10 Homework: Week 1 Solution
10.1 Question 1 (2 Points):
The HIDEG field refers to the highest degree that a person achieved at the time they first entered the MEPS survey.
Here is what the codebook lists for all possible values and record counts:
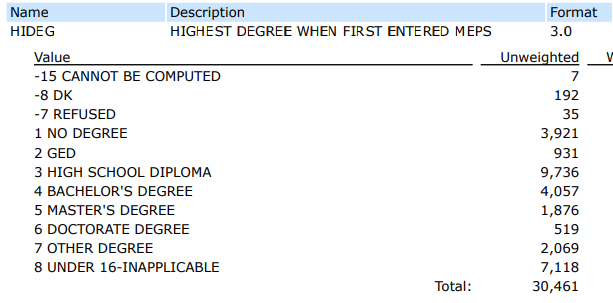
To verify the record count of the “HIDEG” field values, I would use the group_by() function to group by HIDEG, and then I would use the summarize() function along with n() to count the number of records for each value of HIDEG.
It would look like this:
10.2 Question 2 (2 Points):
person_sum_by_race <- meps %>%
dplyr::group_by(RACETHX) %>%
dplyr::summarize(person_weight_sum = sum(PERWT18F)) %>%
ungroup() %>%
dplyr::mutate(percent = 100 * round(person_weight_sum / sum(person_weight_sum),3))
person_sum_by_race
#> # A tibble: 5 × 3
#> RACETHX person_weight_sum percent
#> <chr> <dbl> <dbl>
#> 1 ASIAN ONLY 19307379. 5.9
#> 2 BLACK ONLY 40104475. 12.3
#> 3 HISPANIC 60229788. 18.5
#> 4 OTHER OR MULTIPLE 11920040. 3.7
#> 5 WHITE ONLY 194766207. 59.7Confirming with codebook:
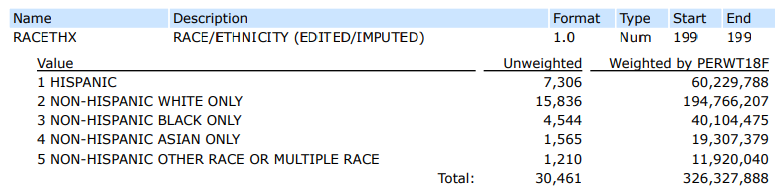
Many solutions could be accepted, but here is one example that would confirm that the MEPS weighted distribution of race is representative of the U.S. population.

10.3 Question 3 (5 Points):
hw_df_1 <- meps %>%
dplyr::select(DUPERSID, AGE42X, ADBMI42) %>%
dplyr::filter(AGE42X >= 0) %>%
dplyr::arrange(desc(ADBMI42)) %>%
dplyr::slice(1:10) %>%
dplyr::mutate(BMI_2 = round(ADBMI42, 0))
head(hw_df_1)
#> # A tibble: 6 × 4
#> DUPERSID AGE42X ADBMI42 BMI_2
#> <chr> <dbl> <dbl> <dbl>
#> 1 2290648101 56 71.1 71
#> 2 2328264101 31 70.8 71
#> 3 2326867103 32 70.5 70
#> 4 2327052101 24 70.2 70
#> 5 2327273101 53 69.7 70
#> 6 2328953102 22 69.1 6910.4 Question 4 (6 Points):
hw_df_2 <- meps %>%
dplyr::filter(POVCAT18 %in% c("POOR/NEGATIVE", "LOW INCOME")) %>%
dplyr::select(POVCAT18, RTHLTH42, TOTEXP18) %>%
dplyr::filter(RTHLTH42 != "INVALID") %>%
dplyr::group_by(POVCAT18, RTHLTH42) %>%
dplyr::summarize(
total_med_exp = sum(TOTEXP18),
person_count = n(),
average_med_exp = total_med_exp / person_count,
average_med_exp_alternative = mean(TOTEXP18),
median_med_exp = median(TOTEXP18),
median_med_exp_alternative = quantile(TOTEXP18, probs = 0.50),
percentile_75 = quantile(TOTEXP18, probs = 0.75)
)
head(hw_df_2)
#> # A tibble: 6 × 9
#> # Groups: POVCAT18 [2]
#> POVCAT18 RTHLTH42 total_med_exp person_count
#> <chr> <chr> <dbl> <int>
#> 1 LOW INCOME EXCELLENT 2087391 1171
#> 2 LOW INCOME FAIR 6073514 485
#> 3 LOW INCOME GOOD 8355011 1338
#> 4 LOW INCOME POOR 3360914 132
#> 5 LOW INCOME VERY GOOD 4391685 1320
#> 6 POOR/NEGATIVE EXCELLENT 2581576 1515
#> # … with 5 more variables: average_med_exp <dbl>,
#> # average_med_exp_alternative <dbl>,
#> # median_med_exp <dbl>, median_med_exp_alternative <dbl>,
#> # percentile_75 <dbl>- Note the multiple ways to calculate the average and median spend
10.5 Question 5 (10 Points):
# This data manipulation section is worth 4 points:
flushot_by_usc_inscov = meps %>%
dplyr::filter(ADFLST42 != "INVALID",
HAVEUS42 != "INVALID") %>%
dplyr::group_by(ADFLST42,
HAVEUS42,
INSCOV18) %>%
dplyr::summarise(person_count = n()) %>%
ungroup() %>%
tidyr::pivot_wider(names_from = ADFLST42, values_from = person_count) %>%
dplyr::mutate(total_count = YES + NO,
percent_flushot = YES / total_count)
head(flushot_by_usc_inscov)
#> # A tibble: 6 × 6
#> HAVEUS42 INSCOV18 NO YES total_count percent_flushot
#> <chr> <chr> <int> <int> <int> <dbl>
#> 1 NO ANY PRIV… 2004 729 2733 0.267
#> 2 NO PUBLIC O… 781 262 1043 0.251
#> 3 NO UNINSURED 919 98 1017 0.0964
#> 4 YES ANY PRIV… 4492 4640 9132 0.508
#> 5 YES PUBLIC O… 2069 2267 4336 0.523
#> 6 YES UNINSURED 484 115 599 0.192
# This plotting section is worth 5 points
# Explanation is worth 1 point
gopher_colors = c("#7A0019", "#FCD131", "#515151")
ggplot(data = flushot_by_usc_inscov,
aes(x = HAVEUS42, y = percent_flushot, fill = INSCOV18)) +
geom_bar(stat = "identity", position = "dodge") +
theme_minimal() +
xlab("Usual Source of Care") +
ylab("Percent Receiving Flu Shot") +
ggtitle("Percent Receiving Flu Shot by USC and Insurance Coverage") +
scale_x_discrete(labels = function(x) str_wrap(x, width = 10)) +
scale_fill_manual(values = gopher_colors) +
scale_y_continuous(labels = scales::percent) +
labs(fill = "Insurance\nCoverage")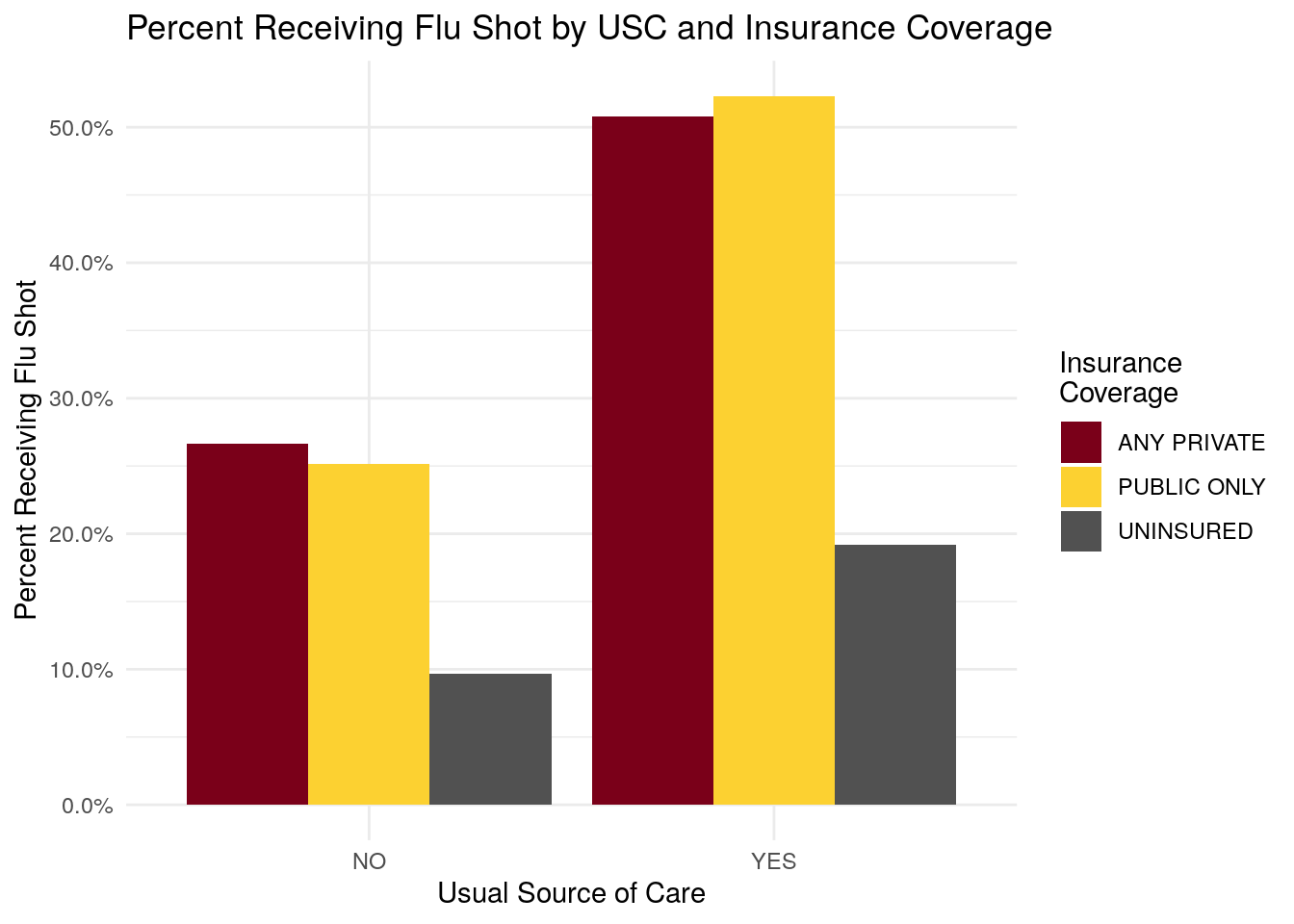
- Individuals with a usual source of care have a higher percentage of members receiving flu shot.
- Individuals with insurance coverage have a higher percentage of members receiving flu shot, as compared to the uninsured.
10.6 Question 6 (11 Points):
10.6.1 Part 1:
# This section using the 'cut' function is worth 5 points
bmi_breaks = c(0, 16, 17, 18.5, 25, 30, 35, 40, Inf)
bmi_labels = c("Underweight - Severe", "Underweight - Moderate", "Underweight - Mild", "Normal", "Overweight", "Obese - I",
"Obese II", "Obese III")
medexp_by_bmi_cut = meps %>%
dplyr::filter(ADBMI42 >= 0) %>%
dplyr::mutate(bmi_buckets = cut(ADBMI42, breaks = bmi_breaks, right = F, labels = bmi_labels)) %>%
dplyr::group_by(bmi_buckets) %>%
dplyr::summarise(Total_Med_Exp = sum(TOTEXP18),
person_count = n()) %>%
ungroup() %>%
dplyr::mutate(average_med_exp = Total_Med_Exp / person_count)
ggplot(data = medexp_by_bmi_cut,
aes(x = bmi_buckets, y = average_med_exp, fill = bmi_buckets)) +
geom_bar(stat = "identity") +
theme_minimal() +
ylab("Average Medical Spend") +
xlab("BMI Range") +
ggtitle("Medical Expenditures by BMI Range") +
scale_fill_brewer(palette = "RdPu") +
scale_x_discrete(labels = function(x) str_wrap(x, width = 10)) +
scale_y_continuous(labels = scales::comma) +
coord_flip() +
labs(fill = "BMI Range")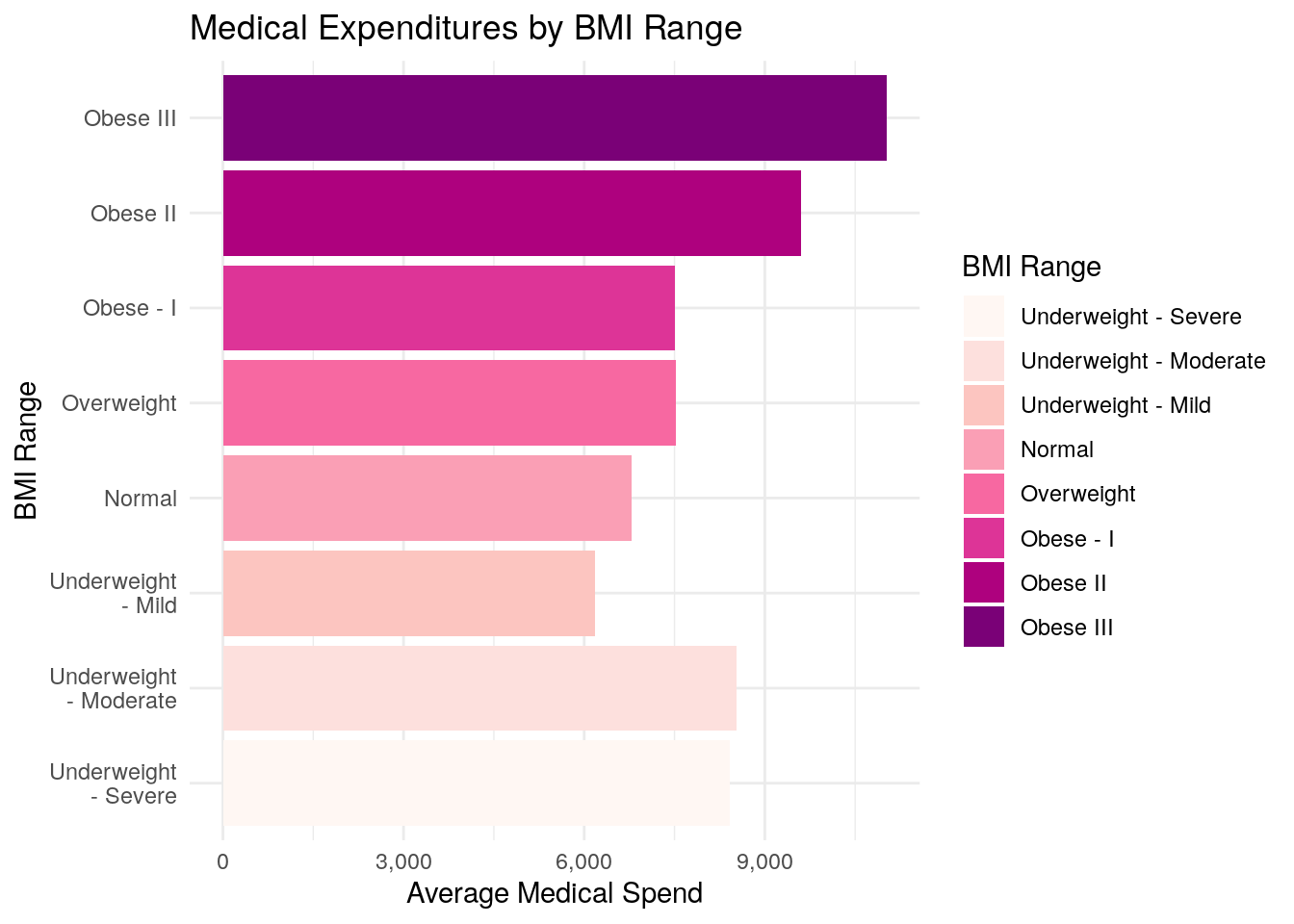
10.6.2 Part 2:
# This section using the 'case_when` function is worth 5 points.
# Explanation below is worth 1 point
medexp_by_bmi_case_when <- meps %>%
dplyr::filter(ADBMI42 >= 0) %>%
dplyr::mutate(
bmi_buckets = case_when(
ADBMI42 >= 0 & ADBMI42 < 16 ~ "Underweight - Severe",
ADBMI42 >= 16 & ADBMI42 < 17 ~ "Underweight - Moderate",
ADBMI42 >= 17 & ADBMI42 < 18.5 ~ "Underweight - Mild",
ADBMI42 >= 18.5 & ADBMI42 < 25 ~ "Normal",
ADBMI42 >= 25 & ADBMI42 < 30 ~ "Overweight",
ADBMI42 >= 30 & ADBMI42 < 35 ~ "Obese - I",
ADBMI42 >= 35 & ADBMI42 < 40 ~ "Obese - II",
ADBMI42 >= 40 ~ "Obese - III"
)
) %>%
dplyr::group_by(bmi_buckets) %>%
dplyr::summarise(Total_Med_Exp = sum(TOTEXP18),
person_count = n()) %>%
ungroup() %>%
dplyr::mutate(average_med_exp = Total_Med_Exp / person_count)
ggplot(data = medexp_by_bmi_case_when,
aes(x = bmi_buckets, y = average_med_exp, fill = bmi_buckets)) +
geom_bar(stat = "identity") +
theme_minimal() +
ylab("Average Medical Spend") +
xlab("BMI Range") +
ggtitle("Average Medical Expenditures by BMI Range") +
scale_fill_brewer(palette = "YlOrRd") +
scale_x_discrete(labels = function(x) str_wrap(x, width = 10)) +
scale_y_continuous(labels = scales::comma) +
coord_flip() +
labs(fill = "BMI Range")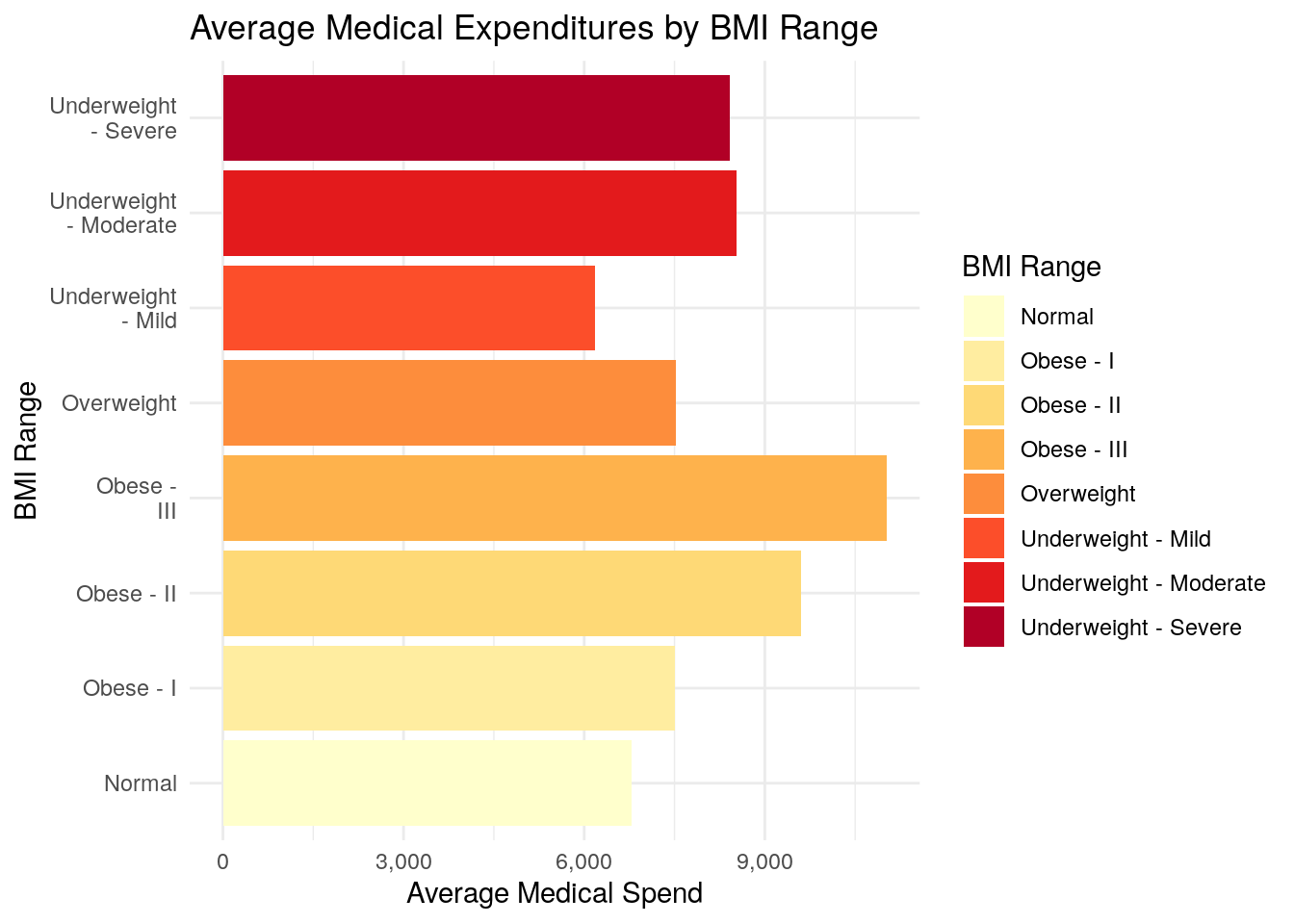
10.6.3 Part 3:
The biggest difference between using the cut() function and the case_when() function is that the cut() function creates a new field that is a “factor” type instead of a “character” type.
Factor variables are special types of fields in R, and when cut() creates the new factor variable, it tells R that there is an inherent order to the BMI classification.
When you use case_when(), R does not know that there is an intended order to the classification schedule.
As a result, when you plot Part 1 using cut(), the bar chart is plotted in the order that you defined the BMI classification, where it progresses from Underweight, to Normal, to Overweight, to Obese - as it should.
When you plot Part 2 using case_when(), R doesn’t know about the inherent order of the BMI classification, so it will plot the bar chart in alphabetical order, which makes the bar chart more difficult to read, especially when you use one of the ‘sequential’ color palettes, as we have.
In other words, using a sequential color palette implies some sort of order to your chart, and so the plot in Part 2 of this question really is not a very good plot.
10.7 Question 7 (5 Bonus Points):
10.7.2 Part 2:
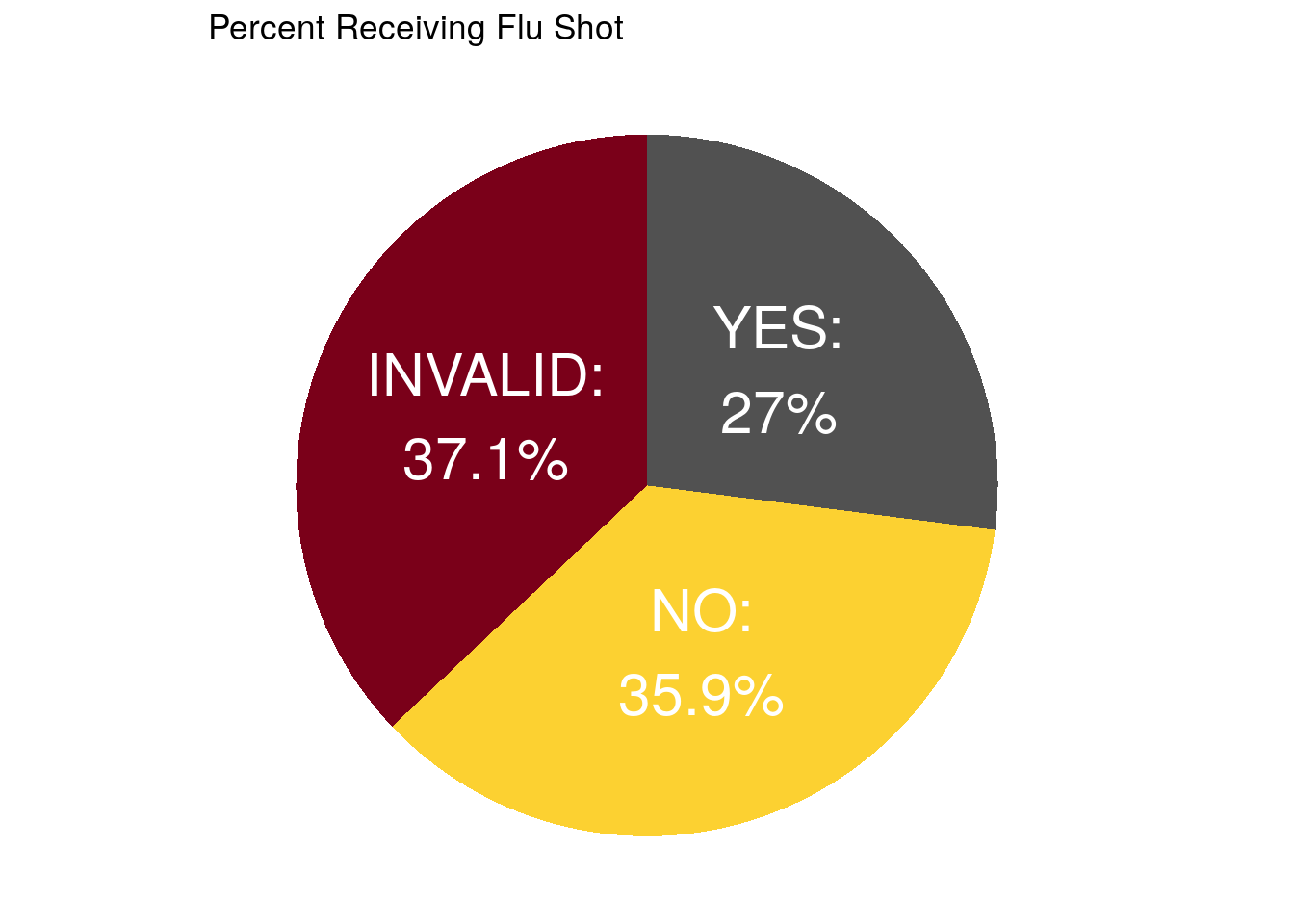
ggplot(data = pie_data,
aes(x = "", y = count, fill = ADFLST42)) +
geom_bar(stat = "identity", width = 1) +
coord_polar("y", start = 0) +
theme_void() +
theme(legend.position="none") +
geom_text(aes(label = paste0(ADFLST42, ":\n", percent, "%")),
position = position_stack(vjust = 0.5),
color = "white",
size = 8) +
# geom_text(aes(y = ypos, label = ADFLST42), color = "white", size=6) +
scale_fill_manual(values = gopher_colors) +
ggtitle("Percent Receiving Flu Shot")