5 Categorical predictor variables
Categorical variables with k (>=2) categories will have k-1 coefficient estimates. The simplest categorical variable is dummy variable with 2 categories 0 and 1 denoting not belonging or belonging to a category. An example of this dummy variable may be smoker with category “no” (equivalent to 1) and “yes” (equivalent to 1). Categorical variables with k categories is treated as k-1 dummy variables with the omitted category treated as reference category.
Categorical variables in R are represented by factor variables and the factor order and reference group may be selected. When performing lm(), R will automatically create dummy variables for each level of factor except for the first level (which is used as the reference group).
6.2.5.1 Example simple linear regression with categorical predictor variable
We will use example data regarding the association of medical costs and patient information. We first load the data and summarize all variables with the “arsenal” package (Heinzen et al. 2021).
# Load example linear regression data
dlm<-read.csv("https://raw.githubusercontent.com/stedy/Machine-Learning-with-R-datasets/master/insurance.csv")
colnames(dlm)<-tolower(colnames(dlm))
str(dlm)## 'data.frame': 1338 obs. of 7 variables:
## $ age : int 19 18 28 33 32 31 46 37 37 60 ...
## $ sex : chr "female" "male" "male" "male" ...
## $ bmi : num 27.9 33.8 33 22.7 28.9 ...
## $ children: int 0 1 3 0 0 0 1 3 2 0 ...
## $ smoker : chr "yes" "no" "no" "no" ...
## $ region : chr "southwest" "southeast" "southeast" "northwest" ...
## $ charges : num 16885 1726 4449 21984 3867 ...#turn character variables into factor variables
dlm[,colnames(dplyr::select_if(dlm, is.character))] <- lapply(dlm[,colnames(dplyr::select_if(dlm, is.character))], as.factor)
#summary of all variables in the dataset
my_controls <- tableby.control(
numeric.stats = c("meansd", "medianq1q3","meanCI","range","Nmiss2"),
cat.stats = c("countpct", "Nmiss2"),
stats.labels = list(
meansd = "Mean (SD)",
medianq1q3 = "Median (Q1, Q3)",
range = "Min - Max",
Nmiss2 = "Missing",
meanCI = "Mean (95%CI)"
)
)
mylabels <-as.list(colnames(dlm))
names(mylabels)<-colnames(dlm)
tabs <- tableby(as.formula(paste("",paste(colnames(dlm),collapse="+"),sep="~")),
data = dlm,
control = my_controls)
kable(summary(tabs,labelTranslations = mylabels, text=TRUE))| Overall (N=1338) | |
|---|---|
| age | |
| - Mean (SD) | 39.207 (14.050) |
| - Median (Q1, Q3) | 39.000 (27.000, 51.000) |
| - Mean (95%CI) | 39.207 (38.454, 39.961) |
| - Min - Max | 18.000 - 64.000 |
| - Missing | 0 |
| sex | |
| - female | 662 (49.5%) |
| - male | 676 (50.5%) |
| - Missing | 0 |
| bmi | |
| - Mean (SD) | 30.663 (6.098) |
| - Median (Q1, Q3) | 30.400 (26.296, 34.694) |
| - Mean (95%CI) | 30.663 (30.336, 30.990) |
| - Min - Max | 15.960 - 53.130 |
| - Missing | 0 |
| children | |
| - Mean (SD) | 1.095 (1.205) |
| - Median (Q1, Q3) | 1.000 (0.000, 2.000) |
| - Mean (95%CI) | 1.095 (1.030, 1.160) |
| - Min - Max | 0.000 - 5.000 |
| - Missing | 0 |
| smoker | |
| - no | 1064 (79.5%) |
| - yes | 274 (20.5%) |
| - Missing | 0 |
| region | |
| - northeast | 324 (24.2%) |
| - northwest | 325 (24.3%) |
| - southeast | 364 (27.2%) |
| - southwest | 325 (24.3%) |
| - Missing | 0 |
| charges | |
| - Mean (SD) | 13270.422 (12110.011) |
| - Median (Q1, Q3) | 9382.033 (4740.287, 16639.913) |
| - Mean (95%CI) | 13270.422 (12620.954, 13919.890) |
| - Min - Max | 1121.874 - 63770.428 |
| - Missing | 0 |
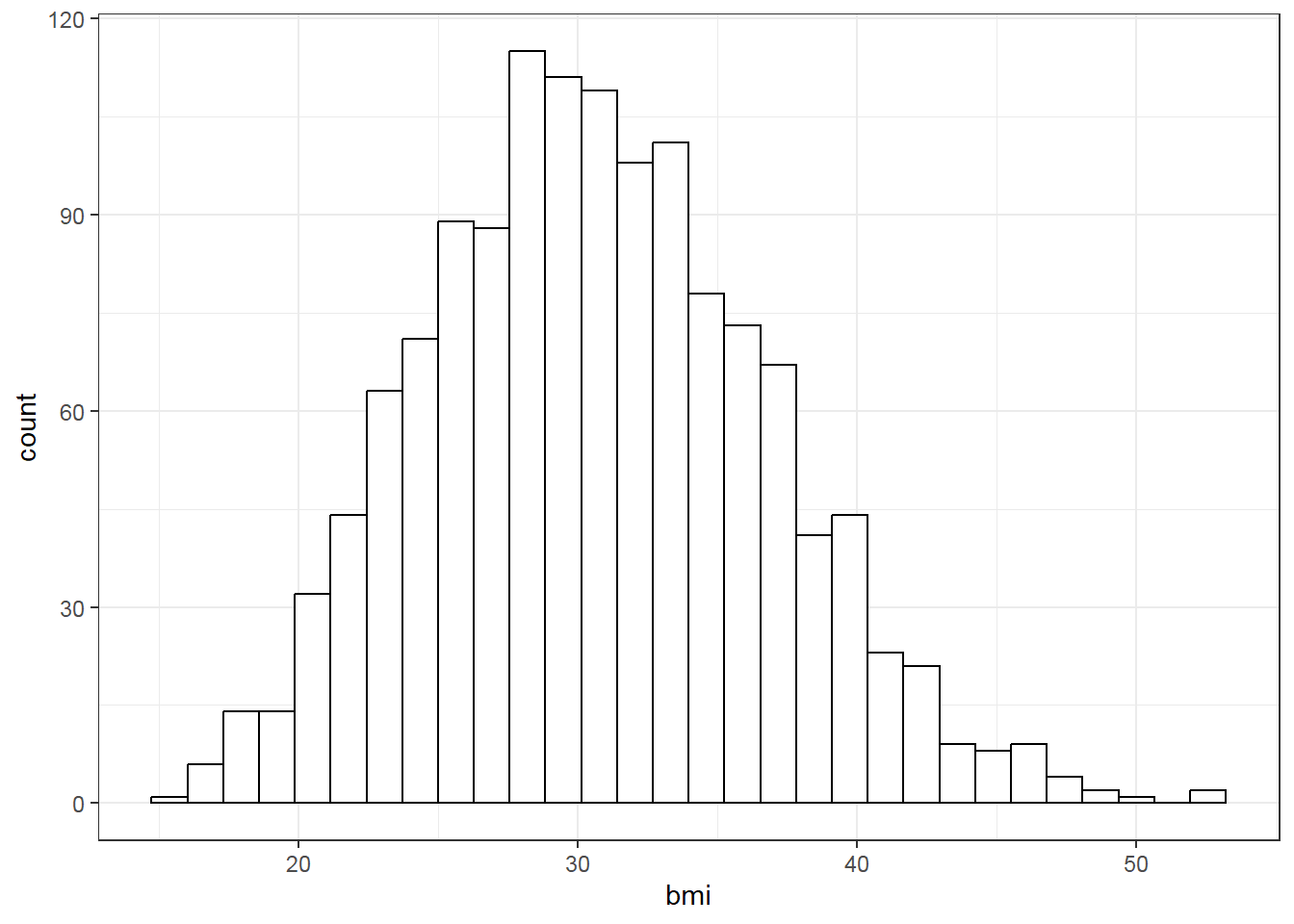
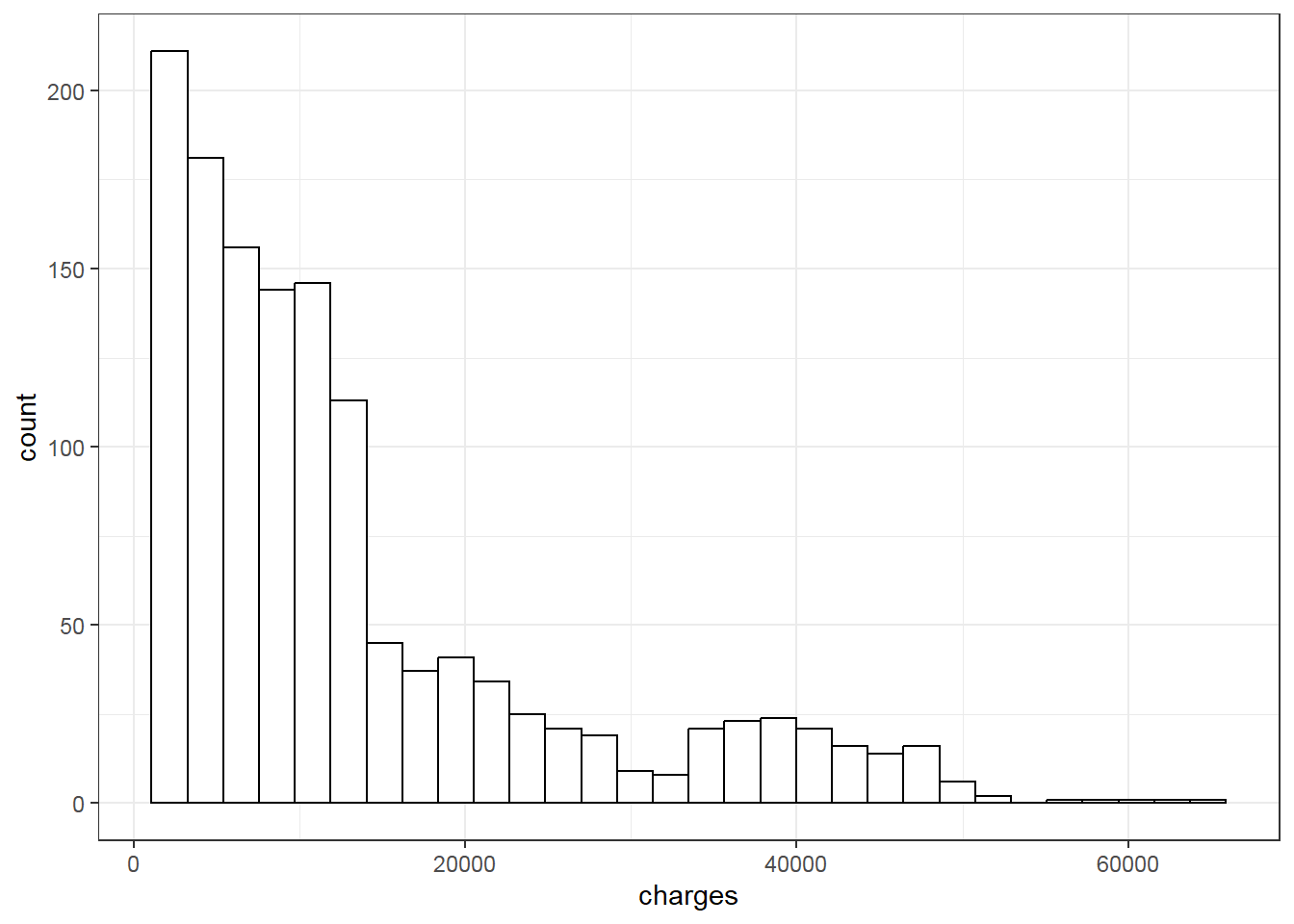
We then examine data distribution with histogram:
# Data distribution plot
vars<-colnames(dplyr::select_if(dlm, is.numeric))
for (i in 1:length(vars)){
print(ggplot(data=dlm, aes(x=dlm[,vars[i]])) +
geom_histogram(colour="black", fill="white") +
theme_bw()+
labs(x=vars[i]))}
We fit an example simple linear model with charges as outcome and smoker as categorical predictor.
##
## Call:
## lm(formula = charges ~ smoker, data = dlm)
##
## Residuals:
## Min 1Q Median 3Q Max
## -19221 -5042 -919 3705 31720
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 8434.3 229.0 36.83 <2e-16 ***
## smokeryes 23616.0 506.1 46.66 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 7470 on 1336 degrees of freedom
## Multiple R-squared: 0.6198, Adjusted R-squared: 0.6195
## F-statistic: 2178 on 1 and 1336 DF, p-value: < 2.2e-16# Add confidence interval to summary
ciflm <- confint(flm)
sumciflm <- cbind(sumflm$coefficients, ciflm)
round(sumciflm, 4)## Estimate Std. Error t value Pr(>|t|) 2.5 % 97.5 %
## (Intercept) 8434.268 229.0142 36.8286 0 7985.002 8883.535
## smokeryes 23615.964 506.0753 46.6649 0 22623.175 24608.752From the output, smoker= no is expected to have charges of 8434.2682979 and smoker = yes is expected to have charges of 2.3615964^{4} higher than smoker= no.
Predicted charges for smokers and non-smokers may be calculated and visualized as below.
predictions <- predict(flm, newdata=dlm, interval="confidence")
new_data <- cbind(dlm, as.data.frame(predictions))
#plot
ggplot(dlm, aes(smoker, charges)) +
geom_point(color="grey32",size=0.75) +
geom_errorbar(data = new_data, aes(x=smoker, ymin=lwr, ymax=upr, color=smoker),
width=0.1) +
geom_point(data = new_data, aes(smoker, fit, color=smoker), size = 3) +
labs(x = "smoker", y = "charges") +
theme_bw()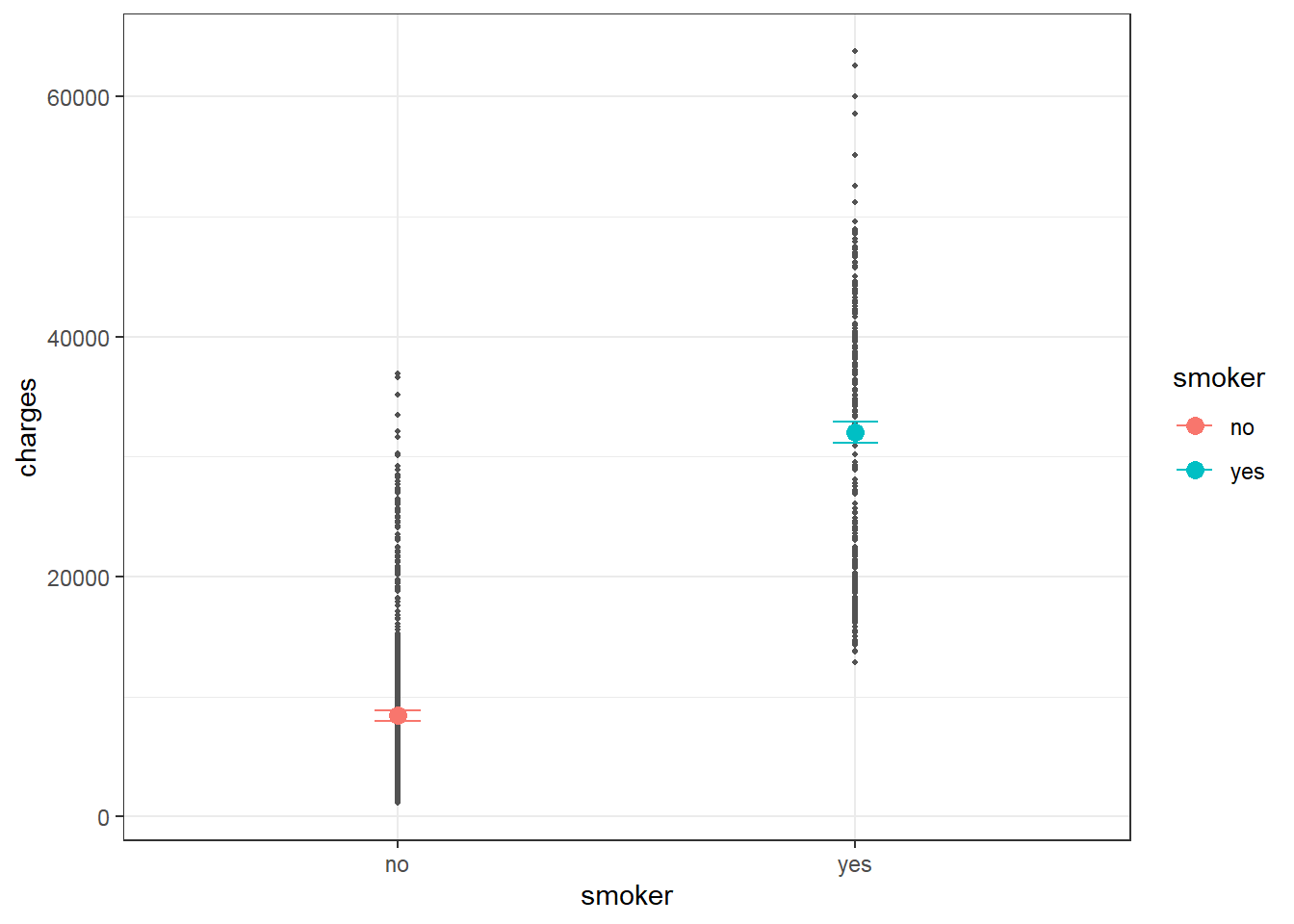
The plot shows clear difference in predicted charges between non-smoker and smoker corresponding to significant coefficient of smoker in the simple linear model.
We fit another example simple linear model with charges as outcome and region as categorical predictor (region has 4 categories).
##
## Call:
## lm(formula = charges ~ region, data = dlm)
##
## Residuals:
## Min 1Q Median 3Q Max
## -13614 -8463 -3793 3385 49035
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 13406.4 671.3 19.971 <2e-16 ***
## regionnorthwest -988.8 948.6 -1.042 0.297
## regionsoutheast 1329.0 922.9 1.440 0.150
## regionsouthwest -1059.4 948.6 -1.117 0.264
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 12080 on 1334 degrees of freedom
## Multiple R-squared: 0.006634, Adjusted R-squared: 0.0044
## F-statistic: 2.97 on 3 and 1334 DF, p-value: 0.03089# Add confidence interval to summary
ciflm.r <- confint(flm.r)
sumciflm.r <- cbind(sumflm.r$coefficients, ciflm.r )
round(sumciflm.r, 4)## Estimate Std. Error t value Pr(>|t|) 2.5 % 97.5 %
## (Intercept) 13406.3845 671.2966 19.9709 0.0000 12089.4724 14723.2966
## regionnorthwest -988.8091 948.6263 -1.0424 0.2974 -2849.7709 872.1526
## regionsoutheast 1329.0269 922.9071 1.4400 0.1501 -481.4805 3139.5343
## regionsouthwest -1059.4471 948.6263 -1.1168 0.2643 -2920.4089 801.5146region = northest is used as reference group and other region categories are turned into 3 dummy variables and are compared with the reference group northest.
From the output, region northest is predicted to have charges of 1.3406385^{4}. Region northwest is predicted to have charges of 988.8091424 lower than region northest. Region southeast is predicted to have charges of 1329.0269212 higher than region northest, and so on. However, the difference between region categories are not significant.
Predicted charges for region may be calculated and visualized as below.
predictions <- predict(flm.r, newdata=dlm, interval="confidence")
new_data <- cbind(dlm, as.data.frame(predictions))
#plot
ggplot(new_data, aes(region, charges)) +
geom_point(color="grey32",size=0.75) +
geom_errorbar(data = new_data, aes(x=region, ymin=lwr, ymax=upr, color=region),
width=0.1) +
geom_point(data = new_data, aes(region, fit, color=region), size = 3) +
labs(x = "region", y = "charges") +
theme_bw()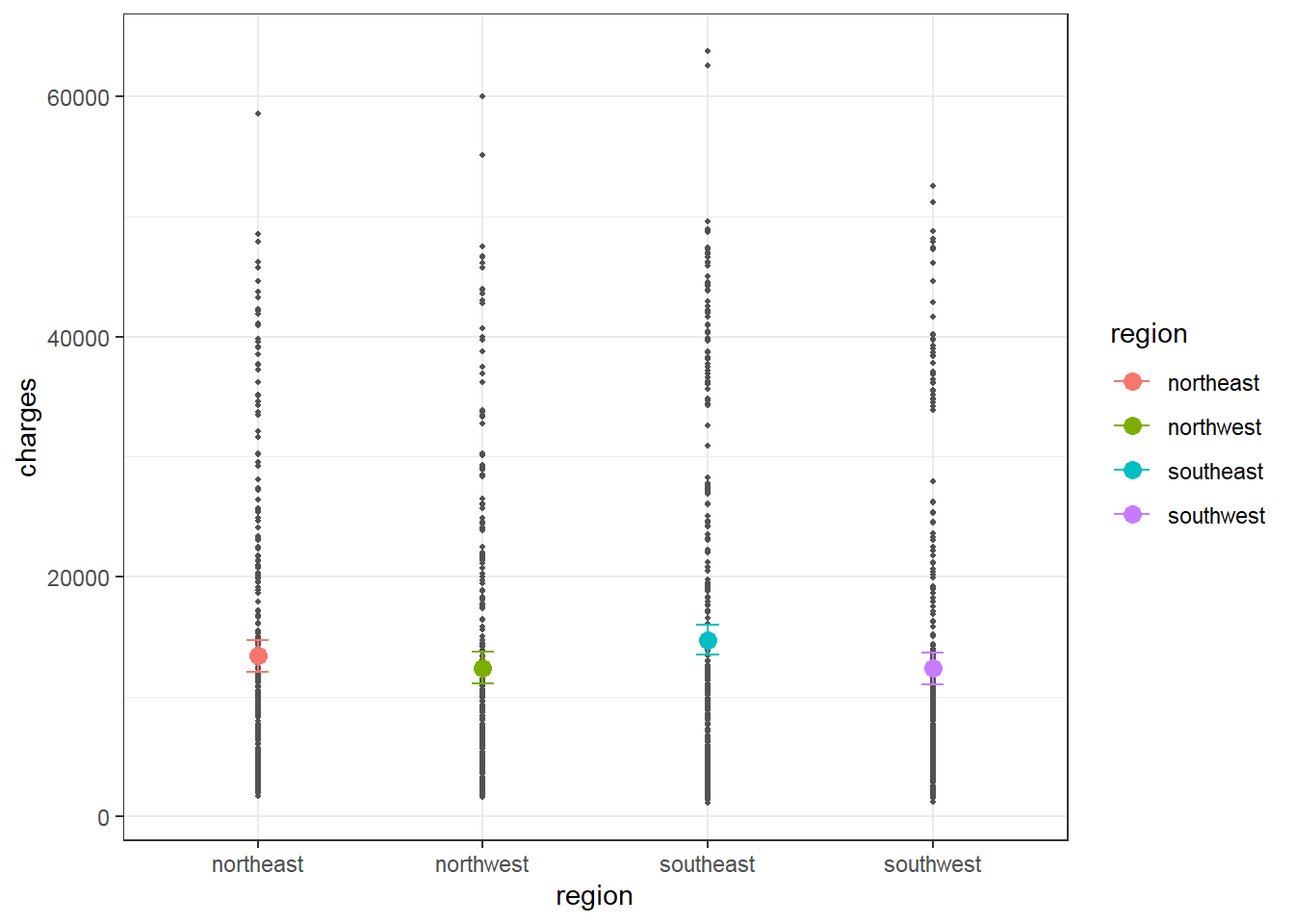
The plot shows similar predicted charges between region categories, which is corresponding to non-significant coefficients of region categories vs. reference category northeast.
Diagnostic plots may be done with residual vs. fitted plot for homoscedasticity and q-q plot for normality of residuals:
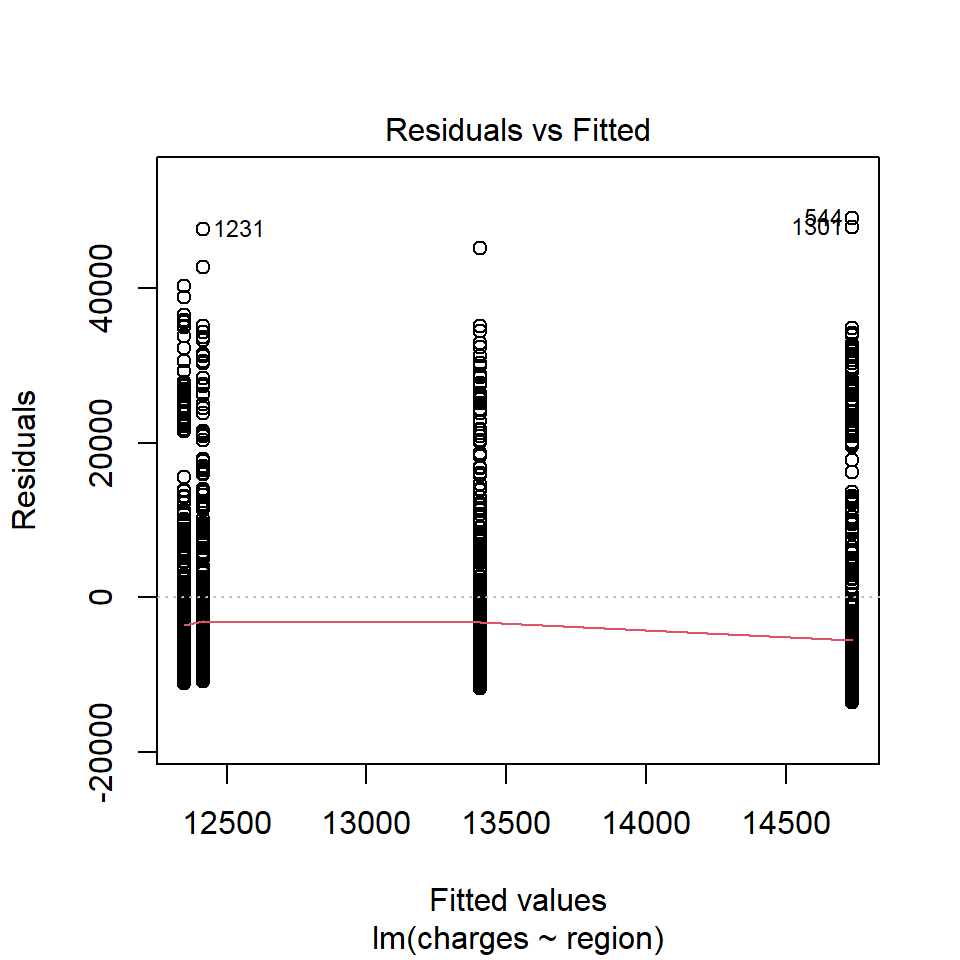
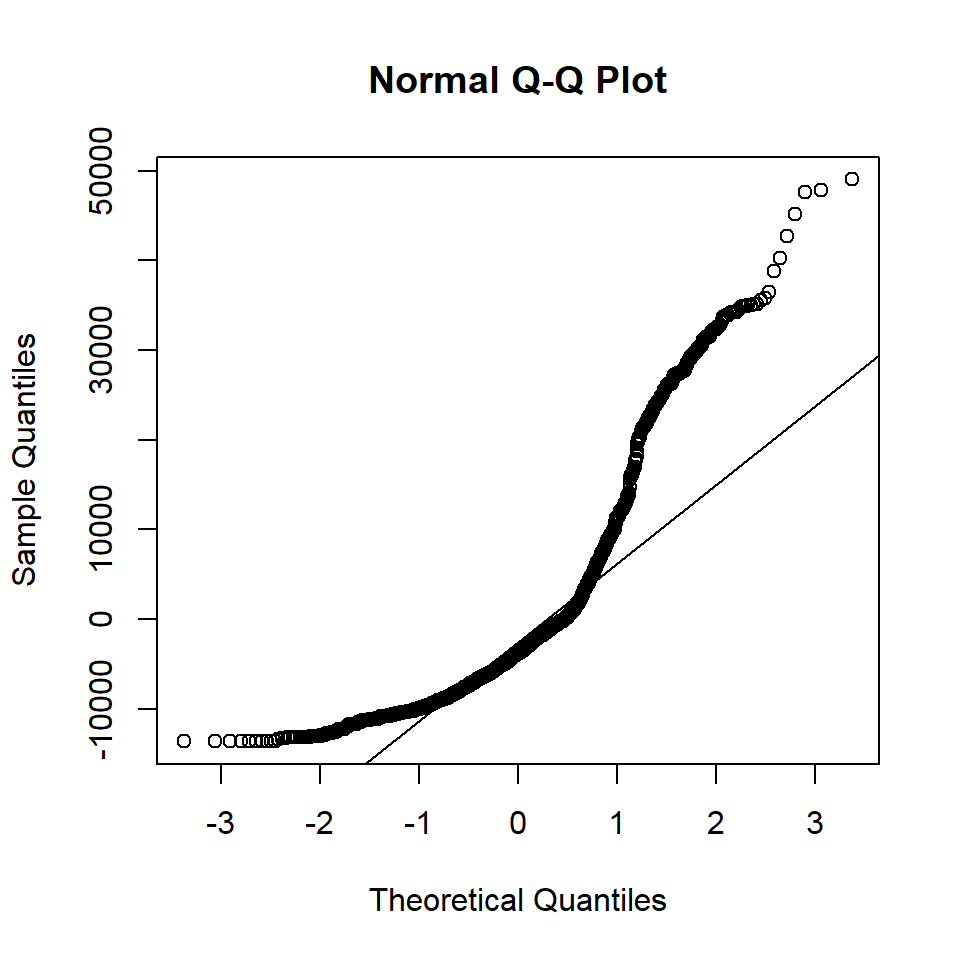
With the above plot, normality assumption seems not met. The histogram of charges also shows skewed distribution. Therefore, one may try logarithm transformation of the outcome charges, refit linear model and re-perform diagnostic plots.
dlm$logcharges<-log(dlm$charges)
flm.lr<-lm(logcharges ~ region, data=dlm)
sumflm.lr<-summary(flm.lr)
sumflm.lr##
## Call:
## lm(formula = logcharges ~ region, data = dlm)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.09965 -0.64192 0.03622 0.62333 1.94064
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 9.16877 0.05106 179.562 <2e-16 ***
## regionnorthwest -0.09903 0.07216 -1.372 0.1701
## regionsoutheast -0.04637 0.07020 -0.660 0.5091
## regionsouthwest -0.13767 0.07216 -1.908 0.0566 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.9191 on 1334 degrees of freedom
## Multiple R-squared: 0.003143, Adjusted R-squared: 0.0009012
## F-statistic: 1.402 on 3 and 1334 DF, p-value: 0.2406# Add confidence interval to summary
ciflm.lr <- confint(flm.lr)
sumciflm.lr <- cbind(sumflm.lr$coefficients, ciflm.lr)
round(sumciflm.lr, 4)## Estimate Std. Error t value Pr(>|t|) 2.5 % 97.5 %
## (Intercept) 9.1688 0.0511 179.5621 0.0000 9.0686 9.2689
## regionnorthwest -0.0990 0.0722 -1.3725 0.1701 -0.2406 0.0425
## regionsoutheast -0.0464 0.0702 -0.6605 0.5091 -0.1841 0.0913
## regionsouthwest -0.1377 0.0722 -1.9079 0.0566 -0.2792 0.0039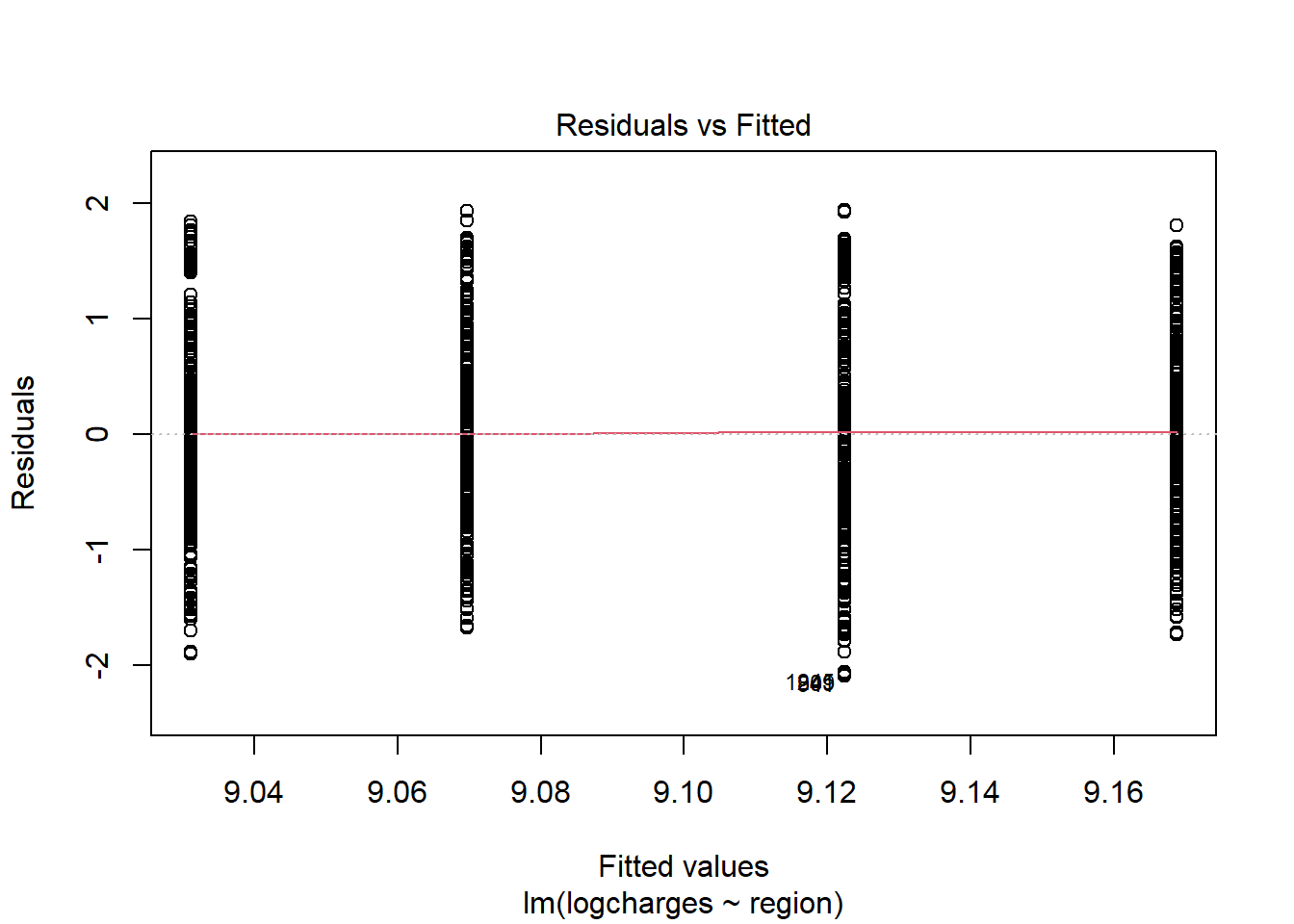
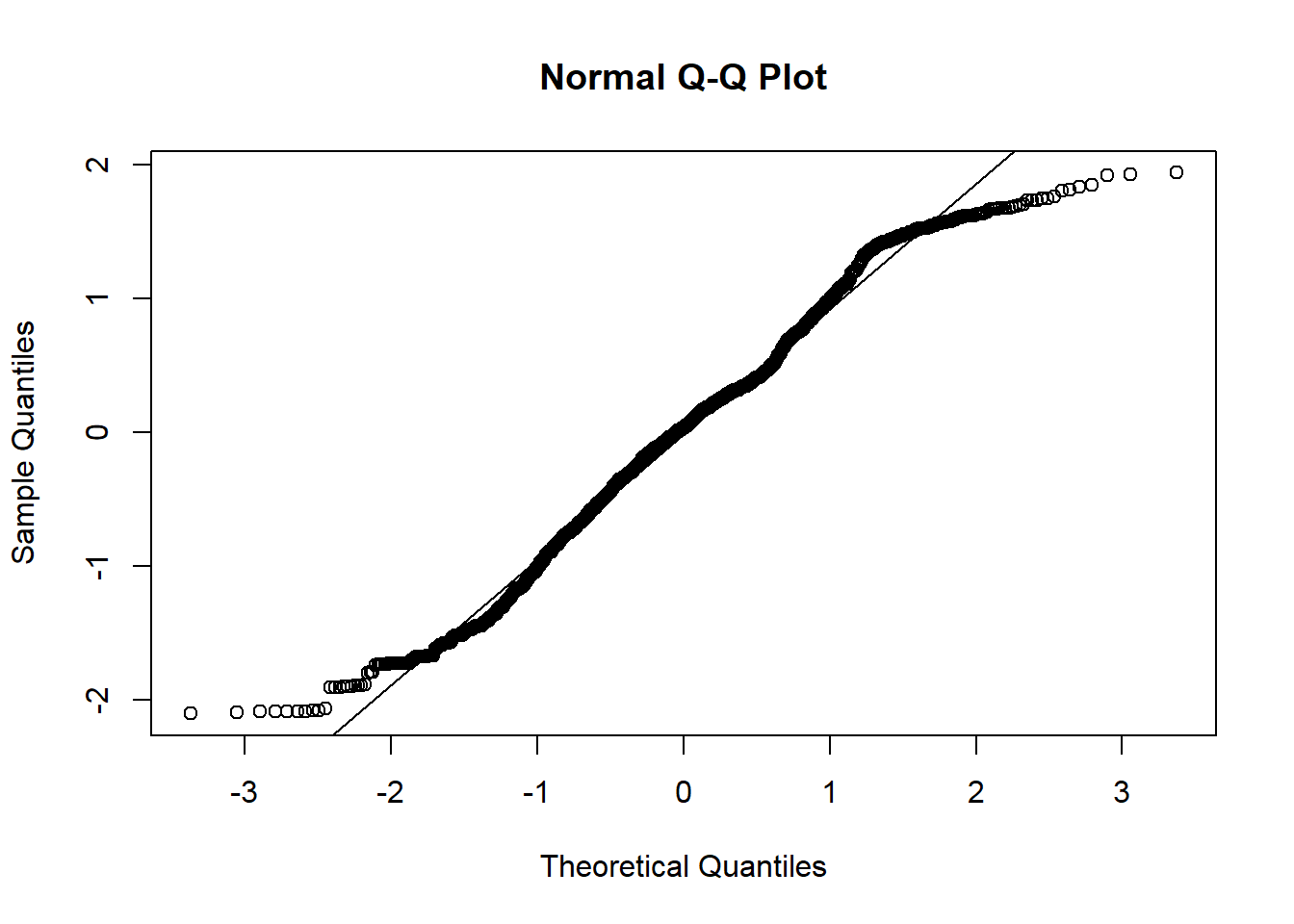
The diagnostic plots look better after logarithm transformation of the outcome charges.
6.3 Multiple linear regression
Multiple linear regression allows the estimation of the effects of multiple predictor variables simultaneously on outcome variable.
For the example medical costs and patient information data above, multiple linear regression may be done by adding several variables as predictors in the linear model, for example bmi and smoker as predictors and logarithm of charges (logcharges) as outcome.
# Simple linear regression model with only bmi as predictor and logarithm charges as outcome
flm.b<-lm(logcharges ~ bmi, data=dlm)
sumflm.b<-summary(flm.b)
sumflm.b##
## Call:
## lm(formula = logcharges ~ bmi, data = dlm)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.48894 -0.63536 0.03136 0.68007 1.95182
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 8.485243 0.127833 66.378 < 2e-16 ***
## bmi 0.020005 0.004089 4.892 1.12e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.9117 on 1336 degrees of freedom
## Multiple R-squared: 0.0176, Adjusted R-squared: 0.01687
## F-statistic: 23.94 on 1 and 1336 DF, p-value: 1.117e-06# Add confidence interval to summary
ciflm.b <- confint(flm.b)
sumciflm.b <- cbind(sumflm.b$coefficients, ciflm.b)
round(sumciflm.b, 4)## Estimate Std. Error t value Pr(>|t|) 2.5 % 97.5 %
## (Intercept) 8.4852 0.1278 66.3778 0 8.2345 8.736
## bmi 0.0200 0.0041 4.8925 0 0.0120 0.028# Simple linear regression model with only smoker as predictor and logarithm charges as outcome
flm.s<-lm(logcharges ~ smoker, data=dlm)
sumflm.s<-summary(flm.s)
sumflm.s##
## Call:
## lm(formula = logcharges ~ smoker, data = dlm)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.7655 -0.4370 0.1237 0.4785 1.7280
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 8.78823 0.02105 417.52 <2e-16 ***
## smokeryes 1.51588 0.04651 32.59 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.6866 on 1336 degrees of freedom
## Multiple R-squared: 0.4429, Adjusted R-squared: 0.4425
## F-statistic: 1062 on 1 and 1336 DF, p-value: < 2.2e-16# Add confidence interval to summary
ciflm.s <- confint(flm.s)
sumciflm.s <- cbind(sumflm.s$coefficients, ciflm.s)
round(sumciflm.s, 4)## Estimate Std. Error t value Pr(>|t|) 2.5 % 97.5 %
## (Intercept) 8.7882 0.0210 417.5206 0 8.7469 8.8295
## smokeryes 1.5159 0.0465 32.5902 0 1.4246 1.6071# Multiple linear regression model with smoker and bmi as predictors and logarithm charges as outcome
flm.sb<-lm(logcharges ~ smoker + bmi, data=dlm)
sumflm.sb<-summary(flm.sb)
sumflm.sb##
## Call:
## lm(formula = logcharges ~ smoker + bmi, data = dlm)
##
## Residuals:
## Min 1Q Median 3Q Max
## -2.17030 -0.40754 0.09871 0.44368 1.75504
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 8.186576 0.095254 85.945 < 2e-16 ***
## smokeryes 1.514765 0.045818 33.061 < 2e-16 ***
## bmi 0.019629 0.003033 6.472 1.36e-10 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.6763 on 1335 degrees of freedom
## Multiple R-squared: 0.4598, Adjusted R-squared: 0.459
## F-statistic: 568.3 on 2 and 1335 DF, p-value: < 2.2e-16# Add confidence interval to summary
ciflm.sb <- confint(flm.sb)
sumciflm.sb <- cbind(sumflm.sb$coefficients, ciflm.sb)
round(sumciflm.sb, 4)## Estimate Std. Error t value Pr(>|t|) 2.5 % 97.5 %
## (Intercept) 8.1866 0.0953 85.9449 0 7.9997 8.3734
## smokeryes 1.5148 0.0458 33.0606 0 1.4249 1.6046
## bmi 0.0196 0.0030 6.4715 0 0.0137 0.0256From the output of multiple linear regression, the coefficients change slightly for both smoker and bmi as compare to those of the simple linear regression models when one of these predictors was used alone.
Interpretation of coefficients:
logchargeswhensmoker= no andbmi= 0 is 8.1865764- After controlling for
bmi,smoker=yes is expected to havelogchargesof 1.514765 higher thansmoker=no. - After controlling for
smoker, each unit increase inbmiis associated with an increase of 0.0196287 inlogcharges.
Observed and predicted values with 95% confidence intevals of the above multiple regression model may be visualized as below:
predictions <- predict(flm.sb, newdata=dlm, interval="confidence")
new_data <- cbind(dlm, as.data.frame(predictions))
#plot
ggplot(new_data, aes(bmi, logcharges, color=smoker)) +
geom_point() +
geom_ribbon(data = new_data, aes(x = bmi, ymin = lwr, ymax = upr, fill=smoker), alpha = 0.5) +
geom_line(data = new_data, aes(bmi, fit, color=smoker), linewidth = 1) +
labs(x = "BMI", y = "log(charges)") +
theme_bw()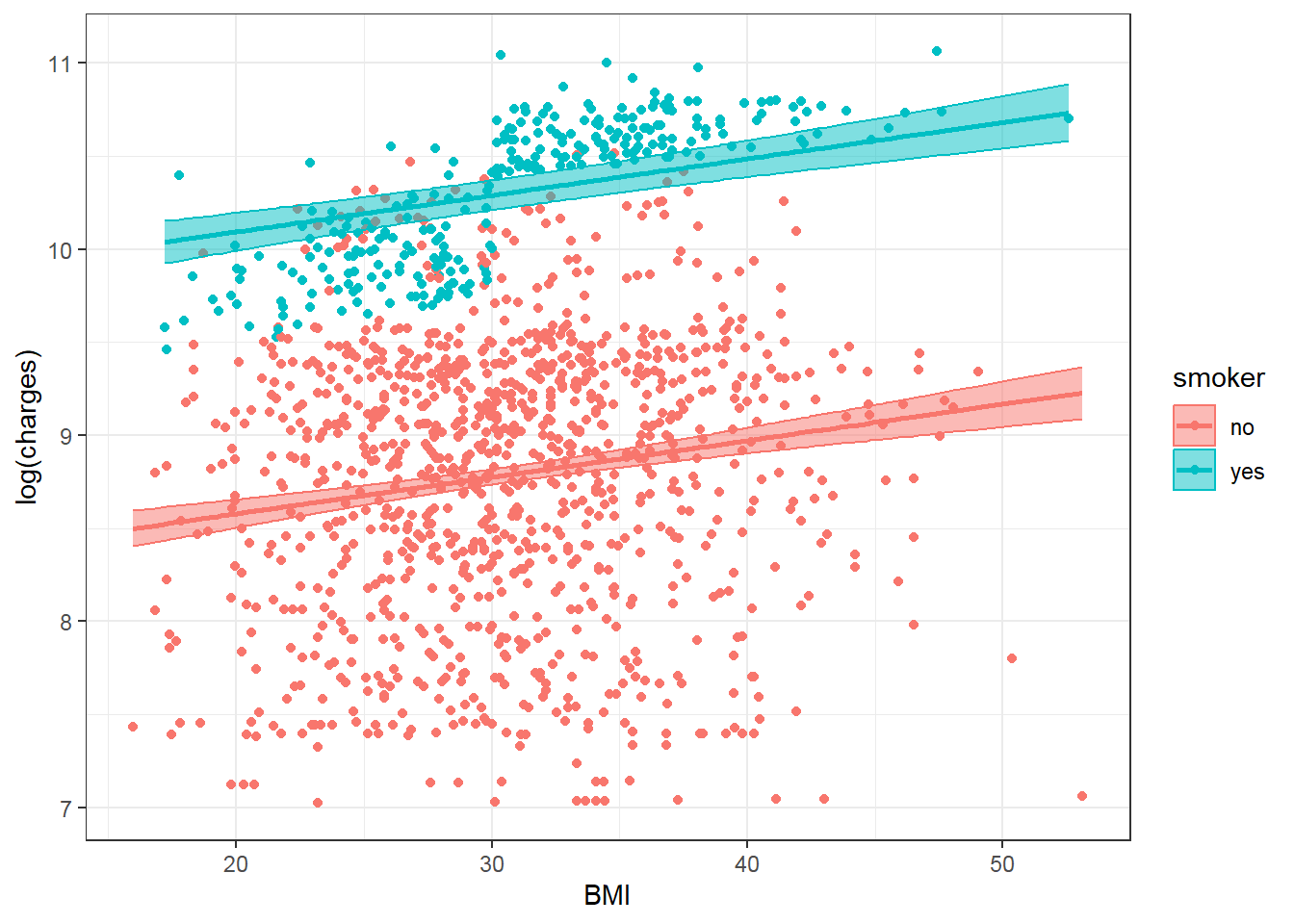
In the above multiple linear regression model with smoker and bmi as predictors, the slopes for logcharges against bmi is the same for smoker = no and smoker = yes. However, the above plot shows the difference in the association of logcharges against bmi between smoker = no and smoker = yes. Thus the multiple linear regression model above may not fit the data well, especially for smoker = yes.
To address this difference, an interaction term between smoker and bmi may be added to the model:
##
## Call:
## lm(formula = logcharges ~ smoker + bmi + smoker * bmi, data = dlm)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.98216 -0.37399 0.07215 0.44881 1.72280
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 8.443124 0.106184 79.514 < 2e-16 ***
## smokeryes 0.342811 0.227399 1.508 0.131911
## bmi 0.011259 0.003399 3.313 0.000949 ***
## smokeryes:bmi 0.038179 0.007259 5.259 1.68e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.6697 on 1334 degrees of freedom
## Multiple R-squared: 0.4708, Adjusted R-squared: 0.4696
## F-statistic: 395.6 on 3 and 1334 DF, p-value: < 2.2e-16# Add confidence interval to summary
ciflm.i <- confint(flm.i)
sumciflm.i <- cbind(sumflm.i$coefficients, ciflm.i)
round(sumciflm.i, 4)## Estimate Std. Error t value Pr(>|t|) 2.5 % 97.5 %
## (Intercept) 8.4431 0.1062 79.5145 0.0000 8.2348 8.6514
## smokeryes 0.3428 0.2274 1.5075 0.1319 -0.1033 0.7889
## bmi 0.0113 0.0034 3.3126 0.0009 0.0046 0.0179
## smokeryes:bmi 0.0382 0.0073 5.2595 0.0000 0.0239 0.0524And re-perform the plot:
predictions <- predict(flm.i, newdata=dlm, interval="confidence")
new_data <- cbind(dlm, as.data.frame(predictions))
#plot
ggplot(new_data, aes(bmi, logcharges, color=smoker)) +
geom_point() +
geom_ribbon(data = new_data, aes(x = bmi, ymin = lwr, ymax = upr, fill=smoker), alpha = 0.5) +
geom_line(data = new_data, aes(bmi, fit, color=smoker), linewidth = 1) +
labs(x = "BMI", y = "log(charges)") +
theme_bw()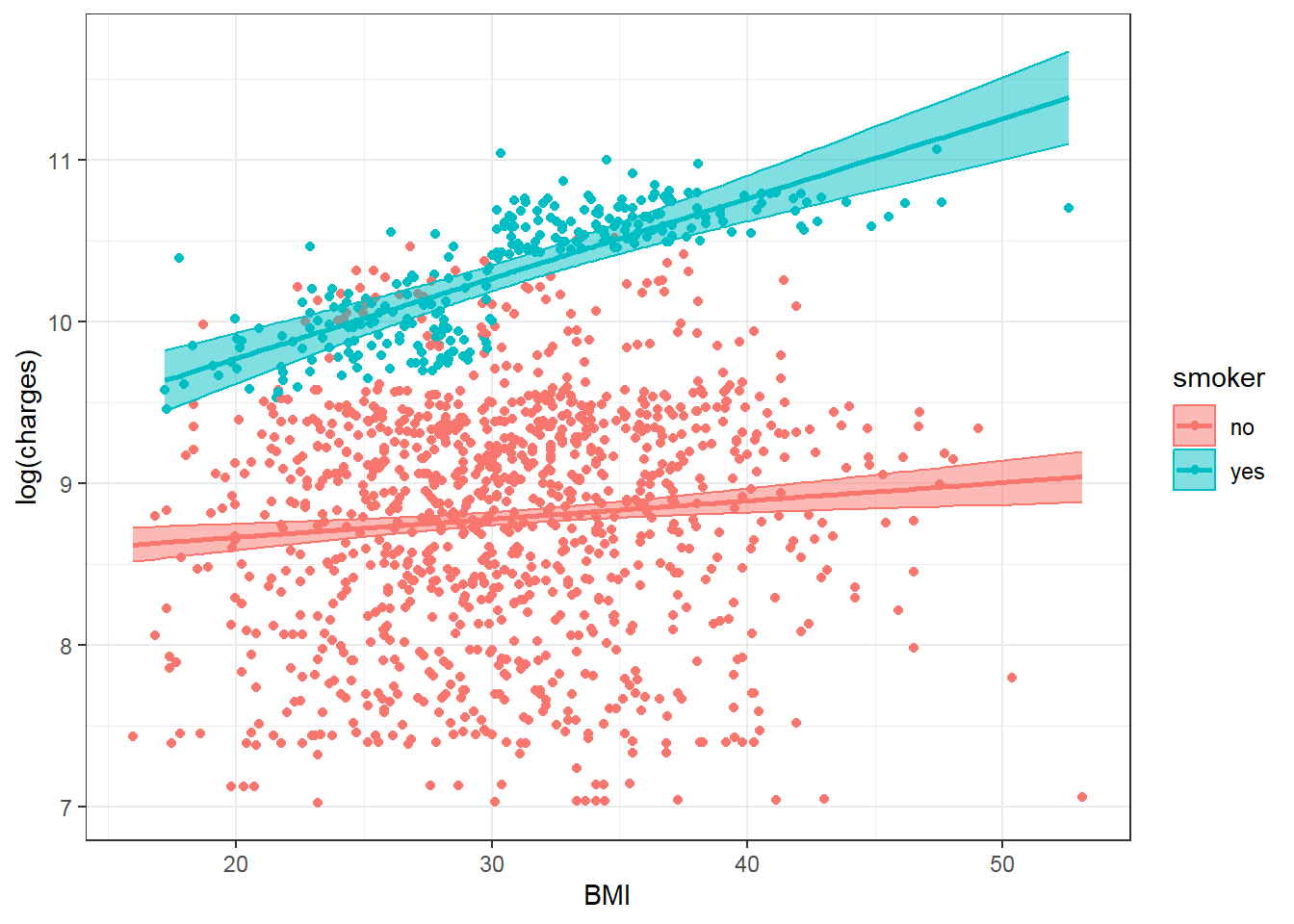
Now the plot shows two different slopes for smoker = no and smoker = yes and the model fits the data better especially for smoker = yes.
The interpretation of coefficients of the multiple linear regression model with interaction term is as below:
logchargeswhensmoker= no andbmi= 0 is 8.443124When
bmi= 0,logchargeswhensmoker= yes is is 0.3428113 higher whensmoker= noWhen
smoker= no, for each unit increase inbmi,logchargesincreases by 0.011259Coefficient for interaction term expresses the change in a variable’s effect when the interacting variable is increased by one-unit:
- The effect of
bmionlogchargesincreases by 0.0381793 (logchargesper one unit ofbmi) insmoker= yes vssmoker= no. - For each unit increase in
bmi, the effect ofsmoker= yes vssmoker= no onlogchargesincreases by 0.0381793.
- The effect of
The effect of bmi for smoker= yes or smoker= no may be calculated by adding the coefficient of bmi 0.011259 to the interaction coefficient, 0.0381793, multiplied by smoker as dummy variable 0, 1:
- bmi.effect (smoker=no) = 0.011259 + 0.0381793 *0
- bmi.effect (smoker=yes) = 0.011259 + 0.0381793 *1
The effect of smoker for any bmi may be calculated by adding the coefficient of smoker 0.3428113 to the interaction coefficient, 0.0381793and multiplied by bmi:
- smoker.effect (bmi=15) = 0.3428113 + 0.0381793 *15
6.4 Reference materials
The following resources have been adapted for this chapter:
Linear Regression Hands on R tutorial by Princeton University Library
The dataset used in some examples of this chapter are example data regarding the association of medical costs and patient information: may be downloaded from Github.