Simple Linear Regression
Setup
intall libraries
Explore relationships
Flow and area
m1<-lm(as.numeric(pp_area) ~ cur_in_mean, data = buf)
plot(buf$cur_mean, as.numeric(buf$pp_area), xlab="mean circuitscape current", ylab="pinch-point area")
abline(m1, col="red")
##
## Call:
## lm(formula = as.numeric(pp_area) ~ cur_in_mean, data = buf)
##
## Residuals:
## Min 1Q Median 3Q Max
## -55786 -14917 -13455 -7521 611100
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 240277.0 124790.7 1.925 0.0548 .
## cur_in_mean -1044.6 584.6 -1.787 0.0747 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 54310 on 446 degrees of freedom
## Multiple R-squared: 0.007107, Adjusted R-squared: 0.004881
## F-statistic: 3.192 on 1 and 446 DF, p-value: 0.07466Flow difference and area
m2<-lm(as.numeric(pp_area) ~ cur_diff, data = buf)
plot(buf$cur_diff, as.numeric(buf$pp_area), xlab="Difference between inner and outer current", ylab="pinch-point area")
abline(m2, col="red")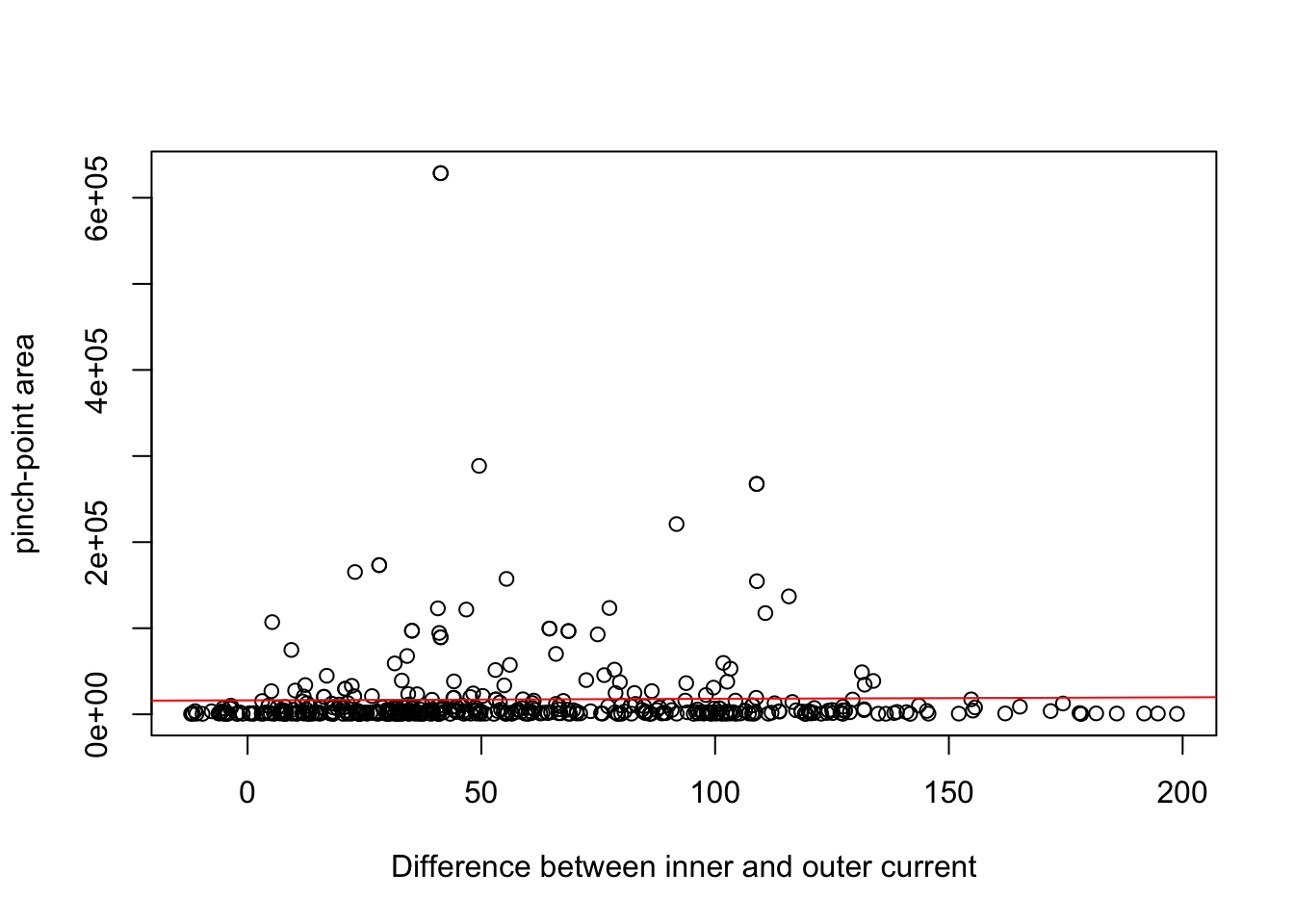
##
## Call:
## lm(formula = as.numeric(pp_area) ~ cur_diff, data = buf)
##
## Residuals:
## Min 1Q Median 3Q Max
## -19382 -15992 -14607 -8060 611518
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 16313.83 4146.23 3.935 9.66e-05 ***
## cur_diff 18.22 56.91 0.320 0.749
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 54500 on 446 degrees of freedom
## Multiple R-squared: 0.0002297, Adjusted R-squared: -0.002012
## F-statistic: 0.1025 on 1 and 446 DF, p-value: 0.749Flow and distance to city center
m4<-lm(as.numeric(dist2cent) ~ cur_in_mean, data = buf)
plot(buf$cur_mean, as.numeric(buf$dist2cent), xlab="mean circuitscape current", ylab="distance to city center (m)")
abline(m4, col="red")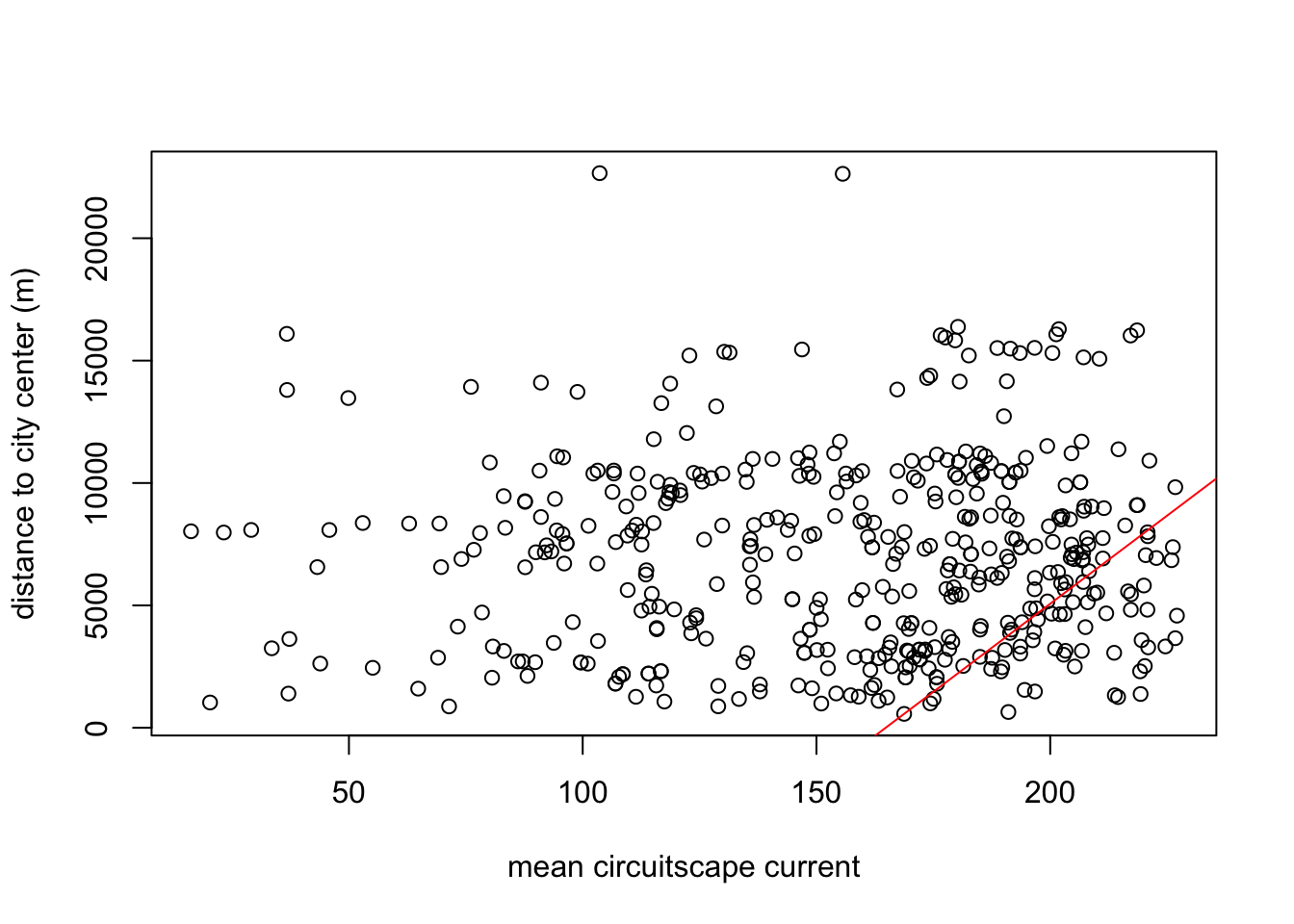
##
## Call:
## lm(formula = as.numeric(dist2cent) ~ cur_in_mean, data = buf)
##
## Residuals:
## Min 1Q Median 3Q Max
## -6659.1 -3158.6 64.4 2605.5 15431.3
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -23728.10 8874.54 -2.674 0.007777 **
## cur_in_mean 143.98 41.58 3.463 0.000586 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3862 on 446 degrees of freedom
## Multiple R-squared: 0.02618, Adjusted R-squared: 0.024
## F-statistic: 11.99 on 1 and 446 DF, p-value: 0.000586Flow difference and distance to city center
m5<-lm(as.numeric(dist2cent) ~ cur_diff, data = buf)
plot(buf$cur_diff, as.numeric(buf$dist2cent), xlab="Difference between inner and outer current", ylab="distance to city center (m)")
abline(m5, col="red")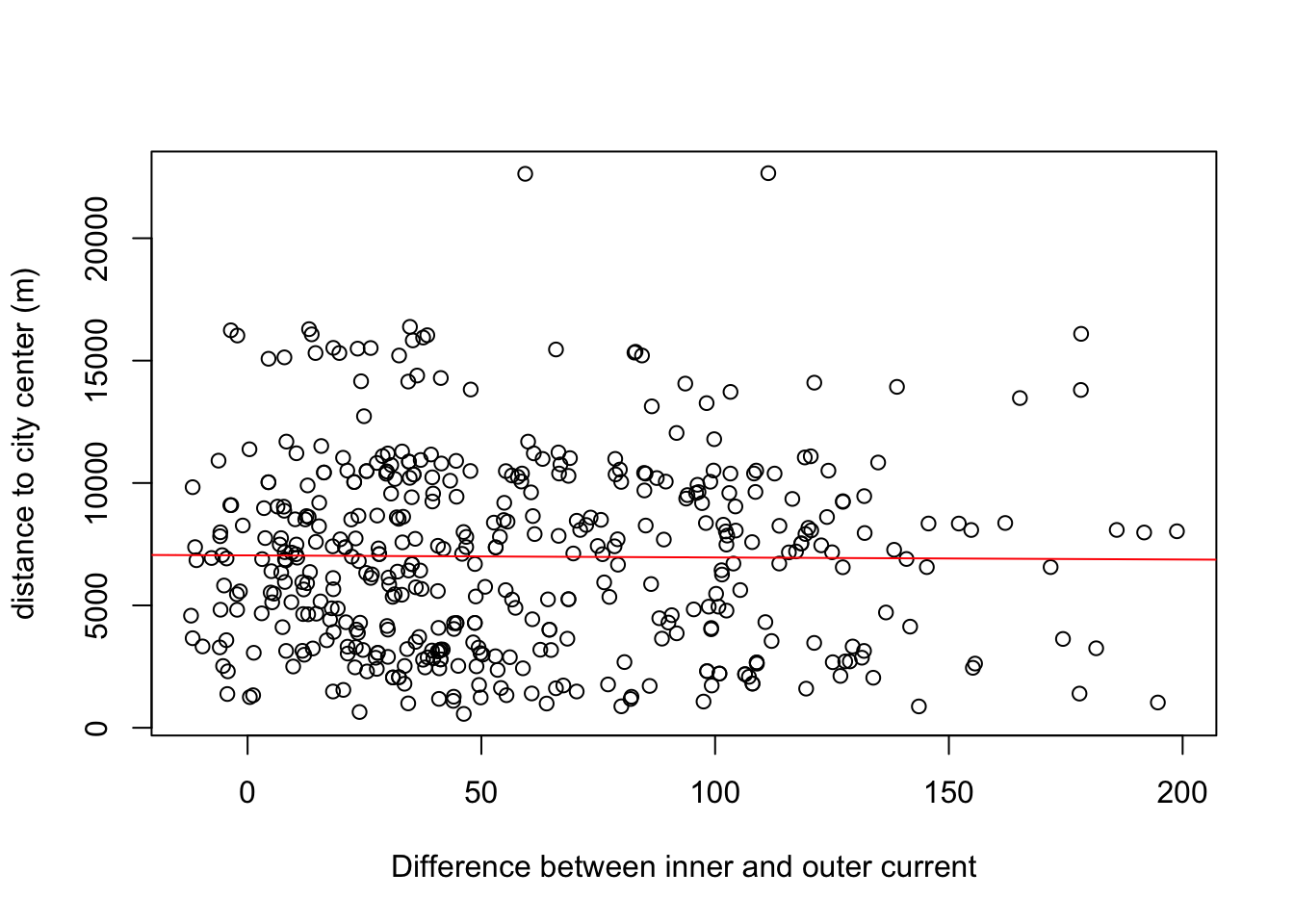
##
## Call:
## lm(formula = as.numeric(dist2cent) ~ cur_diff, data = buf)
##
## Residuals:
## Min 1Q Median 3Q Max
## -6439 -3418 -14 2642 15708
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 7046.0959 297.7544 23.66 <2e-16 ***
## cur_diff -0.8588 4.0871 -0.21 0.834
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3914 on 446 degrees of freedom
## Multiple R-squared: 9.899e-05, Adjusted R-squared: -0.002143
## F-statistic: 0.04415 on 1 and 446 DF, p-value: 0.8337Flow difference and % vegetation cover (buffer)
m3<-lm(PRIMECLASS_VEG~cur_diff, data = buf)
plot(buf$cur_diff, buf$PRIMECLASS_VEG, xlab="Difference between inner and outer current", ylab="Proportion of vegetation cover")
abline(m3, col="red")
##
## Call:
## lm(formula = PRIMECLASS_VEG ~ cur_diff, data = buf)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.61315 -0.09680 0.02327 0.11973 0.36589
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.7883846 0.0134753 58.51 <2e-16 ***
## cur_diff -0.0024433 0.0001852 -13.20 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.1765 on 441 degrees of freedom
## (5 observations deleted due to missingness)
## Multiple R-squared: 0.2831, Adjusted R-squared: 0.2814
## F-statistic: 174.1 on 1 and 441 DF, p-value: < 2.2e-16m3b<-lm(PRIMECLASS_VEG~cur_diff, data = in_pp)
plot(in_pp$cur_diff, in_pp$PRIMECLASS_VEG, xlab="Difference between inner and outer current", ylab="Proportion of vegetation cover")
abline(m3b, col="red")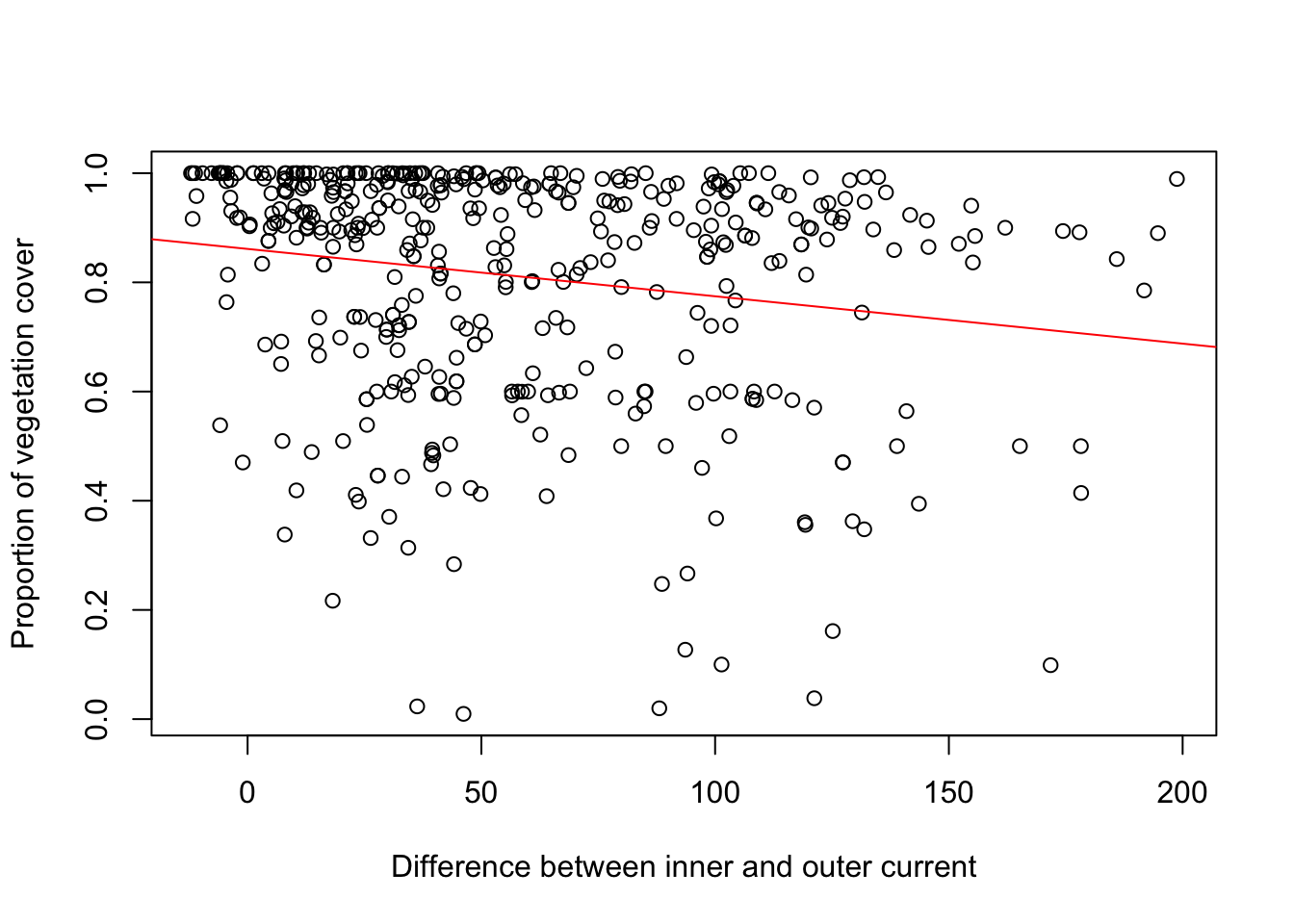
##
## Call:
## lm(formula = PRIMECLASS_VEG ~ cur_diff, data = in_pp)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.8116 -0.1220 0.0807 0.1519 0.3006
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.8612398 0.0160510 53.657 < 2e-16 ***
## cur_diff -0.0008674 0.0002217 -3.912 0.000106 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.2091 on 436 degrees of freedom
## (10 observations deleted due to missingness)
## Multiple R-squared: 0.03391, Adjusted R-squared: 0.0317
## F-statistic: 15.31 on 1 and 436 DF, p-value: 0.0001061Flow difference and % developed (buffer)
m6<-lm(LANDCLAS_DEV~cur_diff, data = buf)
plot(buf$cur_diff, buf$LANDCLAS_DEV, xlab="Difference between inner and outer current", ylab="Proportion of developed land")
abline(m6, col="red")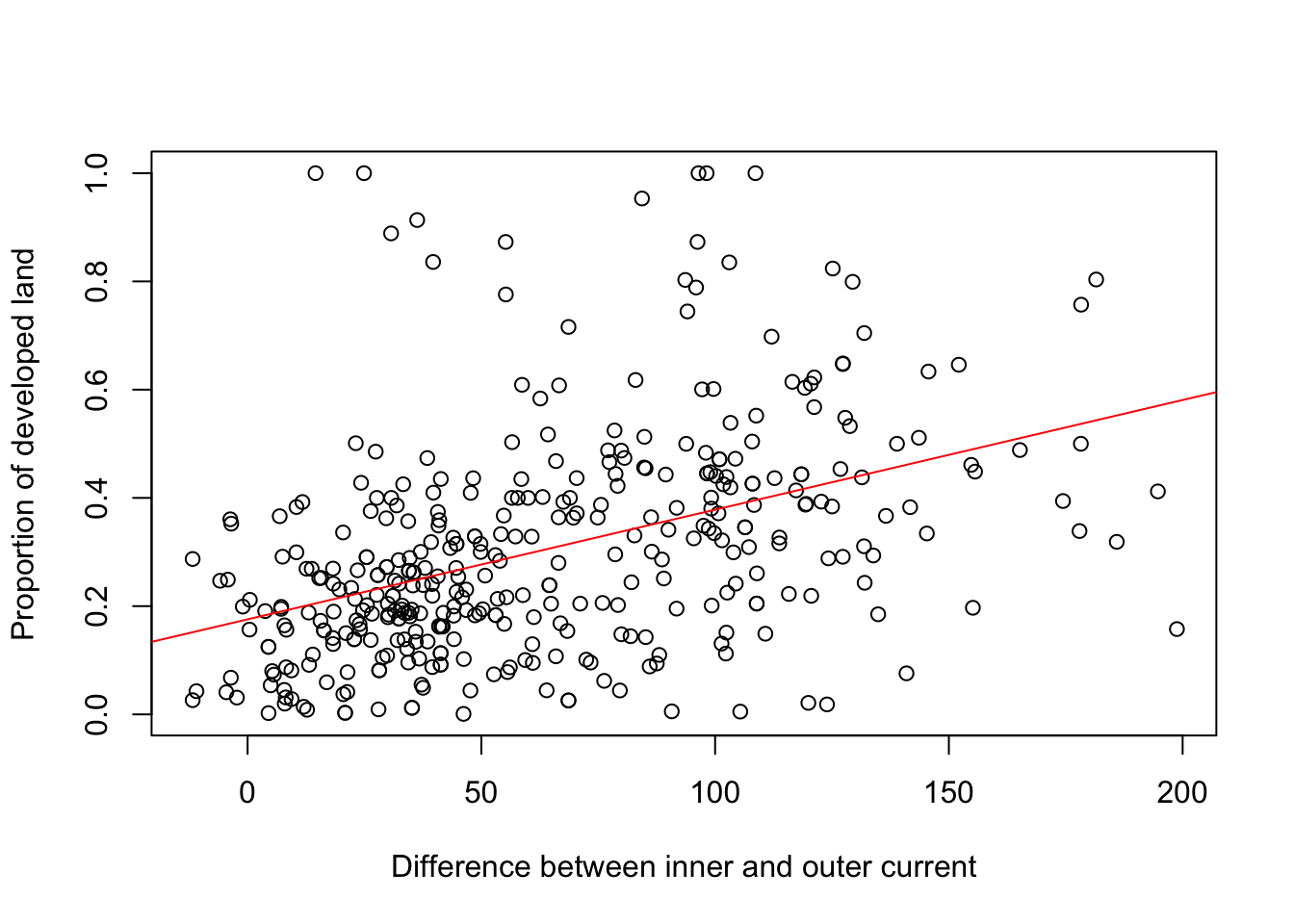
##
## Call:
## lm(formula = LANDCLAS_DEV ~ cur_diff, data = buf)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.42097 -0.11945 -0.02463 0.08490 0.79492
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.1756290 0.0169419 10.367 <2e-16 ***
## cur_diff 0.0020272 0.0002212 9.163 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.1867 on 377 degrees of freedom
## (69 observations deleted due to missingness)
## Multiple R-squared: 0.1822, Adjusted R-squared: 0.18
## F-statistic: 83.97 on 1 and 377 DF, p-value: < 2.2e-16Flow difference and % developed (in pinch-point)
m7<-lm(LANDCLAS_DEV~cur_diff, data = in_pp)
plot(in_pp$cur_diff, in_pp$LANDCLAS_DEV, xlab="Difference between inner and outer current", ylab="Proportion of developed land")
abline(m7, col="red")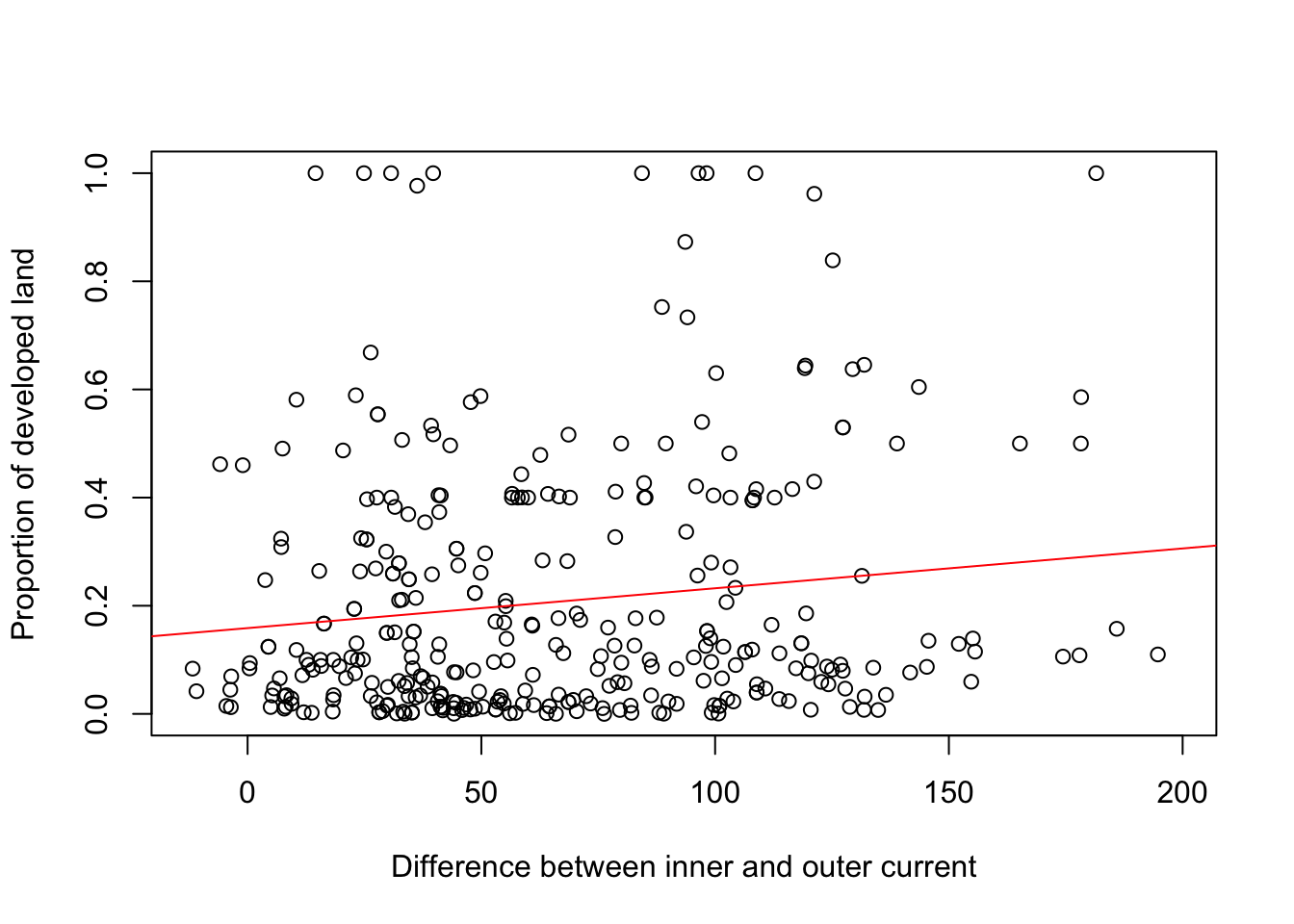
##
## Call:
## lm(formula = LANDCLAS_DEV ~ cur_diff, data = in_pp)
##
## Residuals:
## Min 1Q Median 3Q Max
## -0.25088 -0.16902 -0.09455 0.13828 0.83064
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.1586465 0.0229819 6.903 2.55e-11 ***
## cur_diff 0.0007374 0.0002952 2.498 0.013 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.2331 on 336 degrees of freedom
## (110 observations deleted due to missingness)
## Multiple R-squared: 0.01823, Adjusted R-squared: 0.01531
## F-statistic: 6.239 on 1 and 336 DF, p-value: 0.012974.0.1 Visualize for report
# set up regression plot function
custom_theme<-function(){theme(
panel.background = element_rect(fill = "white",color="#636363"),
axis.line.y = element_line(colour = "#636363"),
axis.text.y = element_text(colour="#636363"),
axis.ticks.y = element_line(colour = "#636363"),
axis.title.y = element_text(colour="#636363"),
axis.text.x = element_text(colour="#636363"),
axis.ticks.x = element_line(colour = "#636363"),
axis.line.x = element_line(colour = "#636363"),
axis.title.x = element_text(colour="#636363"),
legend.title = element_text(colour="#636363"),
legend.text = element_text(colour="#636363"),
legend.key = element_rect(fill = "white"),
panel.grid.major = element_line(size = 0.0, colour="#bdbdbd")
)}
ggplotRegression <- function (fit) {
require(ggplot2)
ggplot(fit$model, aes_string(x = names(fit$model)[2], y = names(fit$model)[1])) +
geom_point() +
stat_smooth(method = "lm", col = "red") +
labs(title = paste("Adj R2 = ",signif(summary(fit)$adj.r.squared, 5),
"Intercept =",signif(fit$coef[[1]],5 ),
" Slope =",signif(fit$coef[[2]], 5),
" P =",signif(summary(fit)$coef[2,4], 5)))+
custom_theme()
}
fit1 <- lm(PRIMECLASS_VEG~cur_diff, data = buf)
fit2 <-lm(LANDCLAS_DEV~cur_diff, data = buf)
a<-ggplotRegression(fit1)
b<-ggplotRegression(fit2)
ggsave(filename = "4_output/regression/veg_curDif.png", a, height=5, width=8)
ggsave(filename = "4_output/regression/dev_curDif.png", b, height=5, width=8)