Chapter 8 Survival Data Analysis with R
Ho Thi Nhan, MD, PhD
8.1 What is survival analysis?
- Survival analysis is the study of survival times and of the factors that influence them.
- Survival analysis investigates the time it takes for an event of interest to occur (time-to-event).
8.1.1 Examples of survival analysis
- Time from birth until death
- Time from entry into a clinical trial until tumor response
- Time from treatment initiation to disease recovery
And much more…
8.1.2 Other application of survival analysis
- Reliability analysis
- Duration analysis
- Event history analysis
- Time-to-event analysis
8.1.3 Survival data
- Response variable is a non-negative discrete or continuous random variable, and represents the time from a well-defined origin to a well-defined event (distinct start time and end time).
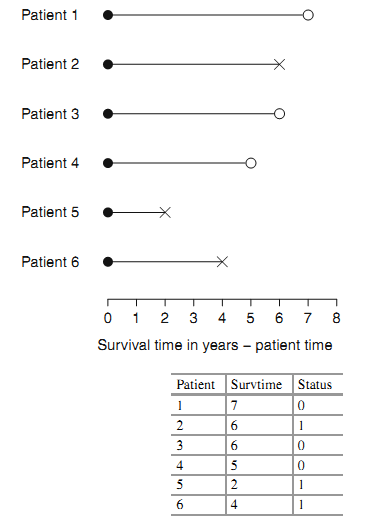
- Survival data is often represented as a pair (\(t_{i}\); \(\delta_{i}\)) where t is the time until endpoint or last follow-up, and \(\delta\) is a 0/1 variable with 0= “subject was censored at t” and 1 = “subject had an event at t”, or in R code as Surv(time, status).
8.1.3.1 Censoring
- Starting or ending events are not precisely observed
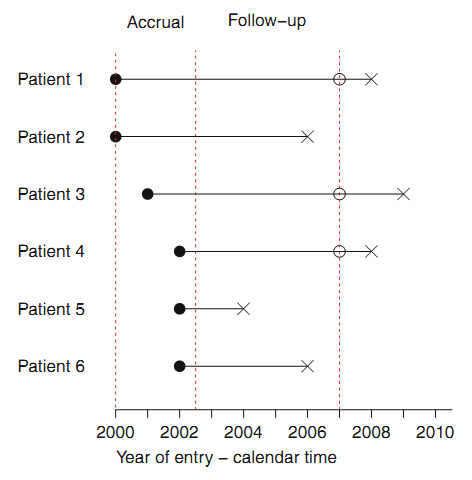
- Right censoring (most common): At the time of an analysis, not everyone will have yet had the event. The final endpoint is only known to exceed a particular value \(\rightarrow\) Incomplete/partial information: if a particular subject was enrolled in a study 2 years ago, and has not yet had an event at the time of analysis, we only know that their time to event is > 2 years.
Right censoring: trial example:
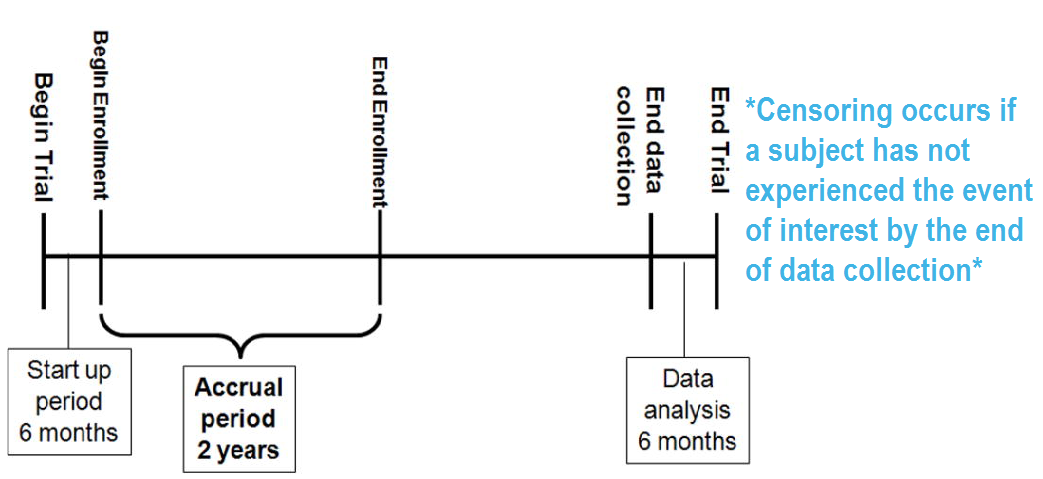
8.1.3.2 Types of censoring
8.1.3.2.1 Three types of censoring:
- Type I: censoring times are pre-specified. E.g. cohort of patients
may start at a specific time, and all followed until a pre-specified
ending time. Patients who have not experienced the event of interest
before the end of the study are then censored at that time.
- Type II: experimental objects are followed until a prespecified fraction have failed. Such a design is rare in biomedical studies, but may be used in industrial settings, where time to failure of a device is of primary interest. E.g. the study stops after 40 out of 100 devices are observed to fail. The remaining 60 devices would then be censored. In this example, the smallest 40% of the ordered failure times are observed, and the remainder are censored.
- Random: Careful attention to the cause of the censoring is essential in order to avoid biased survival estimates.
8.1.3.2.2 Examples of random censoring
- Participant drop-out: If the dropout occurs truly at random, and is unrelated to the disease process, such censoring may not cause any problems with bias in the analysis. But if patients who are near death are more likely to drop out than other patients, serious biases may arise.
- Competing risk (e.g. died of another cause before died of the cause of interest).
- Most common source of random censoring is administrative censoring, which results because some patients in a clinical trial have not yet died at the time the analysis is carried out.
8.1.4 Survival models
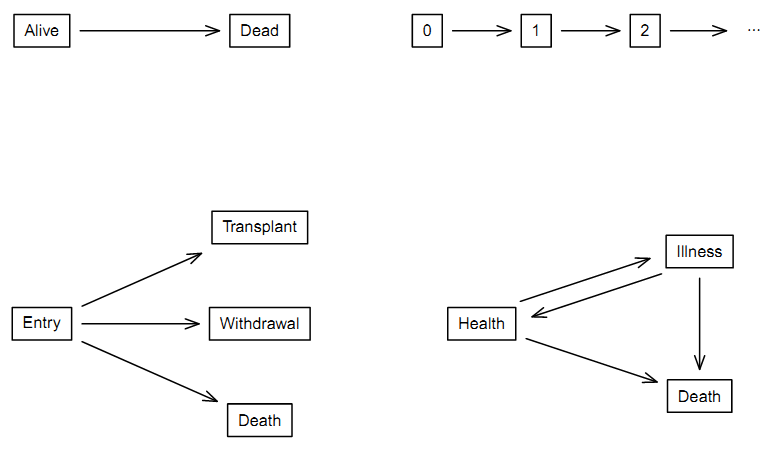
8.1.4.2 2 by 2 division of survival data sets
| One event per subject | Multiple events per subject | |
|---|---|---|
| One event type | 1 | 2 |
| Multiple event type | 3 | 4 |
- 1: “Classic” simple survival with two states of alive and dead
(upper left) \(\rightarrow\) main topic of this module.
- 2: Repeated events of the same type (upper right).
- 3: Competing risk (lower left).
- 4: Illness-death model (lower right).
8.2 Some math definitions
8.2.1 The Hazard and Survival Functions
The survival function defines the probability of surviving up to a point t: \(S(t)=pr(T>t), 0<t<Inf\) .
The hazard function is the instantaneous failure rate: the probability that, given that a subject has survived up to time t, he or she fails in the next small interval of time, divided by the length of that interval (Moore 2016):
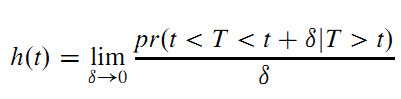
8.2.2 Other Representations of a Survival Distribution
- The probability density function (PDF) is minus the rate of change of the survival function:
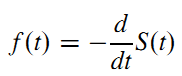
- The hazard at time t is the probability that an event occurs in the neighborhood of time t divided by the probability that the subject is alive at time t. The hazard function is related to the PDF and survival functions by:
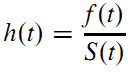
- The cumulative hazard function is defined as the area under the hazard function up to time t:
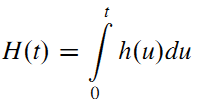
- The survival function may be defined in terms of the hazard function by:
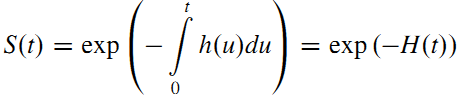
\(\rightarrow\) May compute the survival function corresponding to a hazard function.
8.3 Working with survival data in R
8.3.2 Three main packages
- Essential core package: “survival” (Therneau 2024).
- Package for nice visualization and more: “survminer” (Kassambara, Kosinski, and Biecek 2024)
- Package for working with dates (to create time to event data): “lubridate” (Spinu, Grolemund, and Wickham 2024)
8.3.3 Working with dates
Data will often come with start and end dates rather than pre-calculated survival times.
- Make sure these dates are formatted as dates in R.
- Calculate survival time (time-to-event)
exdate<-read.table("https://stats.idre.ucla.edu/stat/r/examples/asa/hmohiv.csv", sep=",", header = TRUE)
#dates with base R
# calculate the number of days between our two dates and convert it to a numeric value using as.numeric. Then convert to month by dividing by 30.42.
exdateb<-exdate %>%
mutate_at(c("entdate","enddate"), as.Date, format="%m/%d/%Y")%>%
mutate(
ttevent.m=as.numeric(
difftime(enddate,
entdate,
units="days"))/30.42)
head(exdateb)
#with lubridate
exdatel<-exdate%>%
mutate_at(c("entdate","enddate"), mdy)%>%
mutate(
ttevent.m=interval(entdate,enddate)/months(1))
head(exdatel)
#rename
exdatel<-exdatel%>%
rename(status=censor)8.3.4 Package “survival” main functions
8.3.4.1 Surv()
- Surv(time, status): right censored data
- Surv(time, endpoint==‘death’): right censored data, where the status variable is a character or factor
- Surv(t1, t2, status): counting process data
- Surv(t1, ind, type=‘left’): left censoring
- Surv(time, fstat): multiple state data, fstat is a factor
8.3.4.2 coxph() Cox’s proportional hazards model.
- coxph(Surv(time, status) ~ x, data=aml) : standard Cox model
- coxph(Surv(t1, t2, stat) ~ (age + surgery)* transplant): time dependent covariates.
- y <- Surv(t1, t2, stat) coxph(y~strata(inst) * sex + age + treat): Stratified model, with a sepa- rate baseline per institution, and institution specific effects for sex.
- coxph(y ~ offset(x1) + x2) : force in a known term, without estimating a coeffcient for it.
8.3.4.3 cox.zph
Computes a test of proportional hazards for the fitted Cox model.
- zfit <- cox.zph(coxfit); plot(zfit).
8.3.4.4 survreg
Parametric survival models.
- survreg(Surv(time, stat) ~x, dist=“loglogistic”) : Fit a log-logistic distribution.
8.3.4.5 survfit Fit a survival curve.
- survfit(Surv(time, status)): Simple Kaplan-Meier
- survfit(Surv(time, status) ~ rx + sex) : Four groups
- fit <- coxph(Surv(time, stat) ~ rx + sex) survfit(fit, list(rx=1, sex=2)): Predict curv
8.3.4.6 survdiff testing difference between survival curves
One and k-sample versions of the Fleming-Harrington G family. Includes the logrank and Gehan-Wilcoxon as special cases.
- survdiff(Surv(time, status) ~ sex + treat) : Compare the 4 sub-groups formed by sex and treatment combinations.
- survdiff(Surv(time, status) ~ offset(pred)): One-sample test
8.3.5 Survival curves and models
8.3.5.1 Survival model and curve for overall data
## Call: survfit(formula = Surv(time, status) ~ 1, data = lung)
##
## n events median 0.95LCL 0.95UCL
## [1,] 228 165 310 285 363Median survival time
## med_surv
## 1 284Right or wrong? Why?
\(\rightarrow\) Wrong as censoring is ignored.
The correct estimate is:
## Call: survfit(formula = Surv(time, status) ~ 1, data = lung)
##
## n events median 0.95LCL 0.95UCL
## [1,] 228 165 310 285 363Survival as specific time point, for example survival at 1 year:
evy1<-lung %>%
filter(time<=365 & status==1) %>%
summarise(
sum(as.numeric(as.character(status))))
1-evy1/nrow(lung) ## sum(as.numeric(as.character(status)))
## 1 0.8157895Right or wrong? Why?
\(\rightarrow\) Wrong as censoring is ignored.
The correct estimate for 1 year survival is:
## Call: survfit(formula = Surv(time, status) ~ 1, data = lung)
##
## time n.risk n.event survival std.err lower 95% CI upper 95% CI
## 365 65 121 0.409 0.0358 0.345 0.486Kaplan-Meier plot using base R
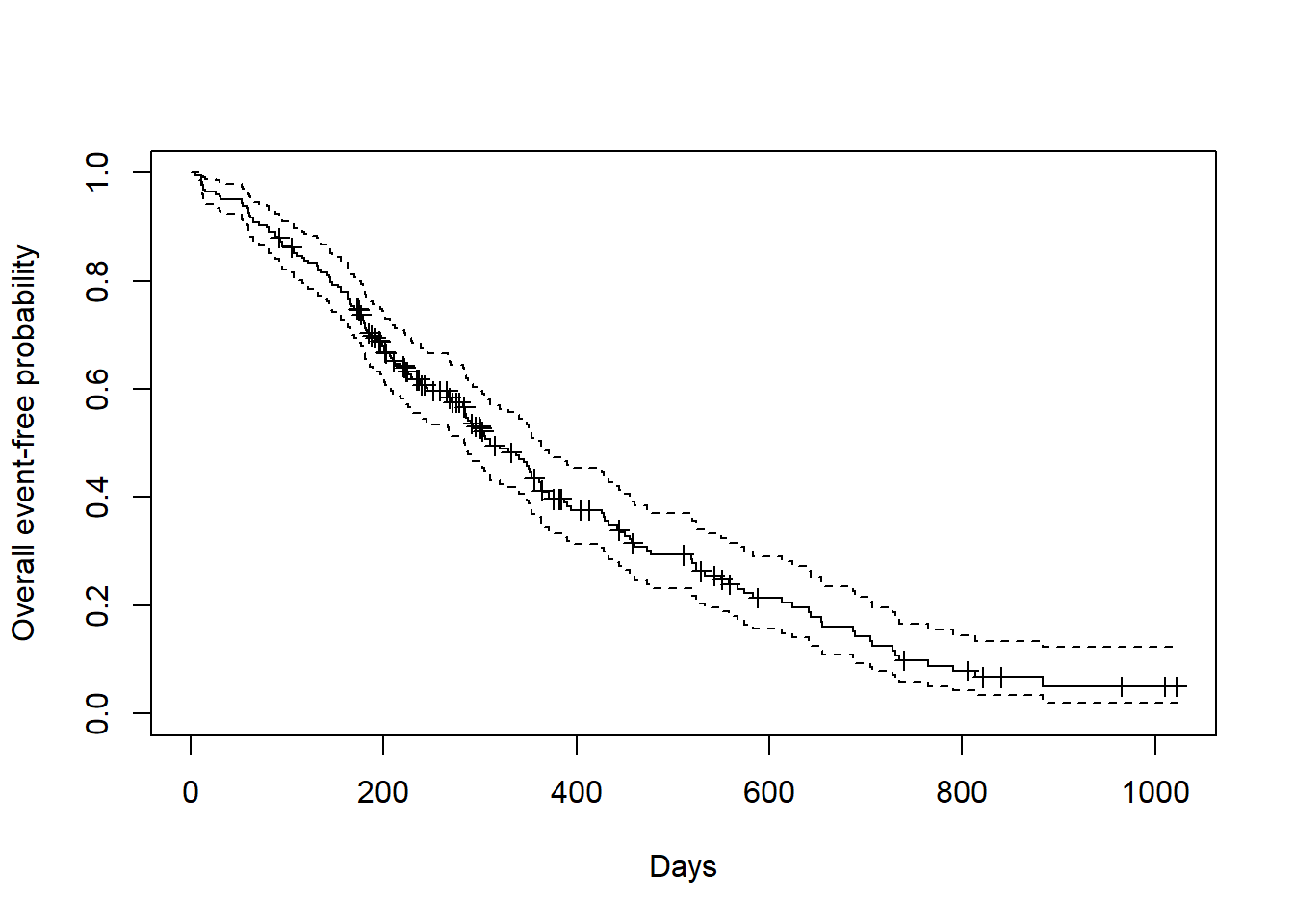
Kaplan-Meier plot using survminer package.
ggsurvplot(
fit=fit1,
risk.table = TRUE,
risk.table.height = 0.3,
xlab = "Days",
ylab = "Overall event-free probability")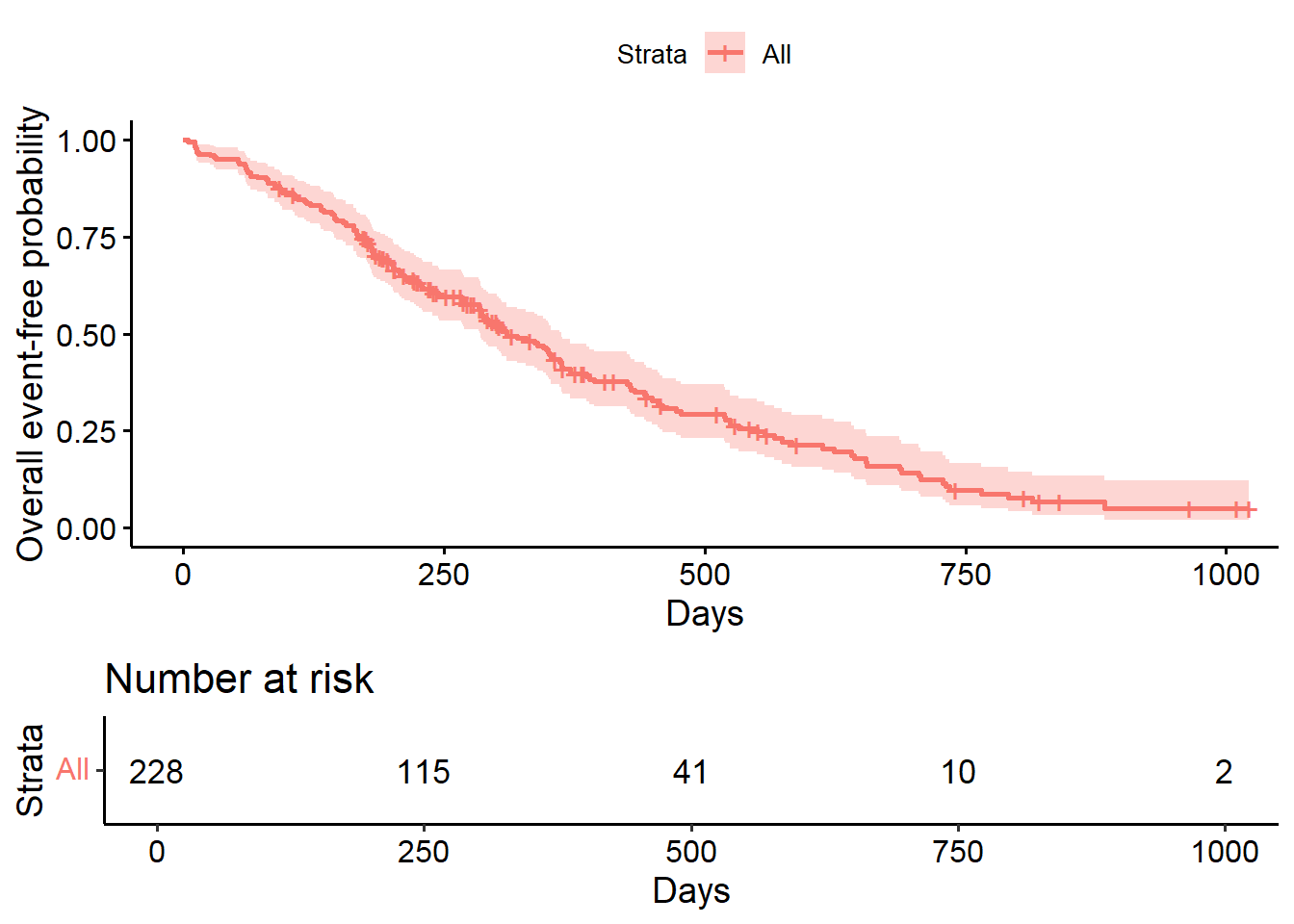
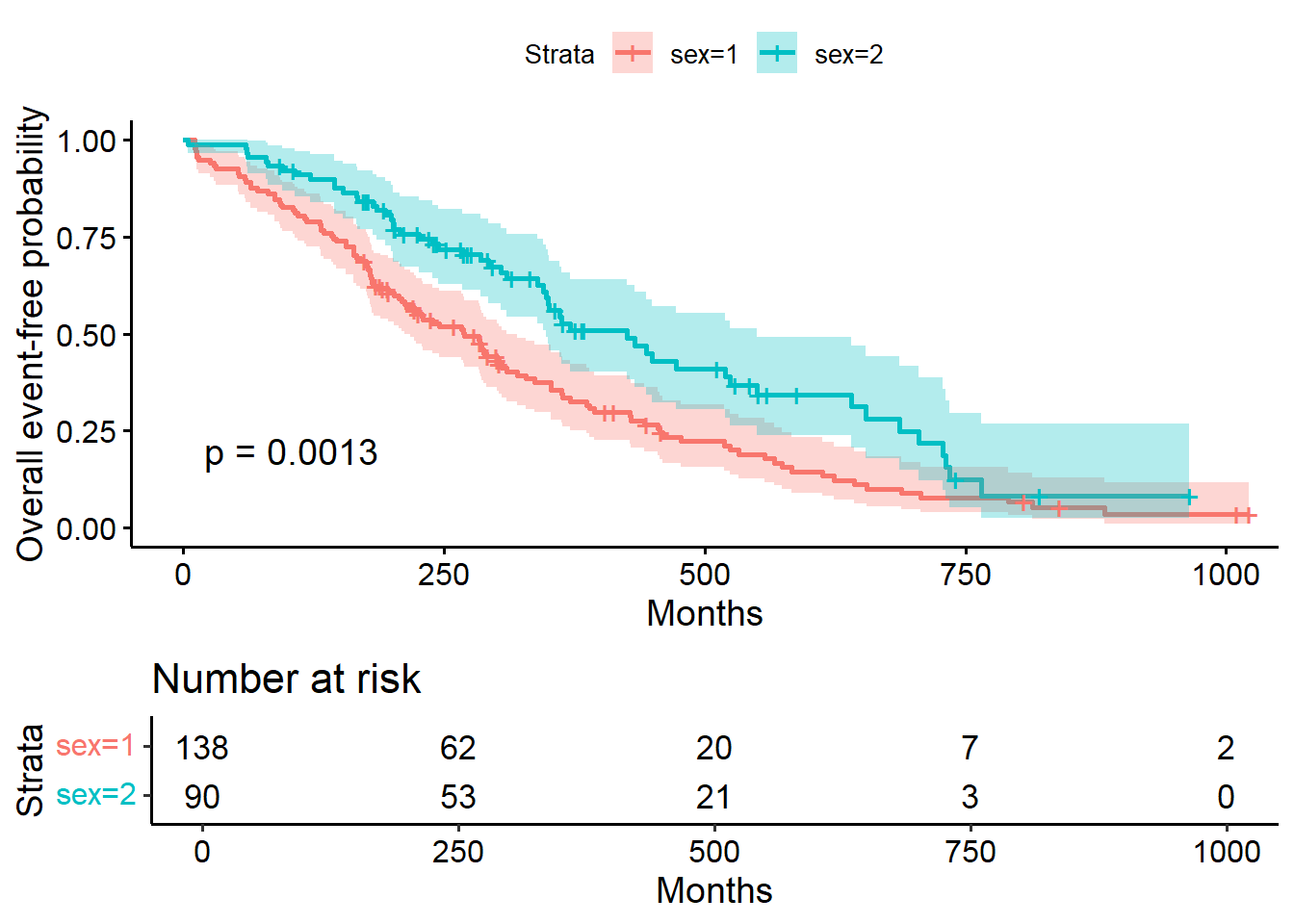
8.3.5.2 Comparing survival times between groups
## Call: survfit(formula = Surv(time, status) ~ sex, data = lung)
##
## n events median 0.95LCL 0.95UCL
## sex=1 138 112 270 212 310
## sex=2 90 53 426 348 5508.3.5.2.1 Testing the difference
The most common way to compare survival times between groups is log-rank test (or Mantel-Haenszel test) which equally weights observations over the entire follow-up time. (rho=0 for log rank test (default); rho=1 for Peto-Wilcoxon test).
The log-rank test is equivalent to the score test from a Cox model with the group as a factor variable.
## Call:
## survdiff(formula = Surv(time, status) ~ sex, data = lung)
##
## N Observed Expected (O-E)^2/E (O-E)^2/V
## sex=1 138 112 91.6 4.55 10.3
## sex=2 90 53 73.4 5.68 10.3
##
## Chisq= 10.3 on 1 degrees of freedom, p= 0.001## Call:
## survdiff(formula = Surv(time, status) ~ sex, data = lung, rho = 1)
##
## N Observed Expected (O-E)^2/E (O-E)^2/V
## sex=1 138 70.4 55.6 3.95 12.7
## sex=2 90 28.7 43.5 5.04 12.7
##
## Chisq= 12.7 on 1 degrees of freedom, p= 4e-048.3.5.3 The Cox regression model
- Semi-parametric model to fit univariable and multivariable regression models for survival outcomes.
- Some key assumptions of the model: non-informative censoring and proportional hazards.
- Most commonly used for single event data (one event per subject)
- Proportional hazards models differ from other types of regression: there is no intercept term (intercept term, if any, absorbed into the baseline hazard).
- \(\psi\) indexes the difference between the two survival distributions via the Lehmann alternative: \(S_{1}(t)=[S_{0}(t)]^{\psi}\) . This relationship can be re-expressed in terms of the hazard functions, yielding proportional hazards assumption: \(h_{1}(t)=\psi h_{0}(t)\).
- The model can be extended to include covariate information in a vector z as follows: \(\psi=e^{z\beta}\) (Moore 2016).
## Call:
## coxph(formula = Surv(time, status) ~ sex, data = lung)
##
## coef exp(coef) se(coef) z p
## sex -0.531 0.588 0.167 -3.18 0.0015
##
## Likelihood ratio test=10.6 on 1 df, p=0.00111
## n= 228, number of events= 165## Call:
## coxph(formula = Surv(time, status) ~ sex, data = lung)
##
## n= 228, number of events= 165
##
## coef exp(coef) se(coef) z Pr(>|z|)
## sex -0.5310 0.5880 0.1672 -3.176 0.00149 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## exp(coef) exp(-coef) lower .95 upper .95
## sex 0.588 1.701 0.4237 0.816
##
## Concordance= 0.579 (se = 0.021 )
## Likelihood ratio test= 10.63 on 1 df, p=0.001
## Wald test = 10.09 on 1 df, p=0.001
## Score (logrank) test = 10.33 on 1 df, p=0.001## Analysis of Deviance Table
## Cox model: response is Surv(time, status)
## Terms added sequentially (first to last)
##
## loglik Chisq Df Pr(>|Chi|)
## NULL -749.91
## sex -744.59 10.634 1 0.001111 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1## Analysis of Deviance Table
## Cox model: response is Surv(time, status)
## Model 1: ~ sex
## Model 2: ~ 1
## loglik Chisq Df Pr(>|Chi|)
## 1 -744.59
## 2 -749.91 10.634 1 0.001111 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 18.3.5.3.1 Hazard ratios
- HR represents the ratio of hazards between two groups at any particular point in time.
- Interpreted as the instantaneous rate of occurrence of the event of interest in those who are still at risk for the event. It is not a risk, though it is commonly interpreted as such
- \(HR = exp(\beta)\) (exponentiation of coefficients of Cox model output)
- A HR < 1 indicates reduced hazard of event whereas a HR > 1 indicates an increased hazard of event. So our HR = 2.28 (drug) implies that around 2.28 times as many of those using drug are having the event as compared to those not using drug, at any given time.
8.3.5.3.2 Multivariate Cox model
## Call:
## coxph(formula = Surv(time, status) ~ sex + age, data = lung)
##
## coef exp(coef) se(coef) z p
## sex -0.51322 0.59857 0.16746 -3.06 0.0022
## age 0.01705 1.01719 0.00922 1.85 0.0646
##
## Likelihood ratio test=14.1 on 2 df, p=0.000857
## n= 228, number of events= 165## Call:
## coxph(formula = Surv(time, status) ~ sex + age, data = lung)
##
## n= 228, number of events= 165
##
## coef exp(coef) se(coef) z Pr(>|z|)
## sex -0.513219 0.598566 0.167458 -3.065 0.00218 **
## age 0.017045 1.017191 0.009223 1.848 0.06459 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## exp(coef) exp(-coef) lower .95 upper .95
## sex 0.5986 1.6707 0.4311 0.8311
## age 1.0172 0.9831 0.9990 1.0357
##
## Concordance= 0.603 (se = 0.025 )
## Likelihood ratio test= 14.12 on 2 df, p=9e-04
## Wald test = 13.47 on 2 df, p=0.001
## Score (logrank) test = 13.72 on 2 df, p=0.0018.3.5.3.3 Stratified Cox model
- Adding one or more strata terms to the model formula.
- Each subject is compared only to subjects within their own stratum for computing the partial likelihood, and then the final results are summed over the strata.
- Variable included as a stratum is adjusted for in the most general way, and not having an estimate of its effect.
- Common use of strata: to adjust for the enrolling institution in a multi-center study.
# Example
cfit <- coxph(Surv(time, status) ~ age + sex + wt.loss, data=lung)
#summary(cfit)
cfits <- coxph(Surv(time, status) ~ age + sex + wt.loss + strata(inst),
data=lung)
#summary(cfits)
# compare between simple and stratified models
round(cbind(simple= coef(cfit), stratified=coef(cfits)), 4)## simple stratified
## age 0.0201 0.0235
## sex -0.5210 -0.5160
## wt.loss 0.0008 -0.00178.3.6 Model selection & Interpretation
8.3.6.1 Partial Likelihood Hypothesis Tests
Three forms of test for H0: \(\beta\) = 0:
- Wald test: commonly used test, from output: Z= \(\hat{\beta}\)/s.e.(\(\hat{\beta}\))
- Score test: equivalent to the log-rank test
- Likelihood ratio test: key advantage of this test over the other two is that it is invariant to monotonic transformations of \(\beta\).
8.3.6.2 Hypothesis Testing for Nested Models
- Survival Model: log(\(\psi\))=\(z_{1i}\)\(\beta_{1}\)+\(z_{2i}\)\(\beta_{2}\)+…+\(z_{ki}\)\(\beta_{k}\). Where \(\beta_{k}\) is the log hazard ratio.
#anova command shows likelihood ratio tests for each term in a model, added sequentially
anova(cfit)## Analysis of Deviance Table
## Cox model: response is Surv(time, status)
## Terms added sequentially (first to last)
##
## loglik Chisq Df Pr(>|Chi|)
## NULL -680.39
## age -677.78 5.2273 1 0.022235 *
## sex -673.06 9.4268 1 0.002138 **
## wt.loss -673.06 0.0150 1 0.902592
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 18.3.6.3 Model selection approaches
Large number of potential factors \(\rightarrow\) need to prune the model so that only necessary covariates are included.
Step-wise:
- Forward: univariate model for each covariate \(\rightarrow\) choose ones with smallest p-values to fit models with two covariates \(\rightarrow\) common critical p-values are 5% and 10%.
- Backward: fit all variables \(\rightarrow\) remove one by one with largest
p-value
- Problems: 1) multiple comparisons: p-values from one stage to another are not the same; 2) P-values only valid for nested models.
8.3.6.4 The Akaike Information Criterion (AIC) for Comparing Non-nested Models
AIC=\(-2*l(\hat{\beta})+2*k\), where:
- \(-2*l(\hat{\beta})\) is goodness of fit (smaller the better), denotes
the value of the partial log likelihood at the M.P.L.E. for a
particular model.
- k is the number of parameters in the model = measure of complexity.
## [1] 1352.112## [1] 577.052# Using AIC for "step" model selection
#fitall<-coxph(Surv(time, status) ~ age + sex + wt.loss+ inst+ ph.ecog+ ph.karno+ meal.cal, data=lung)
#anova(fitall)
#aicstep<-step(fitall, scope=list(upper=~age +sex + wt.loss+ inst+ ph.ecog+ ph.karno+ meal.cal, lower=~age))
lung.nona<-na.omit(lung)
fitall2<-coxph(Surv(time, status) ~ age + sex + wt.loss+ inst+ ph.ecog+ ph.karno+ meal.cal, data=lung.nona)
aicstep2<-step(fitall2, scope=list(upper=~age +sex + wt.loss+ inst+ ph.ecog+ ph.karno+ meal.cal, lower=~age))## Start: AIC=998.33
## Surv(time, status) ~ age + sex + wt.loss + inst + ph.ecog + ph.karno +
## meal.cal
##
## Df AIC
## - meal.cal 1 996.38
## <none> 998.33
## - wt.loss 1 1000.12
## - ph.karno 1 1001.23
## - inst 1 1002.27
## - sex 1 1005.05
## - ph.ecog 1 1015.67
##
## Step: AIC=996.38
## Surv(time, status) ~ age + sex + wt.loss + inst + ph.ecog + ph.karno
##
## Df AIC
## <none> 996.38
## - wt.loss 1 998.16
## + meal.cal 1 998.33
## - ph.karno 1 999.27
## - inst 1 1000.29
## - sex 1 1003.07
## - ph.ecog 1 1013.83## Call:
## coxph(formula = Surv(time, status) ~ age + sex + wt.loss + inst +
## ph.ecog + ph.karno, data = lung.nona)
##
## coef exp(coef) se(coef) z p
## age 0.013784 1.013880 0.011717 1.176 0.23943
## sex -0.572214 0.564275 0.199575 -2.867 0.00414
## wt.loss -0.014682 0.985425 0.007765 -1.891 0.05866
## inst -0.031333 0.969152 0.013087 -2.394 0.01665
## ph.ecog 0.993069 2.699507 0.229849 4.321 1.56e-05
## ph.karno 0.024618 1.024923 0.011472 2.146 0.03189
##
## Likelihood ratio test=31.85 on 6 df, p=1.74e-05
## n= 167, number of events= 1208.3.7 Checking assumptions for Cox model
- Proportional hazards
- Additivity
- Linearity
8.3.7.1 Check Proportional hazards
Not significant \(\rightarrow\) cannot reject
## chisq df p
## age 0.5077 1 0.48
## sex 2.5489 1 0.11
## wt.loss 0.0144 1 0.90
## GLOBAL 3.0051 3 0.39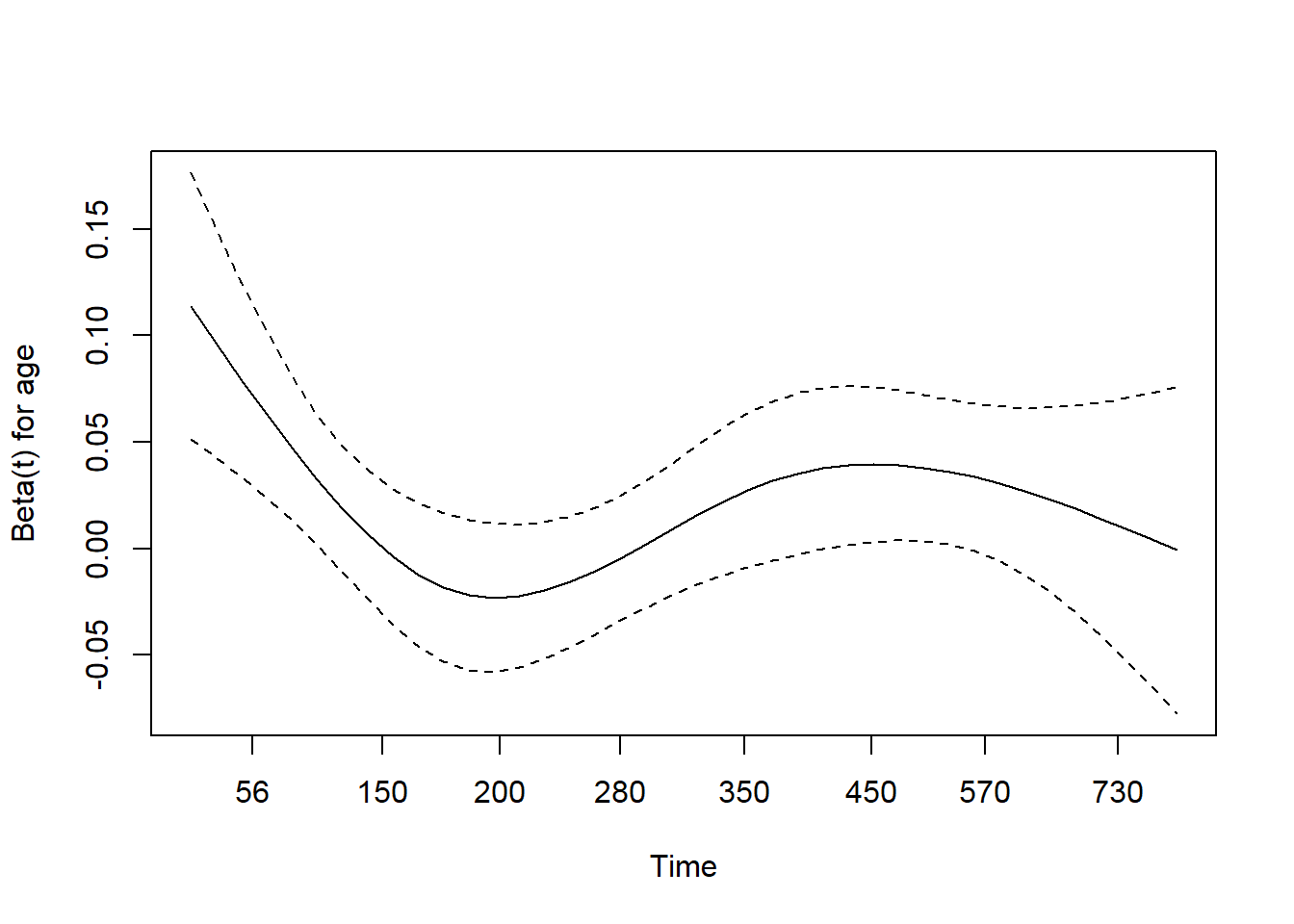
8.3.7.2 Check additivity
Interaction not significant \(\rightarrow\) additivity
## Analysis of Deviance Table
## Cox model: response is Surv(time, status)
## Model 1: ~ age + sex + wt.loss
## Model 2: ~ age + sex + wt.loss + age:sex
## loglik Chisq Df Pr(>|Chi|)
## 1 -673.06
## 2 -672.88 0.3473 1 0.55578.3.7.3 Check linearity
Non-linearity not significant.
## Call:
## coxph(formula = Surv(time, status) ~ pspline(age) + sex + wt.loss,
## data = lung)
##
## coef se(coef) se2 Chisq DF p
## pspline(age), linear 0.02011 0.00931 0.00931 4.66393 1.00 0.0308
## pspline(age), nonlin 3.02601 3.08 0.4018
## sex -0.53145 0.17549 0.17513 9.17105 1.00 0.0025
## wt.loss 0.00056 0.00619 0.00618 0.00818 1.00 0.9279
##
## Iterations: 4 outer, 12 Newton-Raphson
## Theta= 0.799
## Degrees of freedom for terms= 4.1 1.0 1.0
## Likelihood ratio test=18.4 on 6.07 df, p=0.006
## n= 214, number of events= 152
## (14 observations deleted due to missingness)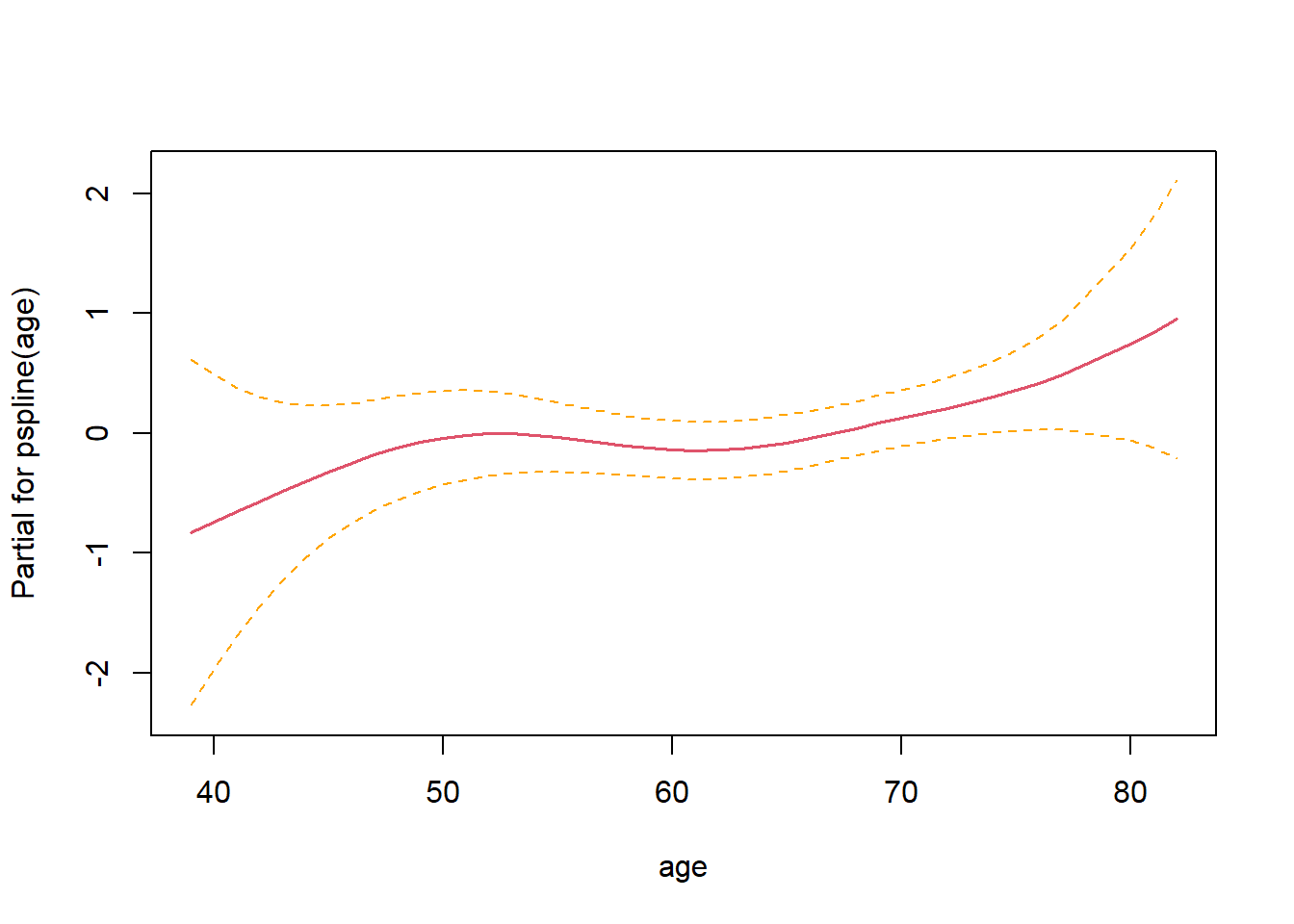
8.3.8 Transform survival curves: plot cumulative events and cumulative hazard function
#plot cumulative event (incidence)
ggsurvplot(fit2, conf.int = TRUE,
fun = "event",
risk.table = TRUE, risk.table.col = "strata")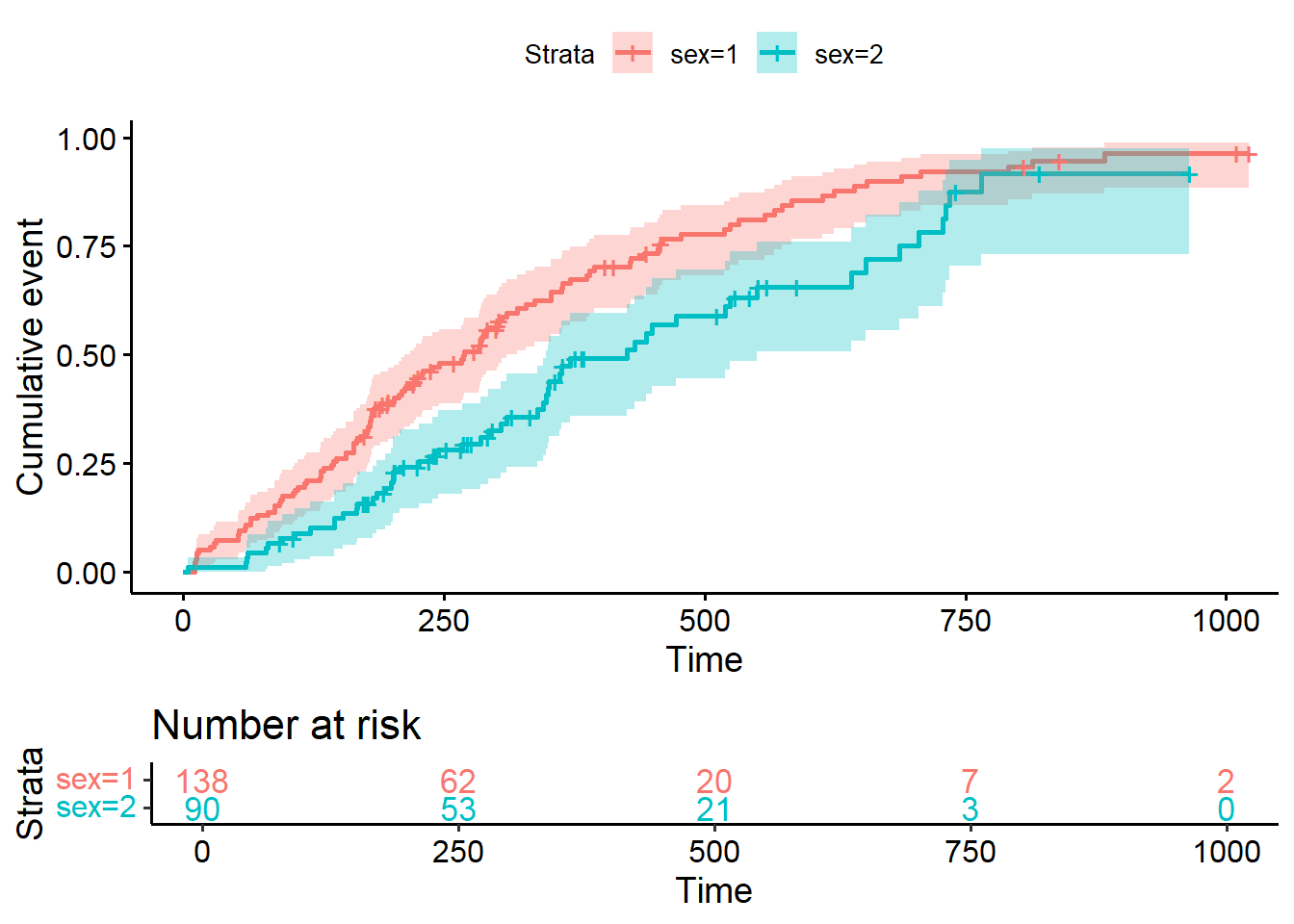
#Plot the cumulative hazard function
ggsurvplot(fit2, conf.int = TRUE,
fun = "cumhaz",
risk.table = TRUE, risk.table.col = "strata")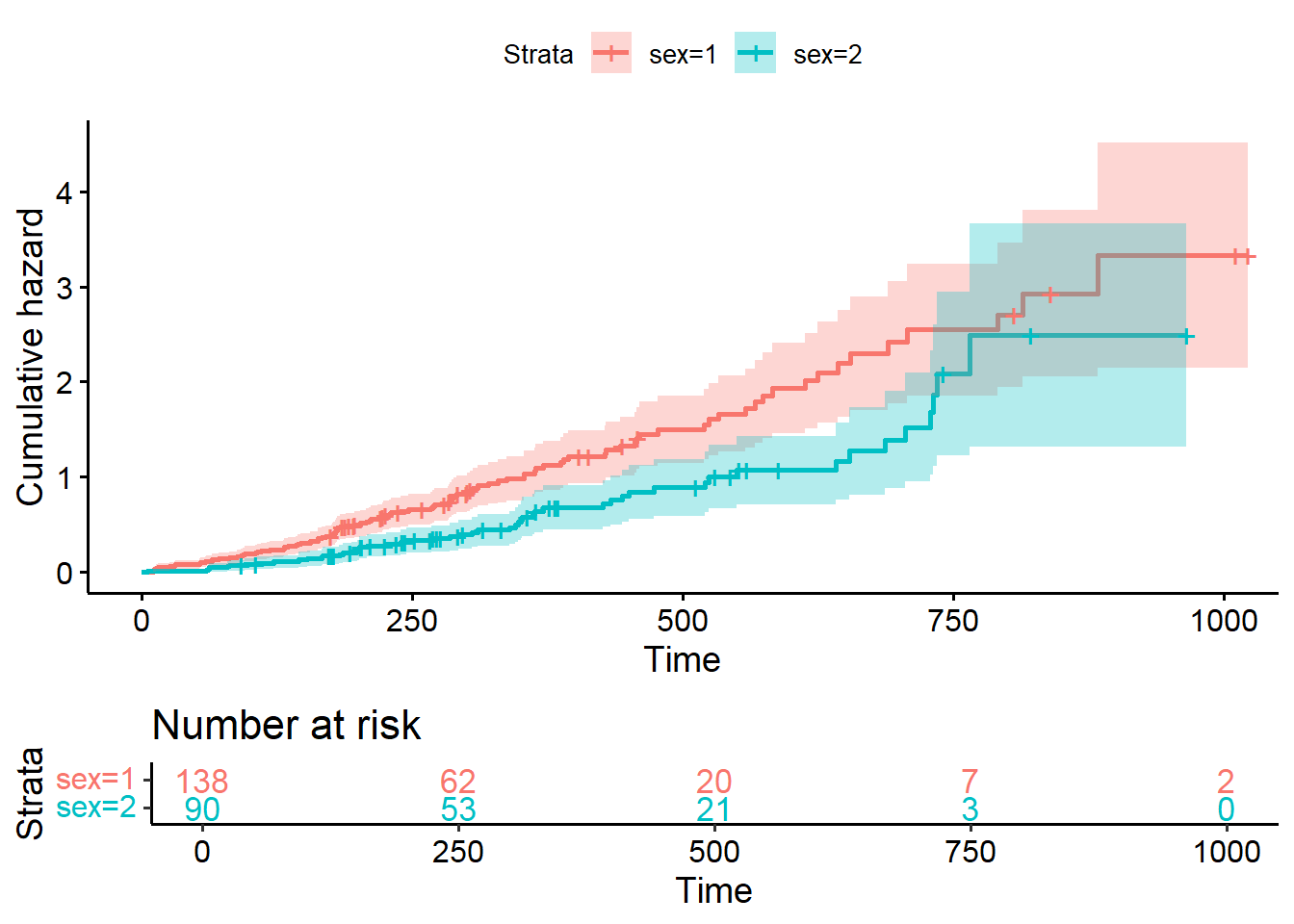
8.3.9 More advanced aspects in survival analysis
8.3.9.1 Time Dependent Covariates
8.3.9.1.1 Background
- Log-rank tests and Cox regression to examine associations between
survival outcomes and covariates of interest at baseline (before
follow-up time for the event begins).
- Covariate that is measured after follow-up time begin \(\rightarrow\) time dependent covariate
8.3.9.1.2 Landmark Analysis Method
- Select a fixed time after baseline as your landmark time. Note: this should be done based on clinical information, prior to data inspection
- Subset population for those followed at least until landmark time. Note: always report the number excluded due to the event of interest or censoring prior to the landmark time.
- Calculate follow-up from landmark time and apply traditional log-rank tests or Cox regression
8.3.9.1.3 Example Heart transplant data
Naive analysis showed patients who received heart transplants lived significantly longer than those who did not.
\(\rightarrow\) Problems:
- Transplant is a time dependent covariate; patients who received a transplant had to live long enough to receive that transplant.
- The analysis only shows that patients who live long enough to receive a transplant have longer lives than patients who don’t live as long (\(\rightarrow\) tautology)
## Call:
## coxph(formula = Surv(futime, fustat) ~ transplant + age + surgery,
## data = jasa)
##
## n= 103, number of events= 75
##
## coef exp(coef) se(coef) z Pr(>|z|)
## transplant -1.71711 0.17958 0.27853 -6.165 7.05e-10 ***
## age 0.05889 1.06065 0.01505 3.913 9.12e-05 ***
## surgery -0.41902 0.65769 0.37118 -1.129 0.259
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## exp(coef) exp(-coef) lower .95 upper .95
## transplant 0.1796 5.5684 0.1040 0.310
## age 1.0607 0.9428 1.0298 1.092
## surgery 0.6577 1.5205 0.3177 1.361
##
## Concordance= 0.732 (se = 0.031 )
## Likelihood ratio test= 45.85 on 3 df, p=6e-10
## Wald test = 47.15 on 3 df, p=3e-10
## Score (logrank) test = 52.63 on 3 df, p=2e-118.3.9.1.4 Landmark analysis example for heart transplant data
- Set a landmark at 30 days
- Divide patients into two groups: patients who receive the intervention prior to the landmark go into the intervention group and those who did not are placed in the comparison group.
- Select patients who lived at least 30 days (79 of the 103 patients lived this long). Of these 79 patients, 33 had a transplant within 30 days, and 46 did not. Of these 46, 30 subsequently had a heart transplant, but we still count them in the “no transplant within 30 days” group.
- Created a variable (we shall call it “transplant30”) which has a
fixed value (that is, it does not change over time) for all patients
in our set of 30-day survivors.
- \(\rightarrow\) there is no difference in survival between those who got a transplant and those who did not.
- Issues: a) When to set the landmark?; b) Exclude a lot of data (only
patients who survive up to the landmark are included in the study);
- all patients (in particular, those in the comparison group) remain in their originally assigned group regardless of what happens in the future, i.e., after the landmark
ind30 <- jasa$futime >= 30
transplant30 <- {{jasa$transplant == 1} & {jasa$wait.time < 30}}
summary(coxph(Surv(futime, fustat) ~ transplant30 + age + surgery, data=jasa, subset=ind30 ))## Call:
## coxph(formula = Surv(futime, fustat) ~ transplant30 + age + surgery,
## data = jasa, subset = ind30)
##
## n= 79, number of events= 52
##
## coef exp(coef) se(coef) z Pr(>|z|)
## transplant30TRUE -0.04214 0.95874 0.28377 -0.148 0.8820
## age 0.03720 1.03790 0.01714 2.170 0.0300 *
## surgery -0.81966 0.44058 0.41297 -1.985 0.0472 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## exp(coef) exp(-coef) lower .95 upper .95
## transplant30TRUE 0.9587 1.0430 0.5497 1.6720
## age 1.0379 0.9635 1.0036 1.0734
## surgery 0.4406 2.2697 0.1961 0.9898
##
## Concordance= 0.618 (se = 0.044 )
## Likelihood ratio test= 9.5 on 3 df, p=0.02
## Wald test = 8.61 on 3 df, p=0.03
## Score (logrank) test = 8.94 on 3 df, p=0.038.3.9.2 Model time depedent covariate
Can be done in the framework of the classical Cox proportional hazards model, but important adjustments to data is required: pre-processing the data into “start-stop” format. Example heart transplant data:
- Divides the time data for patients who had a heart transplant into two time periods, one before the transplant and one after.
- Use tmerge function to create “start-stop” format.
jasaid<-data.frame(id=rownames(jasa), jasa)
# extract some example patients
heart.s<-jasaid[c(2, 5, 10, 12, 28,95), c("id","wait.time","futime","fustat","transplant")]
heart.s ## id wait.time futime fustat transplant
## 2 2 NA 5 1 0
## 5 5 NA 17 1 0
## 10 10 11 57 1 1
## 12 12 NA 7 1 0
## 28 28 70 71 1 1
## 95 95 1 15 1 1dat.td<-tmerge(heart.s, heart.s, id=id, death=event(futime, fustat), transpl=tdc(wait.time))
kable(dat.td) | id | wait.time | futime | fustat | transplant | tstart | tstop | death | transpl |
|---|---|---|---|---|---|---|---|---|
| 2 | NA | 5 | 1 | 0 | 0 | 5 | 1 | 0 |
| 5 | NA | 17 | 1 | 0 | 0 | 17 | 1 | 0 |
| 10 | 11 | 57 | 1 | 1 | 0 | 11 | 0 | 0 |
| 10 | 11 | 57 | 1 | 1 | 11 | 57 | 1 | 1 |
| 12 | NA | 7 | 1 | 0 | 0 | 7 | 1 | 0 |
| 28 | 70 | 71 | 1 | 1 | 0 | 70 | 0 | 0 |
| 28 | 70 | 71 | 1 | 1 | 70 | 71 | 1 | 1 |
| 95 | 1 | 15 | 1 | 1 | 0 | 1 | 0 | 0 |
| 95 | 1 | 15 | 1 | 1 | 1 | 15 | 1 | 1 |
## Call:
## coxph(formula = Surv(tstart, tstop, death) ~ transpl, data = dat.td)
##
## n= 9, number of events= 6
##
## coef exp(coef) se(coef) z Pr(>|z|)
## transpl 0.2846 1.3292 0.9609 0.296 0.767
##
## exp(coef) exp(-coef) lower .95 upper .95
## transpl 1.329 0.7523 0.2021 8.74
##
## Concordance= 0.5 (se = 0.082 )
## Likelihood ratio test= 0.09 on 1 df, p=0.8
## Wald test = 0.09 on 1 df, p=0.8
## Score (logrank) test = 0.09 on 1 df, p=0.88.3.9.3 Repeated events
- Examples: time from treatment to seizure episodes (multiple repeated seizure episodes may occur); infections in transplanted patients
- Need to reformat data to “start, stop” form Example data of Children with chronic granulotomous disease (cgd0).
## Call:
## coxph(formula = Surv(tstart, tstop, status) ~ treat, data = cgd)
##
## n= 203, number of events= 76
##
## coef exp(coef) se(coef) z Pr(>|z|)
## treatrIFN-g -1.0953 0.3344 0.2610 -4.196 2.71e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## exp(coef) exp(-coef) lower .95 upper .95
## treatrIFN-g 0.3344 2.99 0.2005 0.5578
##
## Concordance= 0.629 (se = 0.029 )
## Likelihood ratio test= 20.11 on 1 df, p=7e-06
## Wald test = 17.61 on 1 df, p=3e-05
## Score (logrank) test = 19.42 on 1 df, p=1e-058.4 Practice
Use the dataframe “nafld1” of dataset “ndfld” of the “survival” package:
- Plot the overall survival curve, estimate median survival time, estimate survival at 1.5 year
- Plot the survival curves of males and females, test the difference between the curves
- Examine each of other covariates (age, weight, height, bmi) and check if any covariate significantly affect survival
- Report and provide rationale for the best multivariate Cox model.
8.5 Reference materials
8.5.1 Books, other resources
- Tutorial for the essential R package which is the base of most
packages related to survival analysis:
survival.
- Comprehensive book: Applied Survival Analysis Using R.