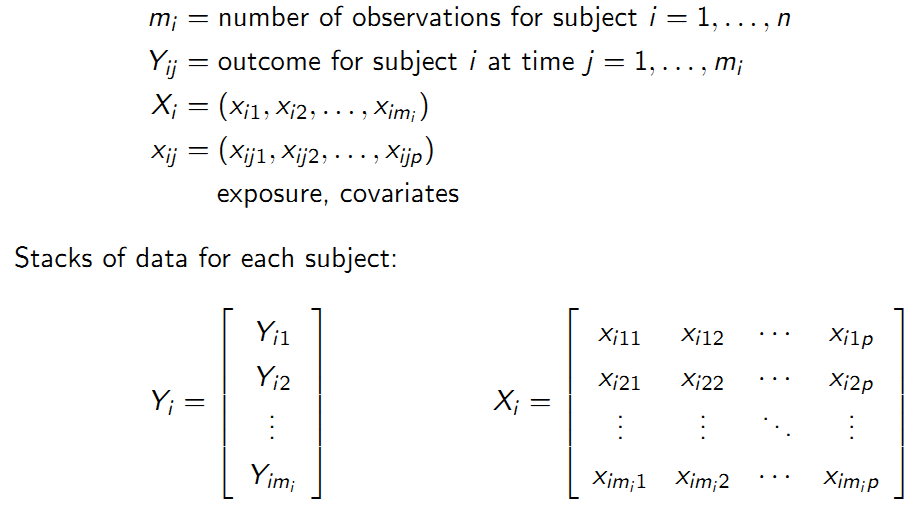
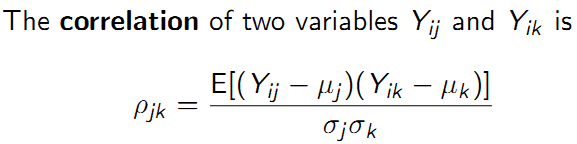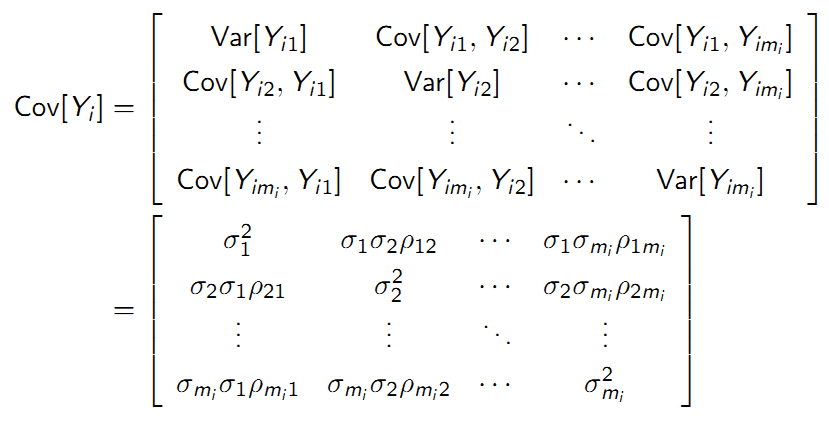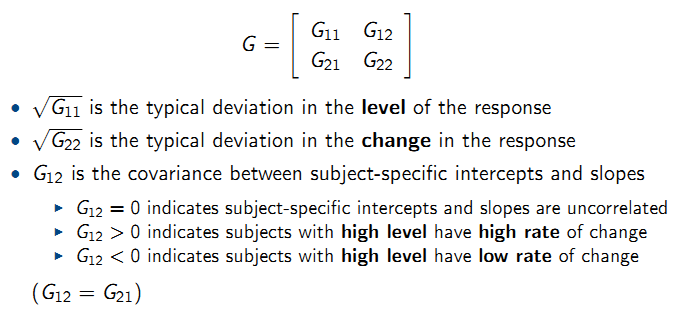Chapter 9 Longitudinal Data Analysis with R
Ho Thi Nhan, MD, PhD
9.1 Longitudinal studies
- Collect data on the same individuals repeatedly over time
9.1.1 Benefits
Advantages of longitudinal studies as compared to cross-sectional studies:
- Record incident events
- Ascertain exposure prospectively
- Separate time effects: cohort, period, age
- Distinguish changes over time within individuals
- Help establish causal effect of exposure on outcome
- Repeated measures over time of the same subjects.
9.1.2 Separate time effects: cohort, period, age
9.1.2.1 Cohort effects
- Differences between individuals at baseline
- “Level”
- Example: Younger individuals begin at a higher level
9.1.3 Distinguish changes over time within individuals
Age can be partitioned into two components:
- Cross-sectional comparison: \(E[Y_{i1}]\) = \(\beta_{0}\) + \(\beta_{C}x_{i1}\)
- Longitudinal comparison: \(E[Y_{ij} - Y_{i1}]\) = \(\beta_{L}(x_{ij}-x_{i1})\) for observation j = 1,…,mi on subject i = 1,…,n.
- Putting these two models together: \(E[Y_{ij}]\) = \(\beta_{0}\) + \(\beta_{C}x_{i1}\) + \(\beta_{L}(x_{ij}-x_{i1})\)
- \(\beta_{L}\) represents the expected change in the outcome per unit change in age for a given subject.
9.1.4 Longitudinal vs. cross-sectional
For comparison of groups A and B:
- Cross-sectional:
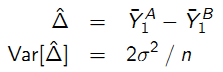
- Longitudinal:
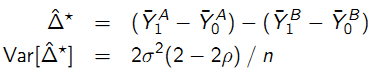
- Benefits of Longitudinal estimate: may be more precise, may reduce bias because each subject “acts as their own control”.
9.2 Example: Bangladesh data
This is a published data from a cohort study of 50 healthy Bangladeshi infants.
#load multiple packages
packages<-c("rio","tidyverse","knitr","lme4","mgcv","geepack","gee","ggplot2","sjPlot","sjlabelled","sjmisc") #,"longCatEDA"
lapply(packages, library, character.only = TRUE)
#import sheet 2 only
sh2<-import("./metamicrobiomeR-master/inst/extdata/QIIME_outputs/Bangladesh/nature13421-s2.xlsx",sheet=2,skip=1)
#Rename column names
oldname<-colnames(sh2)
newname<-c("cohort",
"family.id",
"child.id",
"sample.id",
"age.d",
"age.m",
"whz","waz","haz",
"breast.milk",
"formula",
"solid",
"diarrhea",
"antibiotic.7d",
"medication",
"no.sequence",
"serun.id",
"barcode")
sh2<-sh2 %>% rename_at(vars(oldname), ~newname)
#colnames(sh2)
#remove unused rows
sh2<-sh2 %>%
filter(!is.na(family.id))
#View(sh2)9.3 Options for analysis of change
Does mean change differ across groups?
9.3.1 Pre-post analysis
- Same baseline (e.g.randomization): analysis of post or change
- Different baseline: analysis of post or change adjusting for baseline
=> Simple pre-post data don’t incorporate correlation within individuals.
9.3.2 Longitudinal regression models
GEE and mixed-effects models Gałecki and Burzykowski (2013):
- Exploratory data analysis
- Regression model specification
- Parameter interpretation
- Covariance and correlation
9.4 Exploratory data analysis
9.4.1 Summarize mean and covariance structure
- Summary statistics over time (by groups)
- Individual plots of observed and fitted values
- Empirical covariance structure (variance and correlation)
9.4.2 Main points
- Show as much of the data as possible, rather than only summaries
- Highlight aggregate patterns of potential scientific interest
- Identify both cross-sectional and longitudinal patterns
- Facilitate the identification of unusual individuals or observations
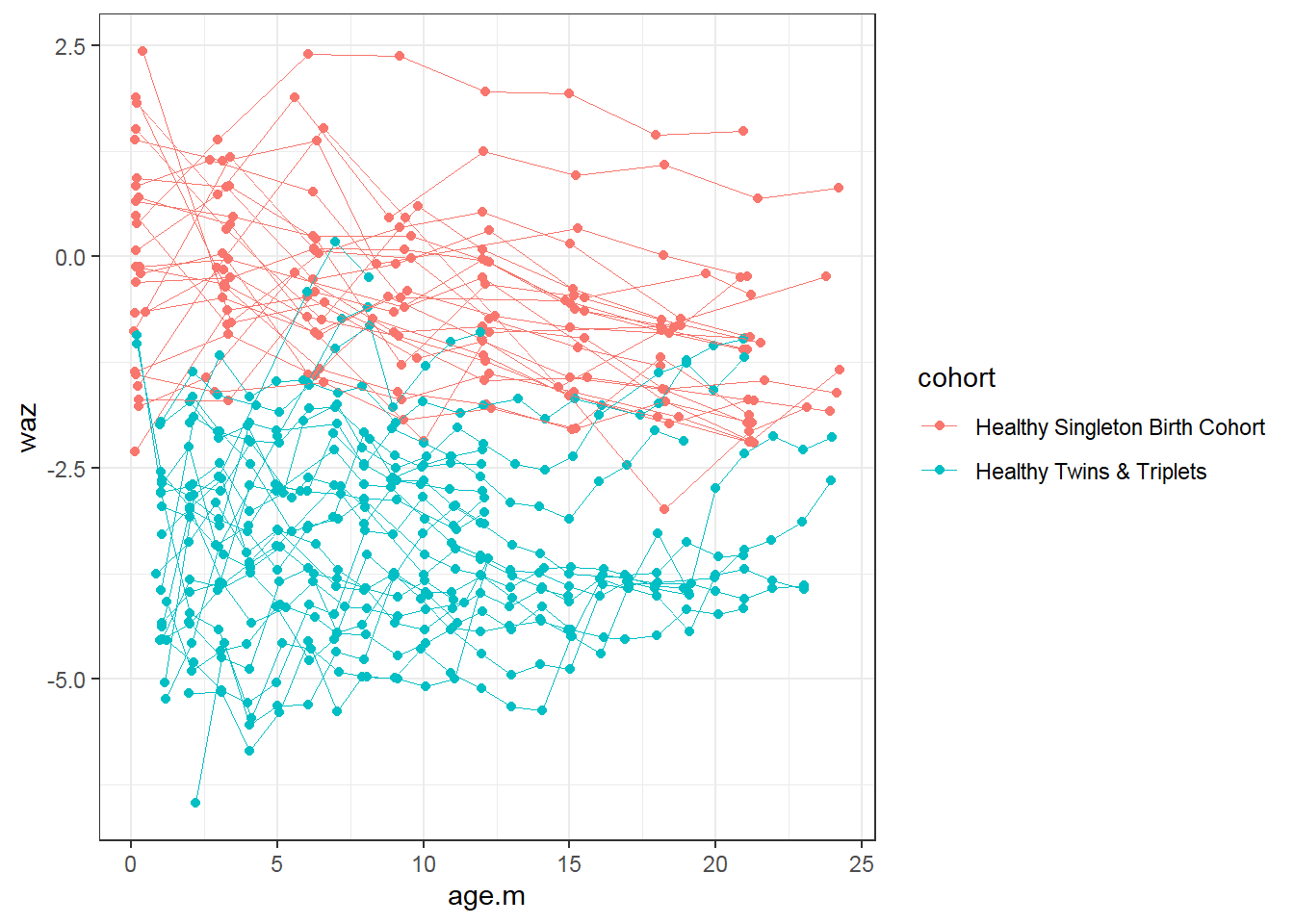
9.4.3 Longitudinal data visualization: Spaghetti plot
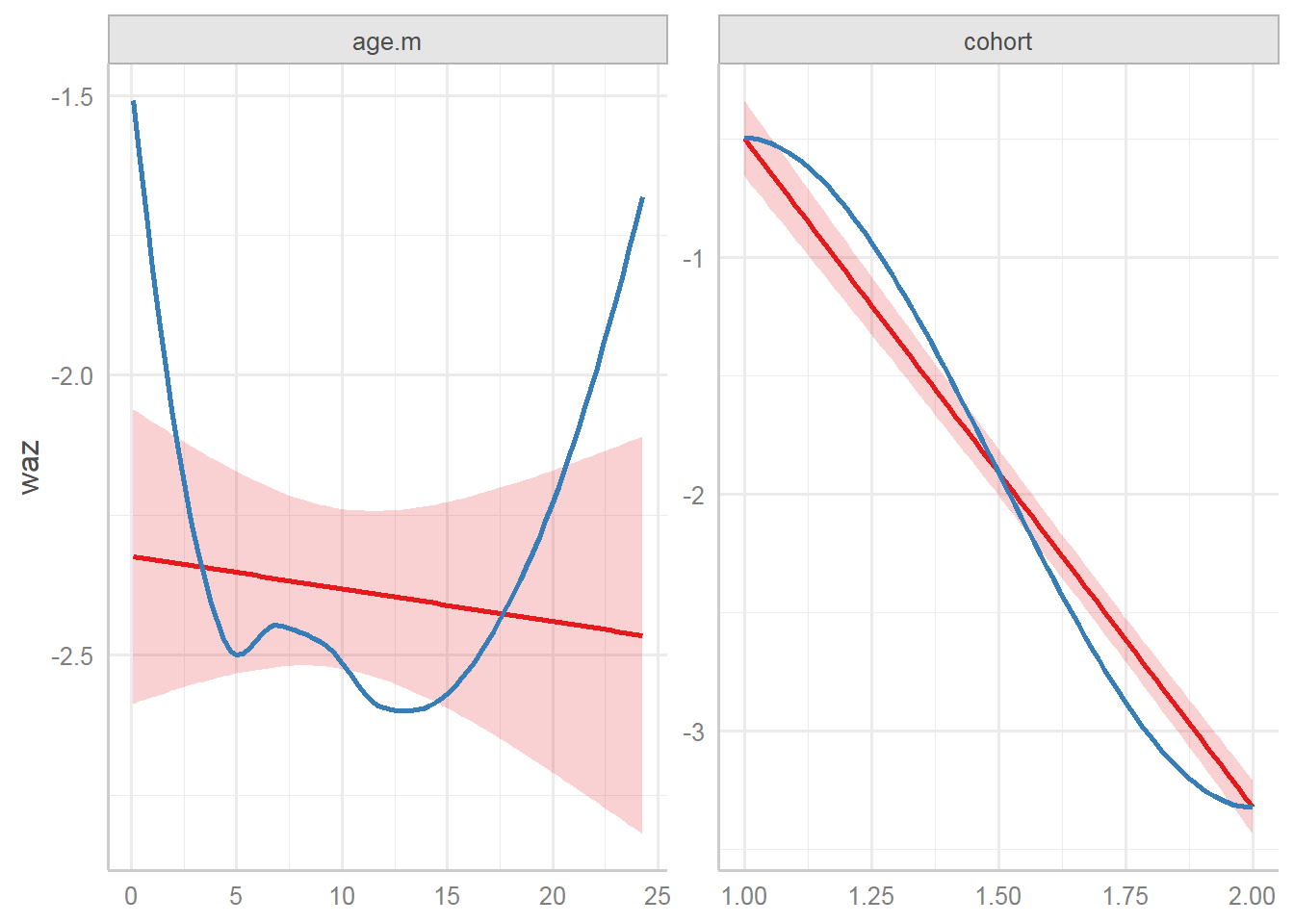
For continuous variables:
sh2 %>%
filter(!is.na(whz)) %>%
ggplot()+
geom_point(aes(x = age.m, y = waz, group = child.id, colour=cohort))+
geom_line(aes(x = age.m, y = waz, group = child.id, colour=cohort),size=0.3)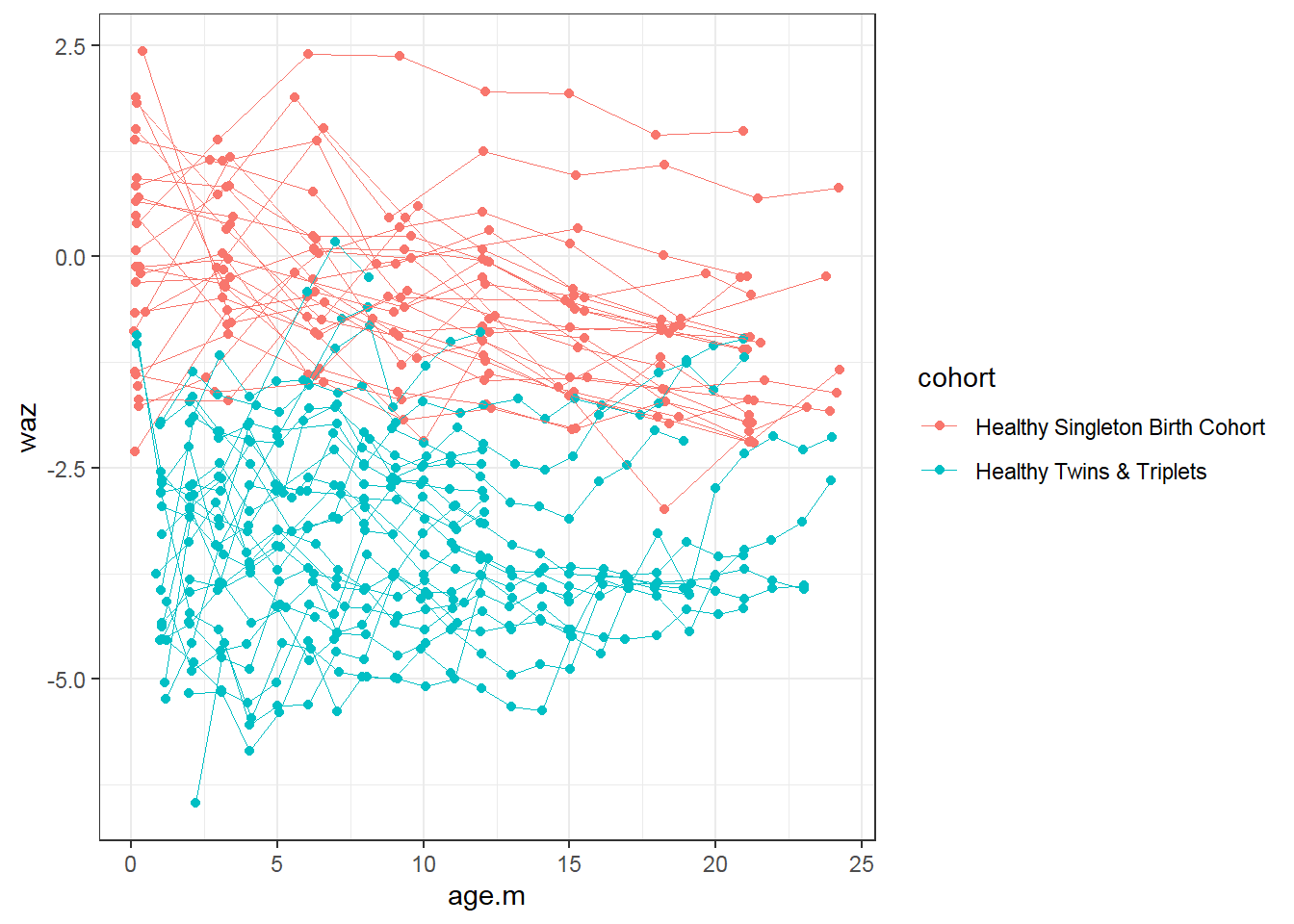
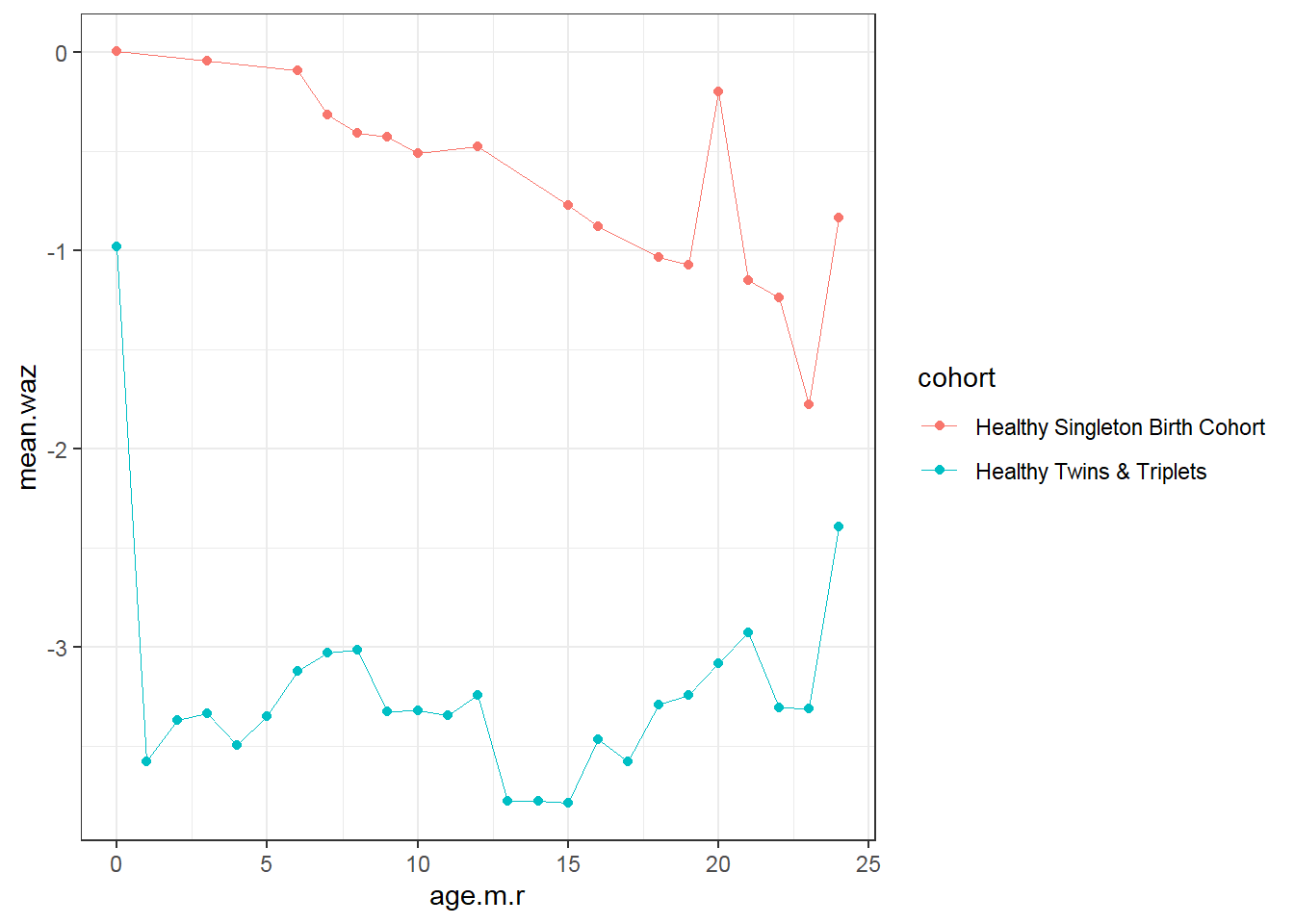
9.4.4 Plot mean
On average…
- Trend: Weight for age z score (waz) decreases over time for singletons; not change in twins-triplets
- Cross-sectional: singletons have larger waz than twins-triplets at every age
- Longitudinal: Decrease in average waz is larger for singletons
d1<-sh2 %>%
filter(!is.na(whz)) %>%
mutate(age.m.r=round(age.m,0)) %>%
group_by(age.m.r, cohort)%>%
summarise(mean.waz=mean(waz,na.rm=T))
d1 %>%
ggplot()+
geom_point(aes(x = age.m.r, y = mean.waz, colour=cohort))+
geom_line(aes(x = age.m.r, y = mean.waz, colour=cohort),size=0.3) 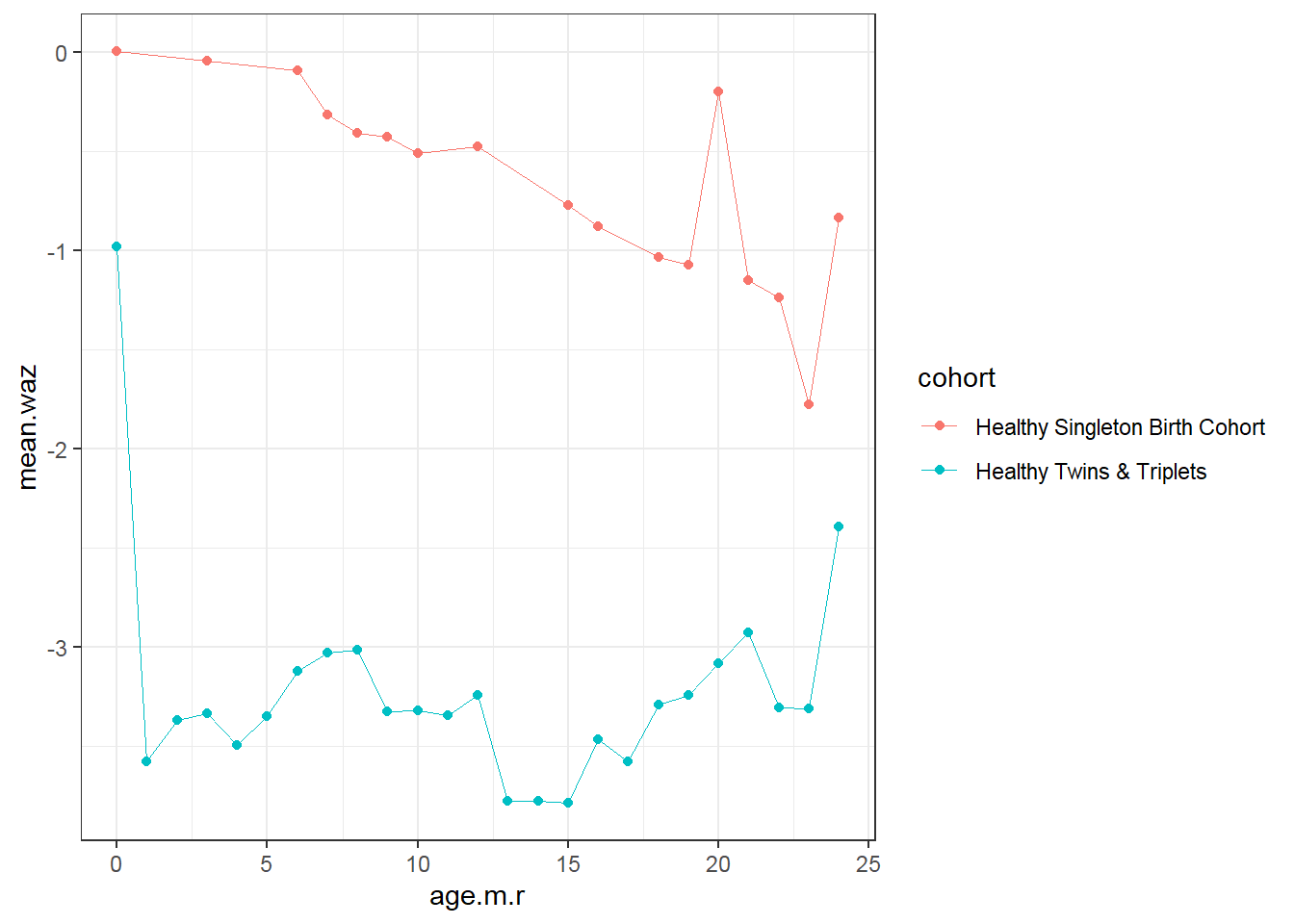
9.4.5 Linear model for time
9.4.5.1 Mean Model
time=age or “age.m” in this data:
\(E[Y_{ij}|age_{ij}]\)= \(\beta_{0} + \beta_{1}age_{ij}\)
Rate of Change - Slope of the curve \(\beta_{1}\) - Constant rate of change
9.4.5.3 Research question
- Rate of change = time slope \(E[waz_{ij}|x_{ij} = (age.m, cohort)]\) = \(\beta_{0}(x_{ij}) + \beta_{1}(x_{ij})time_{ij}\)
##
## Call:
## lm(formula = waz ~ age.m, data = sh2)
##
## Residuals:
## Min 1Q Median 3Q Max
## -4.134 -1.446 -0.114 1.333 4.758
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.323415 0.134893 -17.224 <2e-16 ***
## age.m -0.005831 0.011508 -0.507 0.613
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.742 on 571 degrees of freedom
## (423 observations deleted due to missingness)
## Multiple R-squared: 0.0004494, Adjusted R-squared: -0.001301
## F-statistic: 0.2567 on 1 and 571 DF, p-value: 0.6126- Does the rate of change differ between singletons vs. twins-triplets? \(E[Y_{ij}]\) = \(\beta_{0} + \beta_{1}age_{ij} + \beta_{2}cohort_{i} + \beta_{3}age_{ij}*cohort_{i}\)
##
## Call:
## lm(formula = waz ~ age.m + cohort + age.m * cohort, data = sh2)
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.1604 -0.7730 -0.1336 0.7111 3.4986
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.11060 0.14276 0.775 0.438815
## age.m -0.05676 0.01110 -5.113 4.34e-07 ***
## cohortHealthy Twins & Triplets -3.41607 0.17838 -19.150 < 2e-16 ***
## age.m:cohortHealthy Twins & Triplets 0.05487 0.01469 3.734 0.000207 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.096 on 569 degrees of freedom
## (423 observations deleted due to missingness)
## Multiple R-squared: 0.6055, Adjusted R-squared: 0.6034
## F-statistic: 291.1 on 3 and 569 DF, p-value: < 2.2e-169.4.5.4 Parameter interpretation
- \(\beta_{1}\) = expected waz change (per month) for singletons (reference group)
- \(\beta_{2}\) = expected difference in waz comparing twins_triplets to singletons at first visit (newborn)
- \(\beta_{3}\) = expected difference in waz change (per month) between twins_triplets and singletons
9.4.5.5 Plot with linear regression line
Using “glm” => ignore longitudinal data.
sh2 %>%
filter(!is.na(whz)) %>%
ggplot()+
geom_point(aes(x = age.m, y = waz, group = child.id, colour=cohort))+
geom_line(aes(x = age.m, y = waz, group = child.id, colour=cohort),size=0.3)+
stat_smooth(aes(x = age.m, y = waz, colour=cohort), method = 'glm',size=2)+
geom_point(aes(x = age.m.r, y = mean.waz, colour=cohort), size=4, pch=17,data=d1)+
geom_line(aes(x = age.m.r, y = mean.waz, colour=cohort),size=2,pch=17,data=d1) 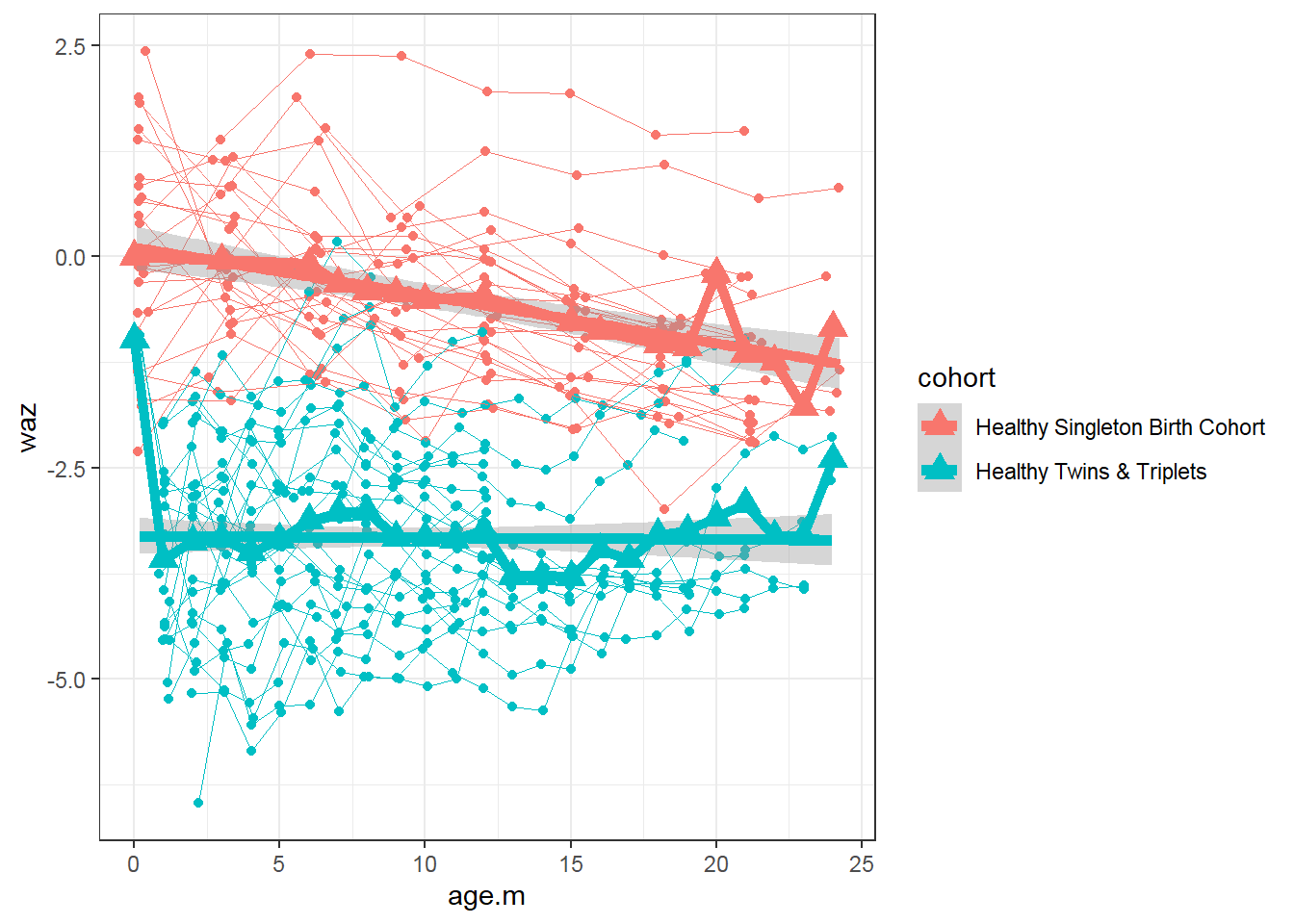
9.4.6 Issues of linear model
The below are not accounted.
9.4.6.1 Dependence & Correlation
Response variables measured on the same subject are correlated. - Observations are dependent or correlated when one variable does predict the value of another variable (waz at month 1 may predict waz at month 2)
9.4.6.2 Variance
The variance measures the average distance that an observation falls away from the mean. The variance of a variable \(Y_{ij}\) (fix time j) is defined as:
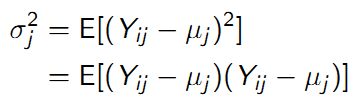
9.4.6.3 Covariance
The covariance measures whether, on average, departures in one variable \(Y_{ij}-\mu_{j}\) ‘go together with’ departures in a second variable \(Y_{ik}-\mu_{k}\).
The covariance of two variables \(Y_{ij}\) and \(Y_{ik}\) is: \(\sigma_{jk}\) = \(E[(Y_{ij}-\mu_{j})(Y_{ik}-\mu_{k})]\)
Simple linear regression covariance:
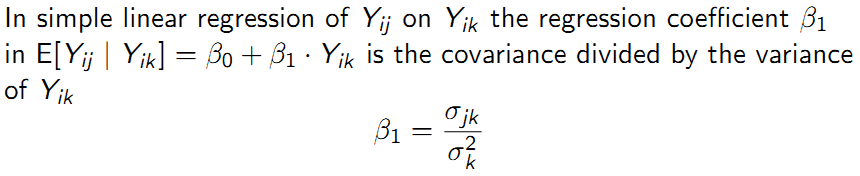
9.5 Generalized estimating equations (GEE)
9.5.1 GEE: example Bangladesh data
Characterize waz change among singletons and twins-triplets:
- Estimate the average waz change among all children
- Estimate the waz change for individual children
- Characterize the degree of heterogeneity across children
- Identify factors that predict waz change
9.5.2 Introduction to GEE
Contrast average outcome values across populations of individuals defined by covariate values, while accounting for correlation.
- Focus on a generalized linear model with regression parameters \(\beta\), which characterize the systemic variation in Y across covariates X.
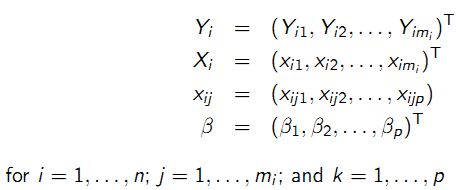
- Longitudinal correlation structure is a nuisance feature of the data.
9.5.3 Assumptions
- Observations are independent across subjects
- Observations may be correlated within subjects
9.5.4 Mean model
Primary focus of the analysis:
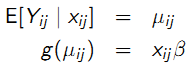
- May correspond to any generalized linear model with link g(.)
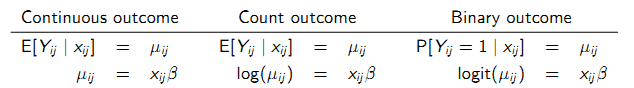
- Characterizes a marginal mean regression model
9.5.5 Marginal mean
Definition: \(\mu_{ij}\) does not condition on anything other than \(x_{ij}\) . Mixed effect and transition model:
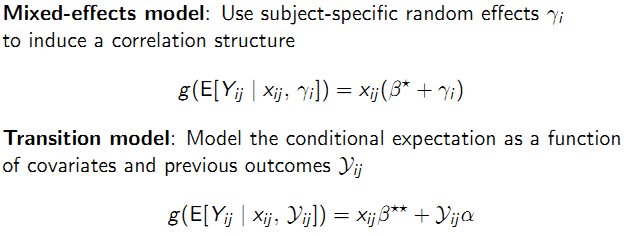
9.5.6 Covariance model
Longitudinal correlation is a nuisance; secondary to mean model of interest.
- Assume a form for variance that may depend on \(\mu_{ij}\) which may also include a scale or dispersion parameter \(\phi\) > 0.
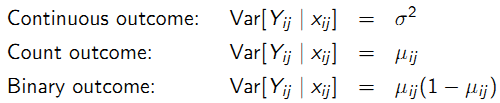
- Select a model for longitudinal correlation with parameters \(\alpha\).
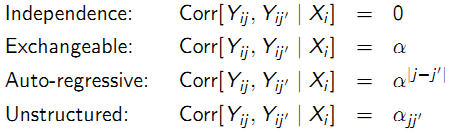
9.5.7 Correlation models
Correlation between any two observations on the same subject:
- Independence: is assumed to be zero (observations over time are independent). Always appropriate with use of robust variance estimator (large n)
- Exchangeable: is assumed to be constant (all observations over time have the same correlation). More appropriate for clustered data
- Auto-regressive: is assumed to depend on time or distance (correlation decreases as a power of how many timepoints apart two observations are). More appropriate for equally-spaced longitudinal data
- Unstructured: is assumed to be distinct for each pair (correlation between all timepoints may be different). Only appropriate for short series (small m) on many subjects (large n)
9.5.8 Semi-parametric
- Specification of a mean model and correlation model does not identify a complete probability model for the outcomes
- The [mean, correlation] model is semi-parametric because it only specifies the first two moments of the outcomes
- Additional assumptions are required to identify a complete probability model and a corresponding parametric likelihood function (GLMM)
Note: Construct an unbiased estimating function to estimate \(\beta\) and generate valid statistical inference, while accounting for correlation.
9.5.9 Generalized estimating equations: Intuition
Generalized estimating equations: Intuition
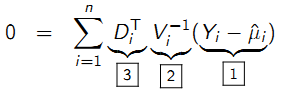
- [1] The model for the mean, \(\mu_{i}(\beta)\), is compared to the observed data, \(Y_{i}\); setting the equations to equal 0 tries to minimize the difference between observed and expected.
- [2] Estimation uses the inverse of the variance (covariance) to weight the data from subject i ; more weight is given to differences between observed and expected for those subjects who contribute more information
- [3] This is simply a ‘change of scale’ from the scale of the mean, \(\mu_{i}(\beta)\), to the scale of the regression coefficients (covariates)
9.5.10 Properties of \(\hat{\beta}\)
- \(\hat{\beta}\) is a consistent estimator for \(\beta\) even if the model for longitudinal correlation is incorrectly specified, i.e. \(\hat{\beta}\) is ‘robust’ to correlation model mis-specification
- However, the variance of \(\hat{\beta}\) must capture the correlation in the data, either by choosing the correct correlation model, or via an alternative variance estimator
- Selecting an approximately correct correlation model will yield a more efficient estimator for \(\beta\), i.e. \(\hat{\beta}\) has the smallest variance (standard error) if the correlation model is correctly specified
9.5.11 GEE: comments
- GEE is specified by a mean model and a correlation model
- A regression model for the average outcome, e.g. linear, logistic;
- A model for longitudinal correlation, e.g. independence, exchangeable
- GEE also computes an empirical variance estimator (aka sandwich, robust, or Huber-White variance estimator)
- Empirical variance estimator provides valid standard errors for \(\hat{\beta}\) even if the correlation model is incorrect, but requires n >= 40 and n >> m.
Note: Although the correlation model does not need to be correctly specified to obtain a consistent estimator for \(\beta\) or valid standard errors for \(\hat{\beta}\), selecting a non-independence or weighted correlation structure permits use of the model-based variance estimator and may provide improved efficiency for \(\hat{\beta}\).
9.5.12 Inference for \(\beta\)
- Wald test
- Note: Likelihood ratio test not available; not relied on a likelihood function
9.5.13 GEE: example Bangladesh data
9.5.13.1 Objectives
Characterize waz change among singletons and twins-triplets:
- Estimate the average waz change among all children
- Estimate the waz change for individual children
- Characterize the degree of heterogeneity across children
- Identify factors that predict waz change
9.5.13.2 Approach
- Examine waz change among twins_triplets and singletons: \(E[Y_{ij}]\) = \(\beta_{0} + \beta_{1}age_{ij} + \beta_{2}cohort{i} + \beta_{3}age_{ij}*cohort_{i}\)
- Consider various specifications for the ‘working’ correlation structure:
- Independence;
- Exchangeable;
- Auto-regressive;
- Unstructured;
- Selection of a working correlation structure should be guided by a prior knowledge and/or exploratory analysis
9.5.13.3 GEE trying different correlation structure
This may be done with “geepack” package (Højsgaard, Halekoh, and Yan 2024):
#library(geepack)
waz.nona<-sh2 %>%
filter(!is.na(waz))
g.i<-geeglm(waz~age.m +cohort +age.m*cohort, id=child.id,corstr="independence", data=waz.nona)
summary(g.i)##
## Call:
## geeglm(formula = waz ~ age.m + cohort + age.m * cohort, data = waz.nona,
## id = child.id, corstr = "independence")
##
## Coefficients:
## Estimate Std.err Wald Pr(>|W|)
## (Intercept) 1.106e-01 4.463e-16 6.141e+28 <2e-16 ***
## age.m -5.676e-02 4.174e-17 1.849e+30 <2e-16 ***
## cohortHealthy Twins & Triplets -3.416e+00 7.189e-17 2.258e+33 <2e-16 ***
## age.m:cohortHealthy Twins & Triplets 5.487e-02 3.328e-18 2.718e+32 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation structure = independence
## Estimated Scale Parameters:
##
## Estimate Std.err
## (Intercept) 1.193 3.975e-17
## Number of clusters: 1 Maximum cluster size: 573g.e<-geeglm(waz~age.m +cohort +age.m*cohort, id=child.id,corstr="exchangeable", data=waz.nona)
summary(g.e)##
## Call:
## geeglm(formula = waz ~ age.m + cohort + age.m * cohort, data = waz.nona,
## id = child.id, corstr = "exchangeable")
##
## Coefficients:
## Estimate Std.err Wald Pr(>|W|)
## (Intercept) 0.1106 NaN NaN NaN
## age.m -0.0568 NaN NaN NaN
## cohortHealthy Twins & Triplets -3.4161 NaN NaN NaN
## age.m:cohortHealthy Twins & Triplets 0.0549 NaN NaN NaN
##
## Correlation structure = exchangeable
## Estimated Scale Parameters:
##
## Estimate Std.err
## (Intercept) 1.19 1.04e-16
## Link = identity
##
## Estimated Correlation Parameters:
## Estimate Std.err
## alpha -0.00175 3.39e-19
## Number of clusters: 1 Maximum cluster size: 573##
## Call:
## geeglm(formula = waz ~ age.m + cohort + age.m * cohort, data = waz.nona,
## id = child.id, corstr = "ar1")
##
## Coefficients:
## Estimate Std.err Wald Pr(>|W|)
## (Intercept) -4.99e-02 1.96e-06 6.47e+08 <2e-16 ***
## age.m -5.45e-02 1.23e-09 1.96e+15 <2e-16 ***
## cohortHealthy Twins & Triplets -3.51e+00 2.38e-06 2.17e+12 <2e-16 ***
## age.m:cohortHealthy Twins & Triplets 8.53e-02 4.48e-08 3.62e+12 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Correlation structure = ar1
## Estimated Scale Parameters:
##
## Estimate Std.err
## (Intercept) 1.23 3.08e-07
## Link = identity
##
## Estimated Correlation Parameters:
## Estimate Std.err
## alpha 0.827 2.13e-07
## Number of clusters: 1 Maximum cluster size: 573#g.u<-geeglm(waz~age.m +cohort +age.m*cohort, id=child.id,corstr="unstructured", data=waz.nona)
#summary(g.u)
#OLS
#=> SE are larger
fit.ols<-lm(waz~age.m +cohort +age.m*cohort,data=waz.nona)
summary(fit.ols)##
## Call:
## lm(formula = waz ~ age.m + cohort + age.m * cohort, data = waz.nona)
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.160 -0.773 -0.134 0.711 3.499
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.1106 0.1428 0.77 0.43882
## age.m -0.0568 0.0111 -5.11 4.3e-07 ***
## cohortHealthy Twins & Triplets -3.4161 0.1784 -19.15 < 2e-16 ***
## age.m:cohortHealthy Twins & Triplets 0.0549 0.0147 3.73 0.00021 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.1 on 569 degrees of freedom
## Multiple R-squared: 0.605, Adjusted R-squared: 0.603
## F-statistic: 291 on 3 and 569 DF, p-value: <2e-169.5.14 GEE summary
9.5.14.1 Main characteristics
- Primary focus of the analysis is a marginal mean regression model that corresponds to any GLM
- Longitudinal correlation is secondary and is treated as a nuisance feature
- Need ‘working’ correlation model
- Semi-parametric: Only the mean and correlation models are specified
- Likelihood ratio test statistics are unavailable; hypothesis testing with GEE uses Wald statistics
- Working correlation model does not need to be correctly specified to obtain a consistent estimator for \(\beta\) or valid standard errors for \(\hat{\beta}\), but efficiency gains are possible if the correlation model is correct
9.6 Generalized linear mixed-effects models (GLMM)
9.6.1 Example data: Bangladesh
Characterize waz change among singletons and twins-triplets:
- Estimate the average waz change among all children
- Estimate the waz change for individual children
- Characterize the degree of heterogeneity across children
- Identify factors that predict waz change
9.6.2 GLMM overview
Contrast outcomes both within and between individuals:
- Assume that each subject has a regression model characterized by subject-specific parameters: a combination of fixed-effects parameters common to all individuals in the population and random-effects parameters unique to each individual subject
- Although covariates allow for differences across subjects, typically cannot measure all factors that give rise to subject-specific variation
- Subject-specific random effects induce a correlation structure
9.6.4 Linear mixed-effects model
For a continuous outcome \(Y_{ij}\):
- Stage 1: Model for response given random effects:
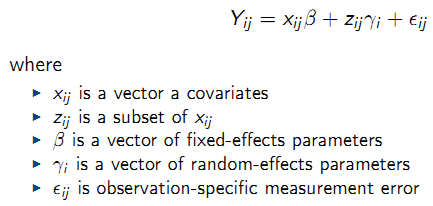
- Stage 2: Model for random effects:
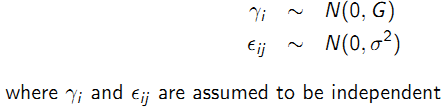
9.6.5 Choices for random effects
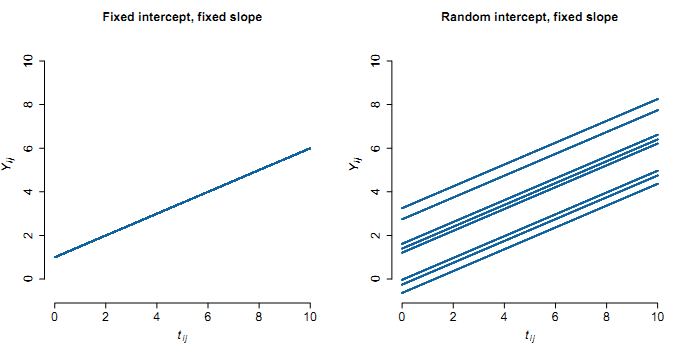
- Random intercepts:
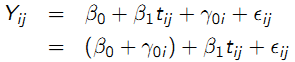
Fixed and Random Intercepts illustration:
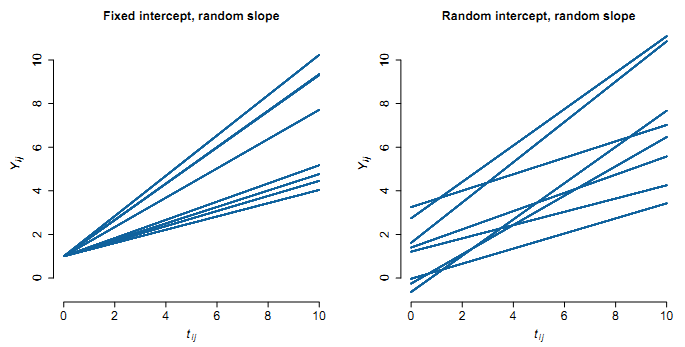
- Random intercepts and slopes:
Random slope illustration:
9.6.8 Generalized linear mixed-effects models
A GLMM is defined by random and systematic components:
- Random: Conditional on \(\gamma_{i}\) the outcomes \(Y_{i} = (Y_{i1},...,Y_{im_{i}})^T\) are mutually independent and have an exponential family density.
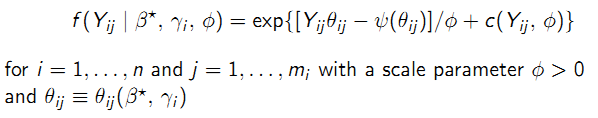
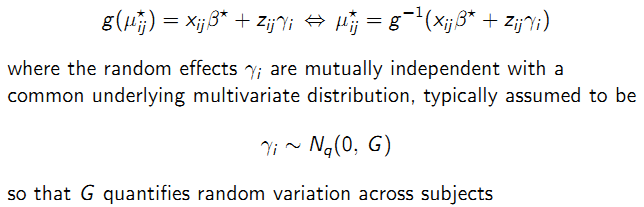
- Systematic: \(\mu_{ij}^\star\) is modeled via a linear predictor containing fixed regression parameters \(\beta^\star\) common to all individuals in the population and subject-specific random effects \(\gamma_{i}\) with a known link function \(g(.)\).
9.6.8.1 Likelihood-based estimation
Likelihood-based estimation of \(\beta\): Requires specification of a complete probability distribution for the data
- Likelihood-based methods are designed for fixed effects, so integrate over the assumed distribution for the random effects:
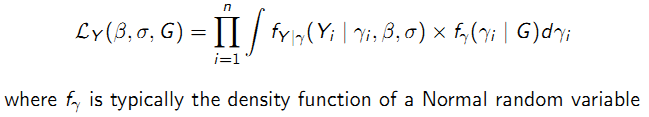
Two likelihood-based approaches to estimation using a GLMM:
- Conditional likelihood: Treat the random effects as if they were fixed parameters and eliminate them by conditioning on their sufficient statistics; does not require a specified distribution for \(\gamma_{i}\).
- Maximum likelihood: Treat the random effects as unobserved nuisance variables and integrate over their assumed distribution to obtain the marginal likelihood for \(\beta\); typically assume \(\gamma_{i}\)~\(N(0; G)\). (lmer and glmer in R package lme4)
9.6.9 Fixed effects vs. random effects
Fixed-effects approach provided by conditional likelihood estimation.
- Comparisons are made within individuals who act as their own control and differences are averaged across all individuals in the sample
- May eliminate potentially large sources of bias by controlling for all stable characteristics of the individuals under study (+)
- Variation across subjects is ignored, which may provide standard error estimates that are too big; conservative inference (-)
- Although controlled for by conditioning, cannot estimate coefficients for covariates that have no within-subject variation (-/+)
Random-effects approach provided by maximum likelihood estimation
- Comparisons are based on within- and between-subject contrasts
- Requires a specified distribution for subject-specific effects; correct specification is required for valid likelihood-based inference (-/+)
- Do not control for unmeasured characteristics because random effects are almost always assumed to be uncorrelated with covariates (-)
- Can estimate effects of within- and between-subject covariates (+)
9.6.9.1 Inference for \(\beta\)
Testing fixed effects in nested linear mixed-effects models:
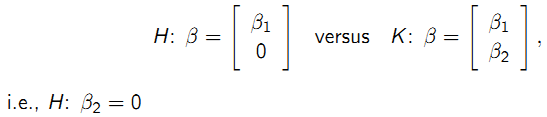
- Likelihood ratio test is valid if ML estimation is used
- Likelihood ratio test may not be valid with other estimation methods
- Wald test is generally valid
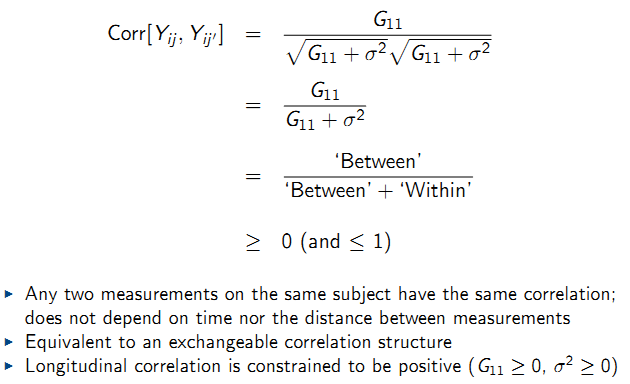
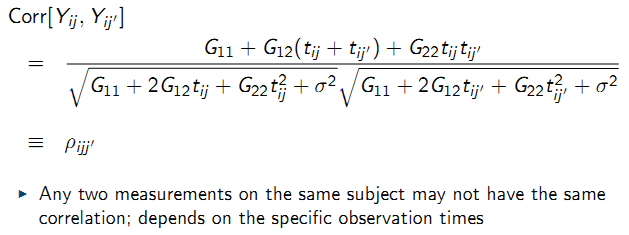
9.6.9.2 Inference for G
Testing whether a random intercept model is adequate:
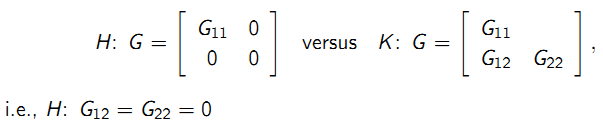
- Adequate covariance modeling is useful for the interpretation of the random variation in the data
- Over-parameterization of the covariance structure leads to inefficient estimation of fixed effects parameters \(\beta\)
- Covariance model choice determines the standard error estimates for \(\hat{\beta}\); correct model is required for correct standard error estimates
9.6.9.3 Assumptions
Valid inference from a linear mixed-effects model relies on:
- Mean model: As with any regression model for an average outcome, need to correctly specify the functional form of \(x_{ij}\beta\) (here also \(z_{ij}\gamma_{i}\). – Included important covariates in the model – Correctly specified any transformations or interactions
- Covariance model: Correct covariance model (random-effects specification) is required for correct standard error estimates for \(\hat{\beta}\).
- Normality: Normality of \(\epsilon_{ij}\) and \(\gamma_{i}\) is required for normal likelihood function to be the correct likelihood function for \(Y_{ij}\).
- \(n\) sufficiently large for asymptotic inference to be valid
These assumptions must be verified to evaluate any fitted model.
9.6.10 Summary
- Mixed-effects models assume that each subject has a regression model characterized by subject-specific parameters; a combination of fixed effects parameters common to all individuals in the population and random subject-specific perturbations
- Likelihood-based estimation and inference requires a complete parametric probability distribution for subject-specific random effects and error terms that must be verified for valid inference
- Estimates for the random effects are available
9.7 Mixed effect model formula in R
Mixed effect model may be fitted in R using the “lme4” package (Bates et al. 2024).
Source: Fitting Linear Mixed-Effects Models Using lme4
9.7.1 Formula
- resp ~ FEexpr + (REexpr1 | factor1) + (REexpr2 | factor2) + …, where FEexpr is an expression determining the columns of the ffxed-effects model matrix, X, and the random-effects terms, (REexpr1 | factor1) and (REexpr2 | factor2), determine both the random-effects model matrix, Z, and the structure of the relative covariance factor, \(\Lambda_{\theta}\). In practice, the number of random-effects terms is typically low.
- Model formulas in R:
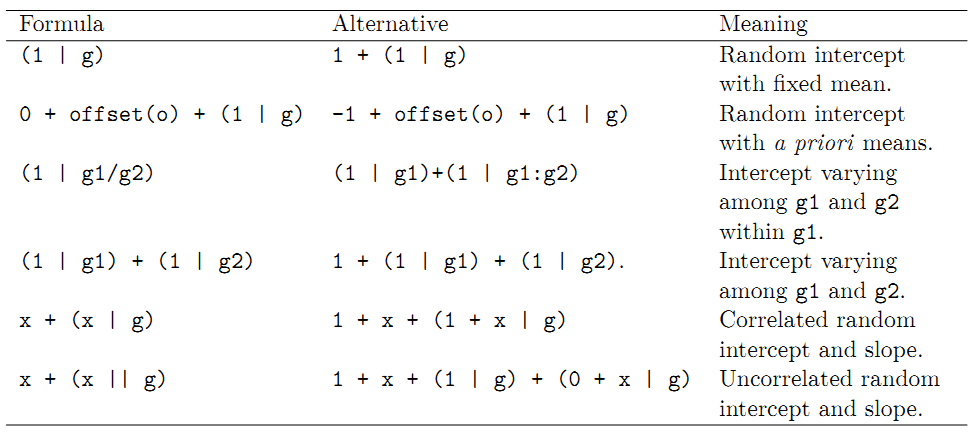
9.7.3 Example Bangladesh data
Characterize waz among singletons and twins_triplets. Consider various specifications for the random effects structure:
- Random intercepts
- Random intercepts and slopes (for age)
Note: selection of a random effects structure should be guided by a priori knowledge and/or exploratory analysis, or specified as relevant to the scientific question of interest
9.7.3.1 GLMM with lme4
Package ‘lmerTest’ to get p-values.
#library(lme4)
#library(lmerTest) #for p-value
waz.nona<-sh2 %>%
filter(!is.na(waz))
r.in<-lmer(waz ~ age.m*cohort+ (1 | child.id), data=waz.nona)
summary(r.in)## Linear mixed model fit by REML ['lmerMod']
## Formula: waz ~ age.m * cohort + (1 | child.id)
## Data: waz.nona
##
## REML criterion at convergence: 1344
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -4.437 -0.565 -0.025 0.552 4.973
##
## Random effects:
## Groups Name Variance Std.Dev.
## child.id (Intercept) 0.721 0.849
## Residual 0.464 0.681
## Number of obs: 573, groups: child.id, 50
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 0.09962 0.19176 0.52
## age.m -0.05498 0.00697 -7.89
## cohortHealthy Twins & Triplets -3.47286 0.26529 -13.09
## age.m:cohortHealthy Twins & Triplets 0.06998 0.00969 7.23
##
## Correlation of Fixed Effects:
## (Intr) age.m chHT&T
## age.m -0.386
## chrtHlthT&T -0.723 0.279
## ag.m:chHT&T 0.277 -0.719 -0.354## Linear mixed model fit by REML ['lmerMod']
## Formula: waz ~ age.m * cohort + (age.m | child.id)
## Data: waz.nona
##
## REML criterion at convergence: 1239
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.998 -0.510 0.007 0.525 4.531
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## child.id (Intercept) 0.9505 0.975
## age.m 0.0126 0.112 -0.44
## Residual 0.2939 0.542
## Number of obs: 573, groups: child.id, 50
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 0.1027 0.2077 0.49
## age.m -0.0549 0.0232 -2.37
## cohortHealthy Twins & Triplets -3.6032 0.2913 -12.37
## age.m:cohortHealthy Twins & Triplets 0.0997 0.0332 3.01
##
## Correlation of Fixed Effects:
## (Intr) age.m chHT&T
## age.m -0.475
## chrtHlthT&T -0.713 0.339
## ag.m:chHT&T 0.332 -0.699 -0.479
## optimizer (nloptwrap) convergence code: 0 (OK)
## Model failed to converge with max|grad| = 0.00410411 (tol = 0.002, component 1)There is a “moderate” correlation (-0.45) between random slopes and intercepts. If a strong correlation is observed, it is “over-parameterized” (too much of parameters”. In such cases, either intercept or slope is implemented as a random effect. We can also designate that intercepts and slopes are determined independently.
## Linear mixed model fit by REML ['lmerMod']
## Formula: waz ~ age.m * cohort + (1 | child.id) + (0 + age.m | child.id)
## Data: waz.nona
##
## REML criterion at convergence: 1248
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -3.369 -0.515 0.002 0.532 4.608
##
## Random effects:
## Groups Name Variance Std.Dev.
## child.id (Intercept) 0.8735 0.935
## child.id.1 age.m 0.0105 0.102
## Residual 0.2990 0.547
## Number of obs: 573, groups: child.id, 50
##
## Fixed effects:
## Estimate Std. Error t value
## (Intercept) 0.1019 0.2003 0.51
## age.m -0.0548 0.0213 -2.58
## cohortHealthy Twins & Triplets -3.5907 0.2807 -12.79
## age.m:cohortHealthy Twins & Triplets 0.0972 0.0305 3.19
##
## Correlation of Fixed Effects:
## (Intr) age.m chHT&T
## age.m -0.081
## chrtHlthT&T -0.714 0.058
## ag.m:chHT&T 0.057 -0.698 -0.086Comparing model with random intercept vs model with random intercept and slope. Random slope is significant.
## Data: waz.nona
## Models:
## r.in: waz ~ age.m * cohort + (1 | child.id)
## r.s: waz ~ age.m * cohort + (age.m | child.id)
## npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
## r.in 6 1337 1363 -662 1325
## r.s 8 1240 1275 -612 1224 100 2 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 19.7.3.2 Get fixed effects and random effects
## (Intercept) age.m
## 0.1027 -0.0549
## cohortHealthy Twins & Triplets age.m:cohortHealthy Twins & Triplets
## -3.6032 0.0997## $child.id
## (Intercept) age.m
## Bgsng7018 0.1813 -0.01865
## Bgsng7035 -1.4131 0.02076
## Bgsng7052 -0.1744 0.02352
## Bgsng7063 0.8686 -0.04558
## Bgsng7071 1.0960 -0.01241
## Bgsng7082 -0.7357 0.04352
## Bgsng7090 0.2326 -0.00878
## Bgsng7096 -0.3517 -0.02508
## Bgsng7106 0.0299 -0.02547
## Bgsng7114 -0.5781 0.03357
## Bgsng7115 0.5440 -0.01088
## Bgsng7128 -1.1239 0.03577
## Bgsng7131 -0.3477 -0.04017
## Bgsng7142 0.5960 -0.06901
## Bgsng7149 -0.0745 0.02303
## Bgsng7150 0.6207 -0.01086
## Bgsng7155 0.2284 -0.07241
## Bgsng7173 -0.9953 0.02941
## Bgsng7177 0.4226 -0.07760
## Bgsng7178 -1.1764 0.07566
## Bgsng7192 -0.9135 -0.00481
## Bgsng7202 0.5255 0.00144
## Bgsng7204 0.1668 0.01788
## Bgsng8064 1.1785 0.08485
## Bgsng8169 1.1933 0.03230
## Bgtw1.T1 0.3700 -0.04224
## Bgtw1.T2 0.7327 -0.09384
## Bgtw10.T1 1.4475 -0.10310
## Bgtw10.T2 0.2469 0.02256
## Bgtw11.T1 0.2106 -0.09735
## Bgtw11.T2 0.6967 -0.06167
## Bgtw12.T1 0.1434 -0.19251
## Bgtw12.T2 0.6882 -0.17715
## Bgtw2.T1 -2.0567 0.02965
## Bgtw2.T2 -0.9605 -0.02606
## Bgtw3.T1 1.6332 -0.02464
## Bgtw3.T2 0.5931 0.02242
## Bgtw4.T1 -0.0146 -0.03925
## Bgtw4.T2 0.3240 -0.07395
## Bgtw4.T3 0.5405 -0.00948
## Bgtw5.T1 -2.0004 0.06607
## Bgtw5.T2 0.2573 -0.10866
## Bgtw6.T1 -0.4264 0.40051
## Bgtw6.T2 -1.2643 0.41722
## Bgtw7.T1 0.2326 -0.11179
## Bgtw7.T2 -0.6600 -0.06444
## Bgtw8.T1 1.5170 0.03427
## Bgtw8.T2 0.8968 -0.00457
## Bgtw9.T1 -1.3654 0.08765
## Bgtw9.T2 -1.7823 0.15036
##
## with conditional variances for "child.id"9.7.3.3 Visualization of results
Three types of plots:
9.7.3.3.1 Coefficients
Estimates:
#install.packages("glmmTMB")
#library(sjPlot)
#library(sjlabelled)
#library(sjmisc)
#library(ggplot2)
theme_set(theme_sjplot())
plot_model(r.s)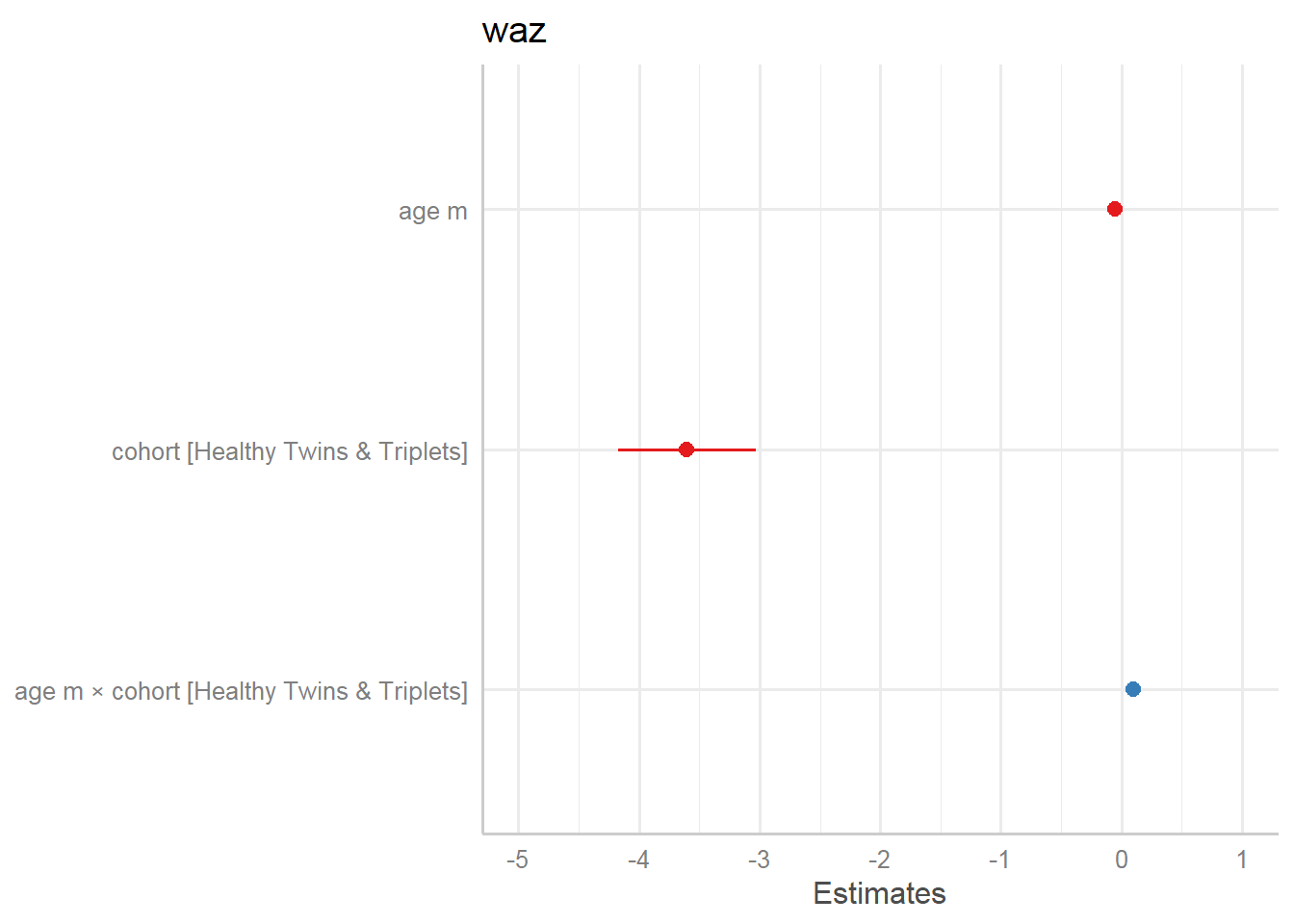
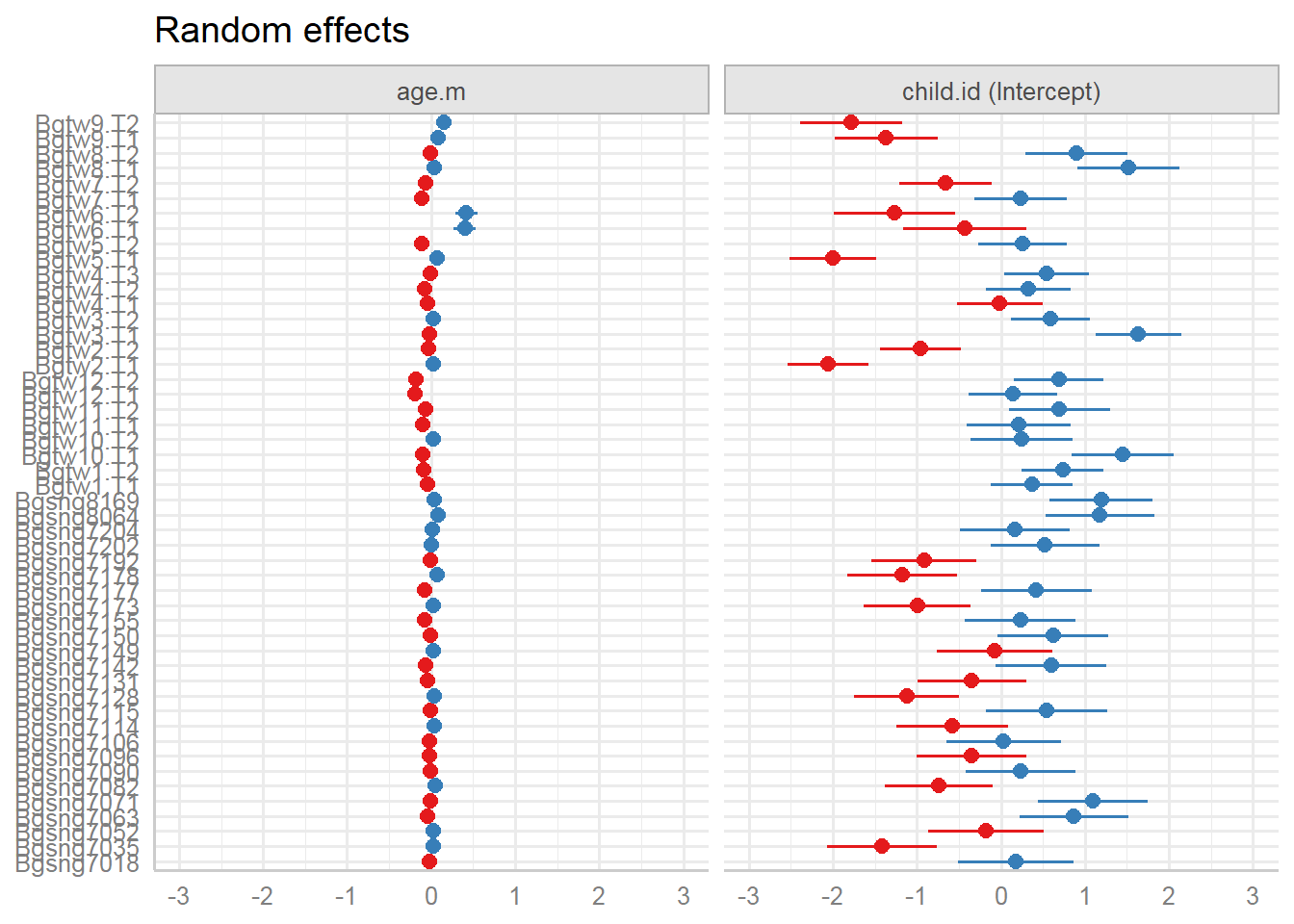
Random effects:
9.7.3.3.2 Marginal effects
Predicted values (marginal effects) for specific model terms:
## $age.m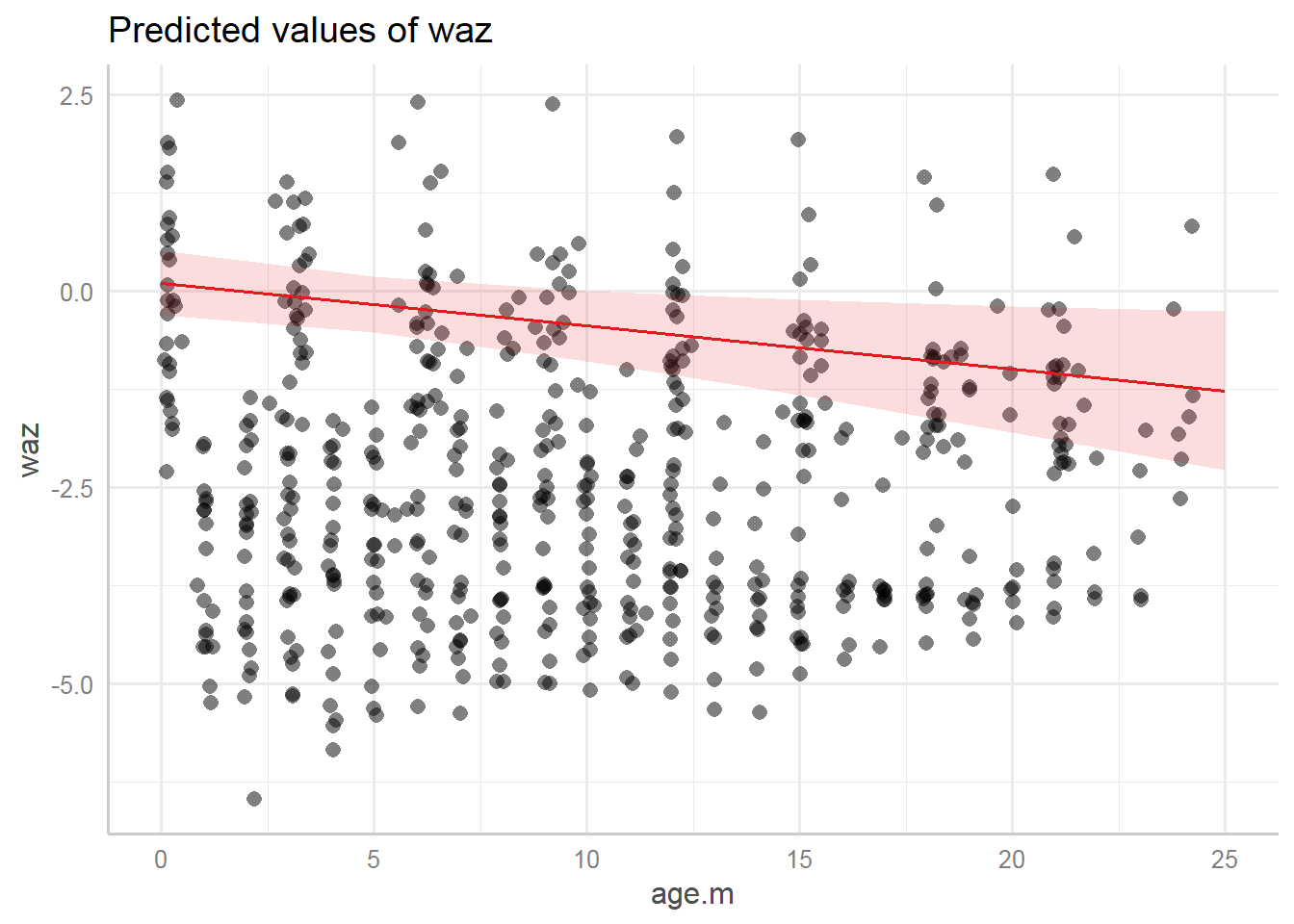
##
## $cohort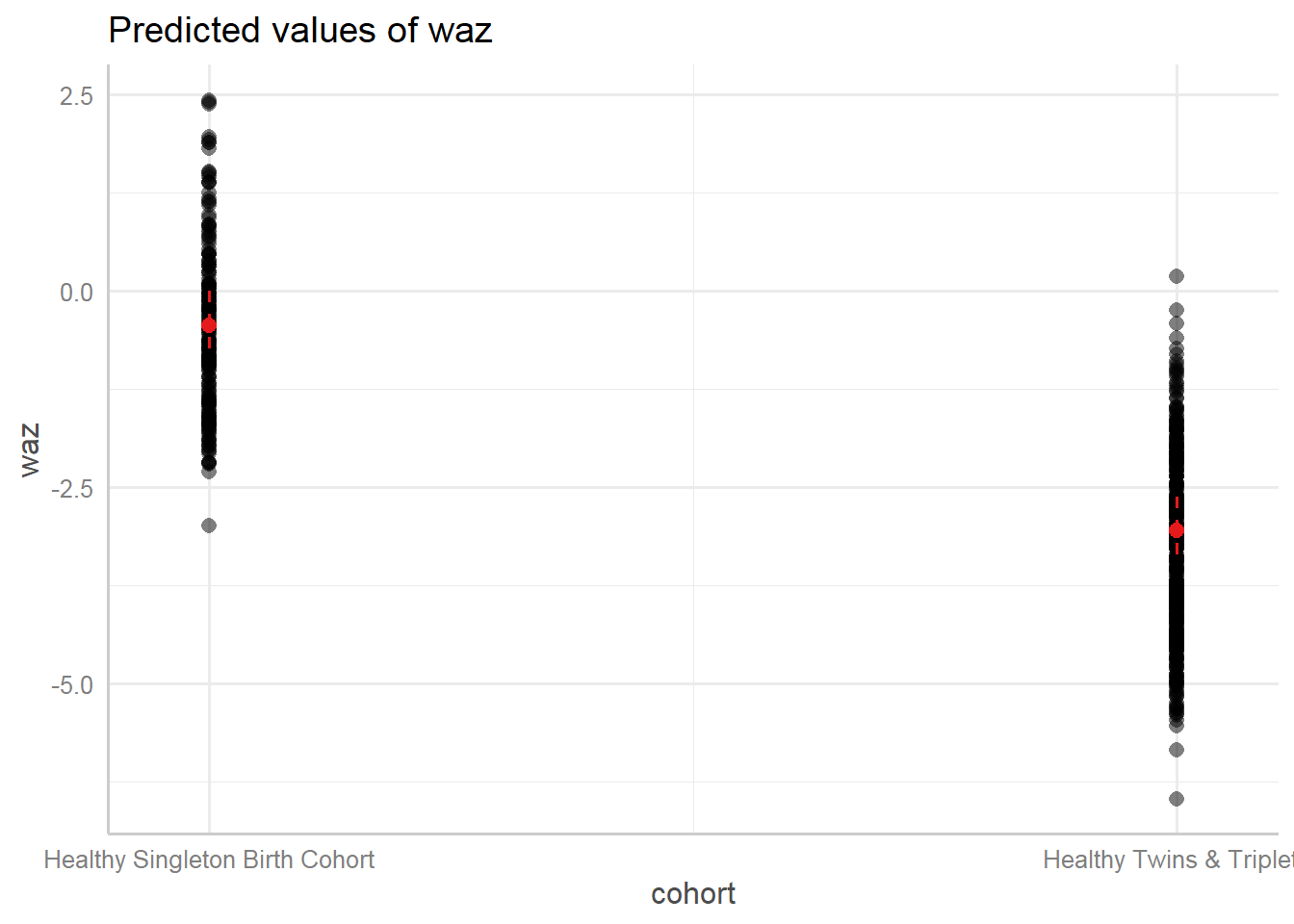
Marginal effects of interaction terms:
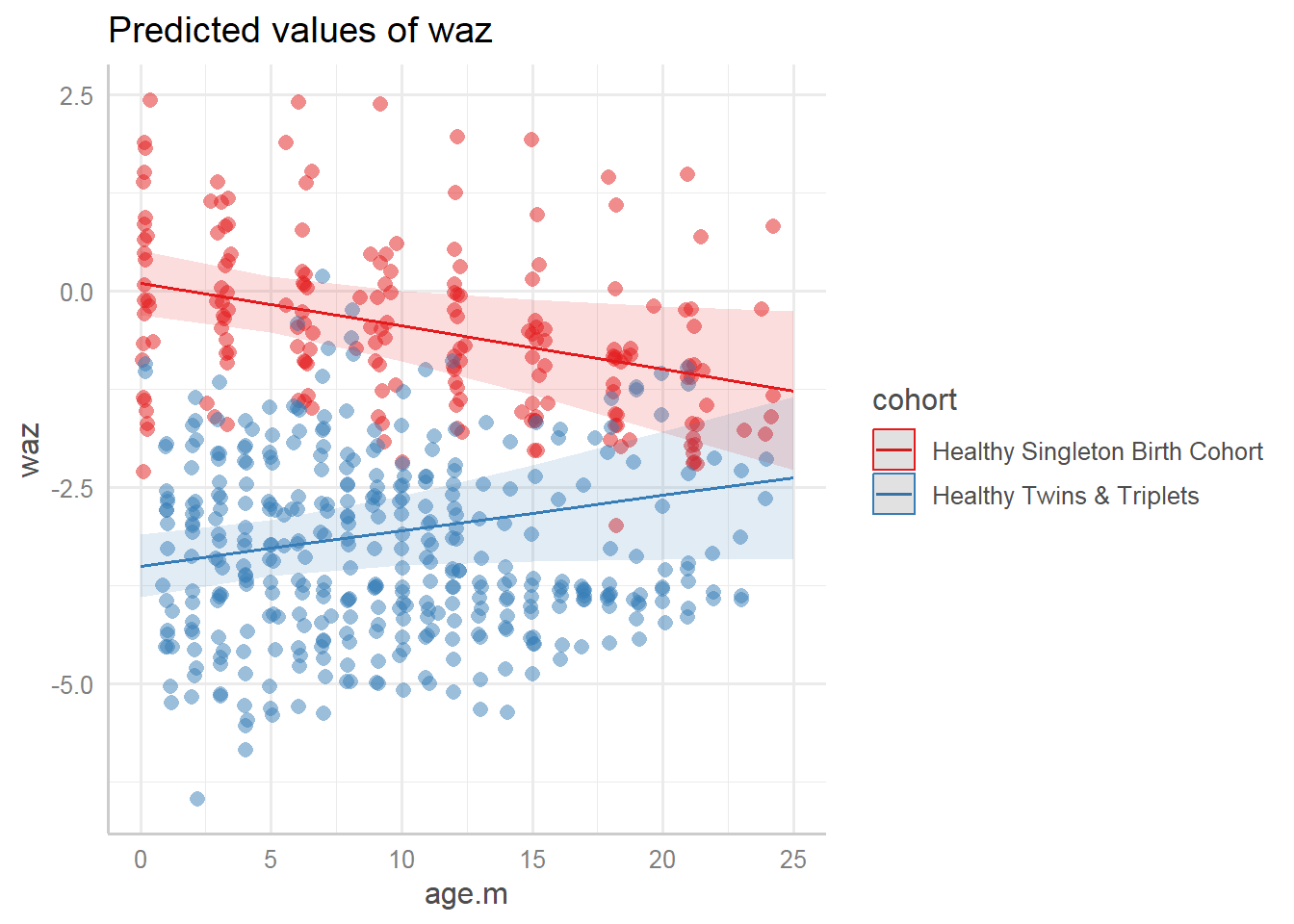
9.7.3.4 Summary of mixed model results
| waz | |||
|---|---|---|---|
| Predictors | Estimates | CI | p |
| (Intercept) | 0.10 | -0.31 – 0.51 | 0.621 |
| age m | -0.05 | -0.10 – -0.01 | 0.018 |
|
cohort [Healthy Twins & Triplets] |
-3.60 | -4.18 – -3.03 | <0.001 |
|
age m × cohort [Healthy Twins & Triplets] |
0.10 | 0.03 – 0.16 | 0.003 |
| Random Effects | |||
| σ2 | 0.29 | ||
| τ00 child.id | 0.95 | ||
| τ11 child.id.age.m | 0.01 | ||
| ρ01 child.id | -0.44 | ||
| ICC | 0.85 | ||
| N child.id | 50 | ||
| Observations | 573 | ||
| Marginal R2 / Conditional R2 | 0.442 / 0.918 | ||
9.8 GEE or Mixed model
Summary of approaches for mixed models and GEE:
Two approaches have different targets for inferences and address subtly different questions about longitudinal change
- GEE: Marginal (population-average) Merely acknowledge the correlation among repeated measurements by robust variance estimation
- Mixed: Conditional (subject-specific) Provide an explanation for the source of correlation at different levels
| Characteristics | Mixed model | GEE |
|---|---|---|
| Focus of interest | Variance component and regression coefficients | Regression coefficients |
| Parameter interpretation | Individual (subject, school, neighborhood) specific | Population average |
| Linear model (estimates equivalent) | Change in the mean outcome for a unit change in subject exposure, keeping the random effect (subject) fixed | Change in the mean outcome for a unit change in subject exposure across all of the subjects observed |
| Binary model (estimates not equivalent) | The log(OR) of an outcome for a unit change in subject exposure, keeping the subject fixed | The log(OR) of an outcome for a unit change in subject exposure across all of the subjects observed |
| Assumption | Correctly specified error distribution. Sensitive to different assumptions about variance and covariance structure, which are usually difficult to validate | Number of subjects should be sufficiently large for robust estimation of standard errors |
9.9 Practice
Use the same example Bangladeshi children dataset as of this material, examine: - The change of haz over age, between cohort and gender - Visualize and interpret the results
Note: use GEE and/or GLMM where applicable.
9.10 Reference materials
This chapter has been adapted from several sources including:
9.10.1 Courses
- [SISCR Course 2016: “Introduction to Longitudinal Data Analysis”” by Benjamin French, PhD (University of Pennsylvania) and Colleen Sitlani, PhD (University of Washington)].