Circuitscape
Import the pinch-point and connectivity polygons derived from the circuitscape flow raster.
- check the crs
- visualize polygons
- clip to the study area (i.e. the North Saskatchewan Central region of the Ribbon of Green)
Import pinch-points
Load spatial data
Load and plot the ribbon of green shape file RoG_Reaches_V1_210301.shp within the city boundary.
# load ribbon of green
RoG_Reaches<-st_read("1_data/external/Ribbon_of_Green_Study_Area_Reaches/Reach_Boundaries/RoG_Reaches_V1_210301/RoG_Reaches_V1_210301.shp")
# check crs
st_crs(RoG_Reaches)
# NAD83_3TM_114_Longitude_Meter_Province_of_Alberta_Canada
RoG_Reaches<-st_transform(RoG_Reaches, crs = st_crs(3776))
# load city boundary
City_Boundary<-st_read("1_data/external/City_Boundary/City_Boundary.shp")
# reproject to match the CRS of RoG_reaches
City_Boundary<-st_transform(City_Boundary, crs = st_crs(3776))
#load road data
v_road<-st_read("1_data/external/roads_data/V_ROAD_SEGMENT_CUR.shp")
# reproject to match the CRS of RoG_reaches
v_road<-st_transform(v_road, crs = st_crs(3776))
# crop roads file by a bounding box of the north saskatchewan central region polygon
v_road_1<-st_crop(v_road, City_Boundary)
#plot shape file
tmap_mode("plot")
m<-tm_shape(City_Boundary)+tm_borders(col = "#636363")+tm_fill(col="#d9d9d9")+
tm_layout(frame=FALSE, legend.text.size=.5, legend.width=2, legend.position=c("left","bottom"),)+
tm_legend(outside=TRUE, frame=FALSE)+
tm_shape(v_road_1)+tm_lines(col="white", lwd = .8)+
tm_shape(RoG_Reaches)+
tm_fill(col="Reach", palette = "Paired", contrast = c(0.26, 1),alpha = .8, title="Ribbon of Green Reaches")+
tm_borders(col = "#636363")
m
bbox_RoG<-st_bbox(RoG_Reaches)%>%
st_as_sfc()
m<-
tm_shape(bbox_RoG)+
tm_borders(col="white")+
tm_shape(City_Boundary)+
tm_borders(col="#636363")+
tm_fill(col="#d9d9d9")+
tm_shape(v_road_1)+
tm_lines(col="white", lwd = .8)+
tm_shape(City_Boundary)+
tm_borders(col="#636363")+
tm_fill(col="#d9d9d9", alpha = 0)+
# tm_legend(outside=TRUE, frame=FALSE)+
tm_shape(RoG_Reaches)+
tm_fill(col="Reach", palette = "Paired", contrast = c(0.26, 1),alpha = .8, title="Ribbon of Green Reaches")+
tm_borders(col="#636363")+
tm_layout(frame=FALSE, legend.text.size=.5, legend.outside = TRUE, legend.position = c("right", "top"))
m
tmap_save(m, "4_output/maps/ribbonGreen_map.png", outer.margins=c(0,0,0,0))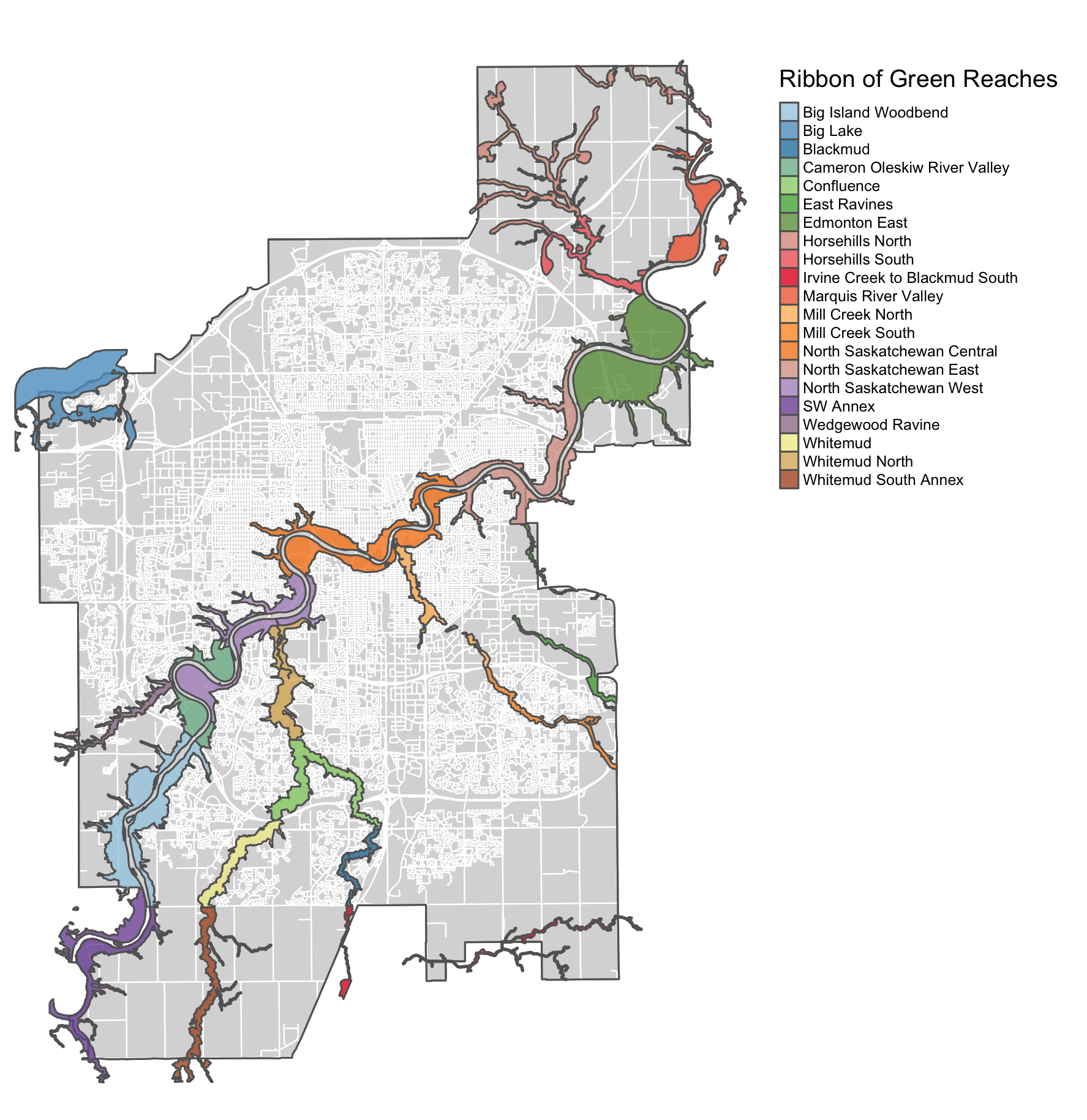
Figure 2.1: Ribbon of Green.
Define study area
For this project I focused on the North Central Region of Saskatchewan region of the ribbon of green
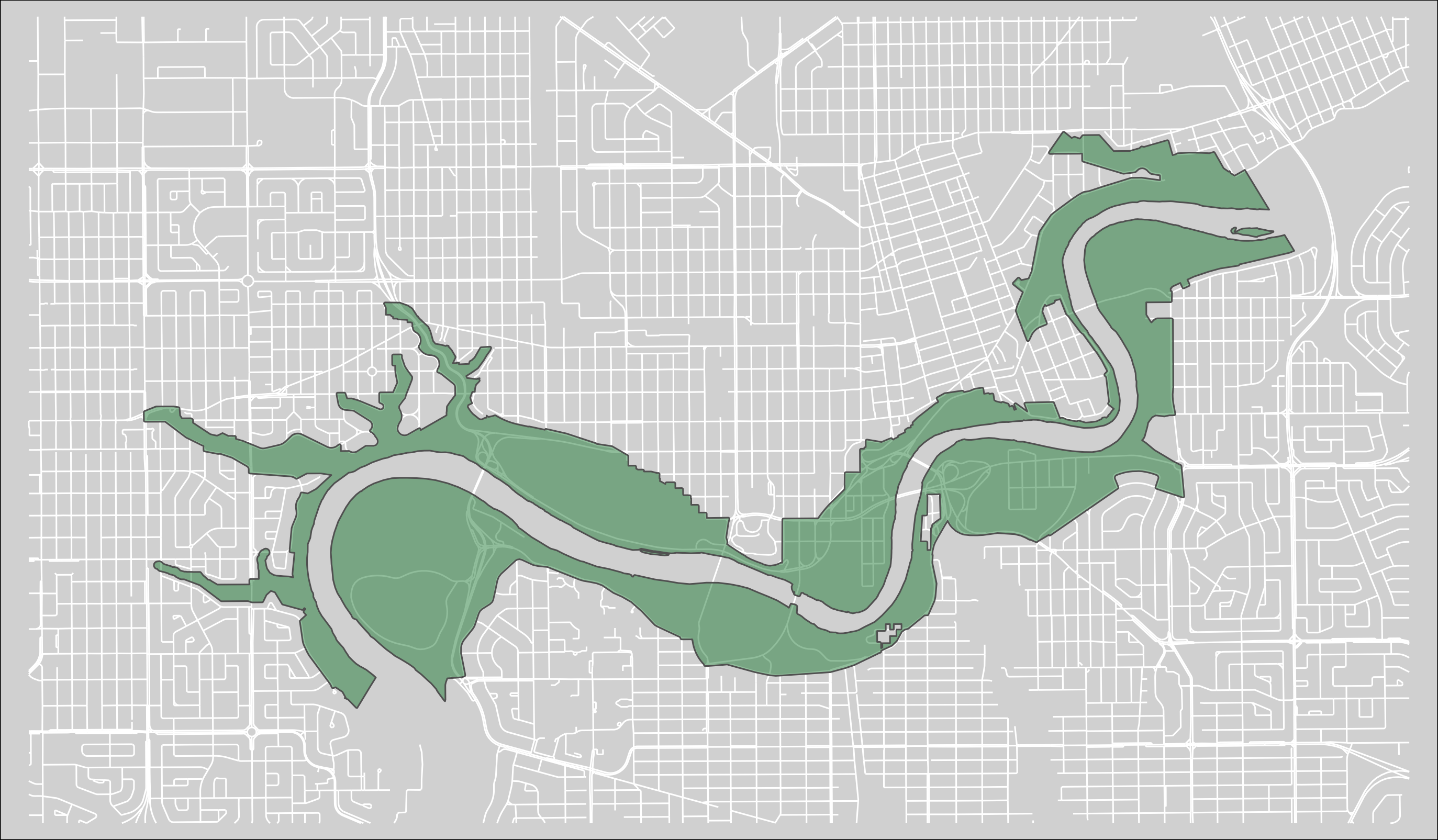
Figure 2.2: The North Saskatchewan Central region of the Ribbon of Green.
Load the connectivity and pinch-point polygons extracted from circuitscape
# Load summer coyote pinchpoints
pp_s<-st_read("1_data/external/Circuitscape/Data/PinchPoints/Data/Coyote_Summer_All_Pinchpoints.shp") %>%
st_buffer(dist=0)
# Load winter coyote pinchpoints
pp_w<-st_read("1_data/external/Circuitscape/Data/PinchPoints/Data/Coyote_Winter_All_Pinchpoints.shp") %>%
st_buffer(dist=0)
# load connetivity polygons
corridors<-st_read("1_data/external/Circuitscape/Vector_Extracted_Corridors/TerrestrialCorridor/TerrestrialCorridor.shp") %>%
st_buffer(dist=0)
corridors<-st_transform(corridors, crs = st_crs(3776))
#filter cooridors polygons to only include terrestrial connectivity polygons
#test<-corridors[corridors$ACoyotCor1=="0",]Clip to study area
pp_s_clip<-st_intersection(pp_s, nscr)
#pp_s_clip<-st_intersection(pp_s, study_area)
pp_w_clip<-st_intersection(pp_w, nscr)
corridors_clip<-st_intersection(corridors, nscr)
#corridors_clip<-st_intersection(corridors, study_area)
save(pp_s_clip, file="1_data/manual/pp_s_clip.rData")
save(pp_w_clip, file="1_data/manual/pp_w_clip.rData")
save(corridors_clip, file="1_data/manual/corridors_clip.rData")
############# plot shape file #############################
# create a bounding box around study area
bbox<-st_bbox(nscr)
# crop roads file by a bounding box of the north saskatchewan central region polygon
v_road_3<-st_crop(v_road, bbox)
tmap_mode("plot")
m3<-
tm_shape(v_road_3)+tm_lines(col="white")+
tm_shape(nscr)+
tm_fill(col="white", alpha=.6)+
tm_borders(col = "#636363")+
tm_shape(corridors_clip)+
tm_fill(col="#238b45", alpha = .5)+
tm_layout(bg.col="#d9d9d9")+
tm_shape(pp_s_clip)+
tm_fill(col="#fb6a4a", alpha = 1)+
tm_borders(col = "#636363", lwd=1)+
tm_layout(legend.outside = FALSE, legend.text.size = .9, legend.bg.color="white", legend.frame=TRUE, legend.frame.lwd=.8)+
tm_add_legend(
type="symbol",
shape=c(22,22, 22),
col=c("#238b45", "#fb6a4a", "white"),
alpha=c(.5, 1, 1),
border.col = "black",
border.lwd=.8,
size=1.5,
labels = c("movement corridors", "summer pinch-points", "N. Saskatchewan Central"))
m3
tmap_save(m3, "4_output/maps/pp_s_map.png", asp=0, outer.margins=c(0,0,0,0))
# library(OpenStreetMap)
# library(tmaptools)
# base<-read_osm(study_area, type="bing")
#
# t<-st_buffer(corridors_clip,10)
# corridors_outline<-st_union(t)
#
m3_b<-
tm_shape(base) + tm_rgb()+
tm_shape(study_area)+
tm_fill(col="white", alpha=.2)+
# tm_shape(v_road_clip)+tm_lines(col="white")+
# tm_shape(nscr)+
# tm_fill(col="white", alpha=.6)+
# tm_borders(col = "#636363")+
# tm_shape(corridors_clip)+
# tm_fill(col="#238b45", alpha = .5)+
# tm_layout(bg.col="#d9d9d9")+
tm_shape(corridors_outline)+
tm_fill(col="#238b45", alpha = .5)+
tm_layout(bg.col="white")+
tm_borders(col="#00441b", lwd=.7)+
tm_shape(pp_s_clip)+
tm_fill(col="#fb6a4a", alpha = 1)+
tm_borders(col = "#fec44f", lwd=.5)+
tm_layout(legend.outside = TRUE, legend.text.size = .9, legend.bg.color="white", legend.frame=TRUE, legend.frame.lwd=.8)+
tm_add_legend(
type="symbol",
shape=c(22,22),
col=c("#238b45", "#fb6a4a"),
alpha=c(.5, 1),
border.col = c("#00441b", "#fec44f"),
border.lwd=.8,
size=1.5,
labels = c("movement corridors", "pinch-points"))
#m3_b
tmap_save(m3_b, "4_output/presentation/pp_s_map_b.png", asp=0, outer.margins=c(0,0,0,0))
#
#
# m3_b<-
# tm_shape(base) + tm_rgb()+
# tm_shape(study_area)+
# tm_fill(col="white", alpha=.2)+
# # tm_shape(v_road_clip)+tm_lines(col="white")+
# # tm_shape(nscr)+
# # tm_fill(col="white", alpha=.6)+
# # tm_borders(col = "#636363")+
# tm_shape(corridors_clip)+
# tm_fill(col="#238b45", alpha = .5)+
# tm_layout(bg.col="#d9d9d9")+
# tm_shape(pp_s_clip)+
# tm_fill(col="#fb6a4a", alpha = 1)+
# tm_borders(col = "#636363", lwd=1)+
# tm_layout(legend.outside = TRUE, legend.text.size = .9, legend.bg.color="white", legend.frame=TRUE, legend.frame.lwd=.8)+
# tm_add_legend(
# type="symbol",
# shape=c(22,22),
# col=c("#238b45", "#fb6a4a"),
# alpha=c(.5, 1),
# border.col = "black",
# border.lwd=.8,
# size=1.5,
# labels = c("movement corridors", "pinch-points"))
# m3_b
#
# tmap_save(m3_b, "4_output/presentation/pp_s_map_b.png", asp=0, outer.margins=c(0,0,0,0))
m4<-
tm_shape(v_road_3)+tm_lines(col="white")+
tm_shape(nscr)+
tm_fill(col="white", alpha=.6)+
tm_borders(col = "#636363")+
tm_shape(corridors_clip)+
tm_fill(col="#238b45", alpha = .5)+
tm_layout(bg.col="#d9d9d9")+
tm_legend(outside= T)+
tm_shape(pp_w_clip)+
tm_fill(col="#3182bd", alpha = 1)+
tm_borders(col = "#636363", lwd=.7)+
tm_layout(legend.outside = FALSE, legend.text.size = .8, legend.bg.color="white", legend.frame=TRUE, legend.frame.lwd=1)+
tm_add_legend(
type="symbol",
shape=c(22,22, 22),
col=c("#238b45", "#3182bd", "white"),
alpha=c(.5, 1, 1),
border.col = "black",
border.lwd=.8,
size=1.5,
labels = c("movement corridors", "winter pinch-points", "N. Saskatchewan Central"))
m4
tmap_save(m4, "4_output/maps/pp_w_map.png", asp=0, outer.margins=c(0,0,0,0))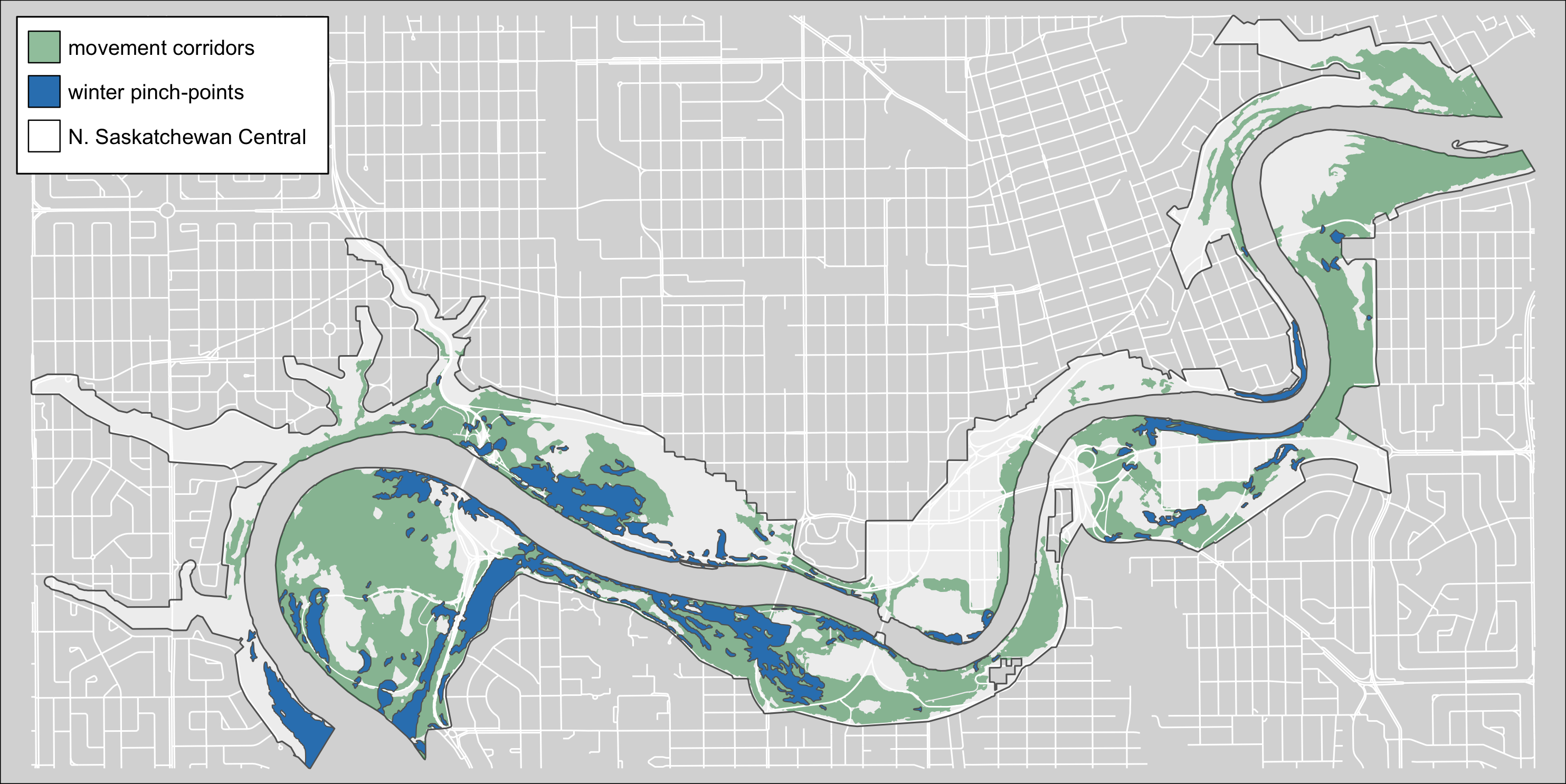
Figure 2.3: Winter terriestrical pinch-points within the North Saskatchewan Central region of the Ribbon of Green.
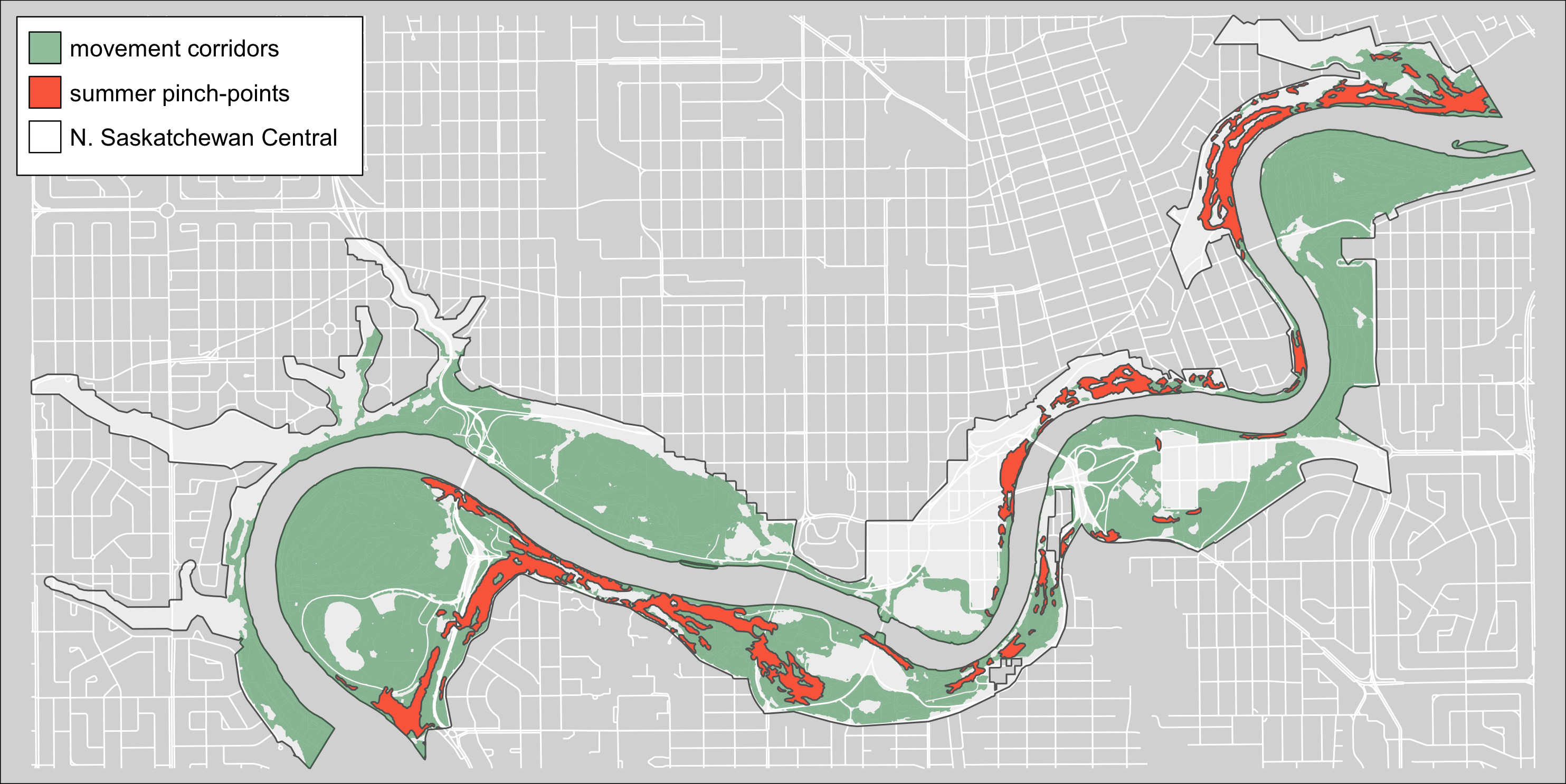
Figure 2.4: Summer terriestrical pinch-points within the North Saskatchewan Central region of the Ribbon of Green.
Cluster pinch-points
There are groups of pinch points very close to each other. This will lead to significant overlap in buffers. Since neighboring pinch-points will likely be caused by similar biophysical features, I decided to group neighboring pinch-points. Below, polygons are clustered based on a user defined threshold. I chose a 50 m threshold.
Filter out small pinch-point polygons
Cluster remaining polygons by proximity
# here is a function that takes an SF object, clusters features within a specified distance threshold, and merges them into a new object
clusterSF <- function(sfpolys, thresh){
dmat = st_distance(sfpolys)
hc = hclust(as.dist(dmat>thresh), method="single")
groups = cutree(hc, h=0.5)
d = st_sf(
geom = do.call(c,
lapply(1:max(groups), function(g){
st_union(sfpolys[groups==g,])
})
)
)
d$group = 1:nrow(d)
d
}
#set a distance threshold to group polygons
t<-25
pp_s_clip_clus<-clusterSF(x_f, set_units(t, m))
save(pp_s_clip_clus, file="3_pipeline/tmp/pp_s_clip_clus.rData")
## plot clustered pinch-points
tmap_mode("plot")
m5<-
tm_shape(v_road_3)+tm_lines(col="white")+
tm_layout(bg.col="#d9d9d9")+
tm_shape(nscr)+
tm_fill(col="white", alpha=.4)+
tm_borders(col = "#636363")+
tm_shape(pp_s_clip_clus)+
tm_fill(col="MAP_COLORS", palette = "Dark2", contrast = c(0.26, 1),alpha = 1, n= nrow(pp_s_clip_clus), legend.show = FALSE)+
tm_borders(col = "#636363")
m5
tmap_save(m5, "4_output/maps/pp_s_clus_map.png", asp=0, outer.margins=c(0,0,0,0))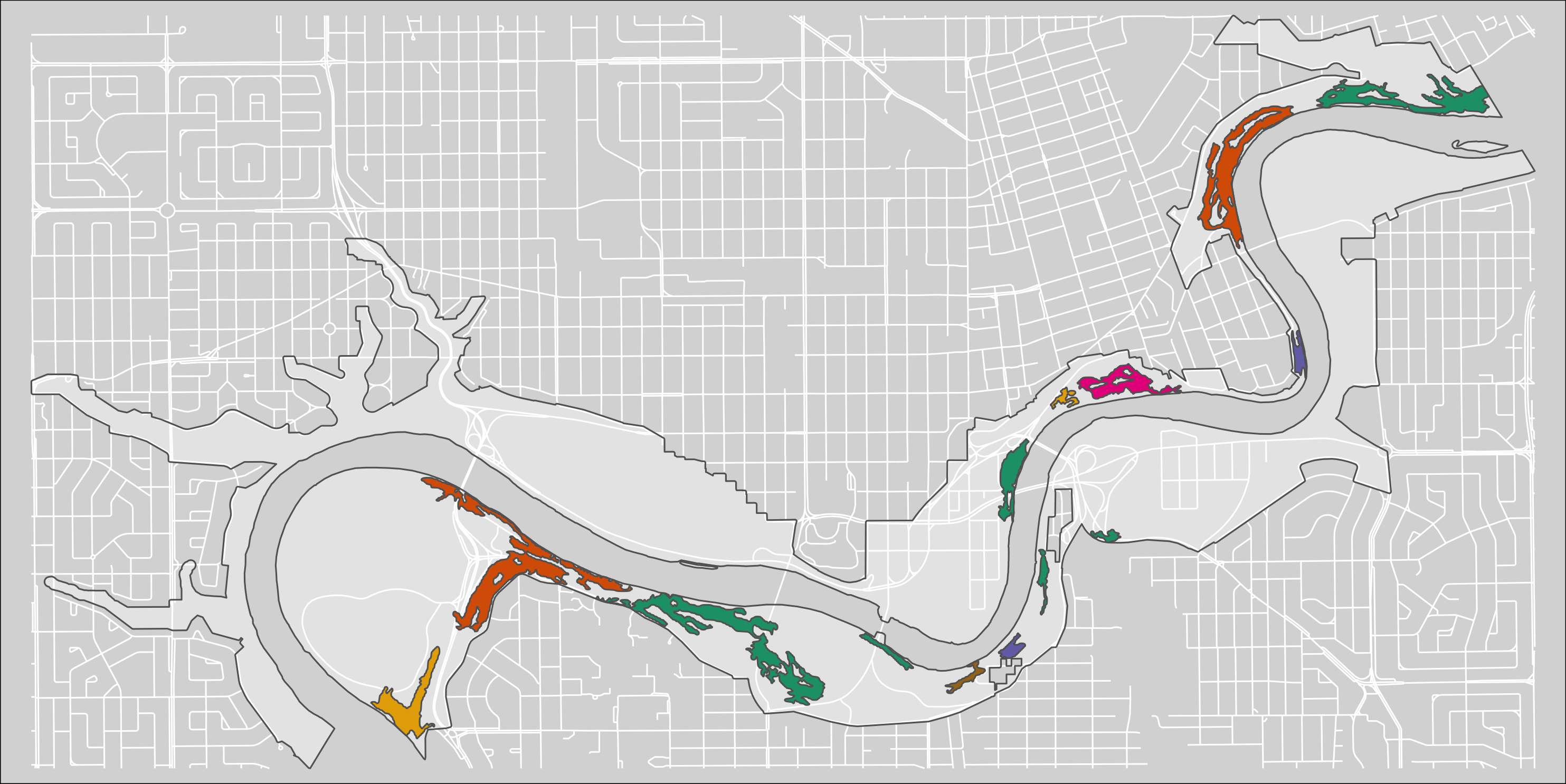
Figure 2.5: Clusters of pinch-points within 50m of each other.
Buffer pinch-points
Create an exterior buffer around pinch-point polygons. We will later characterize features within these buffers. I used a 30 m buffer.
pp_s_clip_clus_buf<-st_buffer(pp_s_clip_clus, dist= 30, singleSide = TRUE, endCapStyle = "ROUND",
joinStyle = "ROUND")
save(pp_s_clip_clus_buf, file="3_pipeline/store/pp_s_clip_clus_buf.rData")
## plot buffered clusters
m6<-
tm_shape(v_road_3)+tm_lines(col="white")+
tm_layout(bg.col="#d9d9d9")+
tm_shape(nscr)+
tm_fill(col="white", alpha=.4)+
tm_borders(col = "#636363")+
tm_shape(pp_s_clip_clus_buf)+
tm_fill(col="MAP_COLORS", palette = "Dark2", contrast = c(0.26, 1),alpha = 1, n= nrow(pp_s_clip_clus), legend.show = FALSE)+
tm_borders(col = "#636363")
m6
tmap_save(m6, "4_output/maps/pp_s_clus_buf_map.png", asp=0, outer.margins=c(0,0,0,0))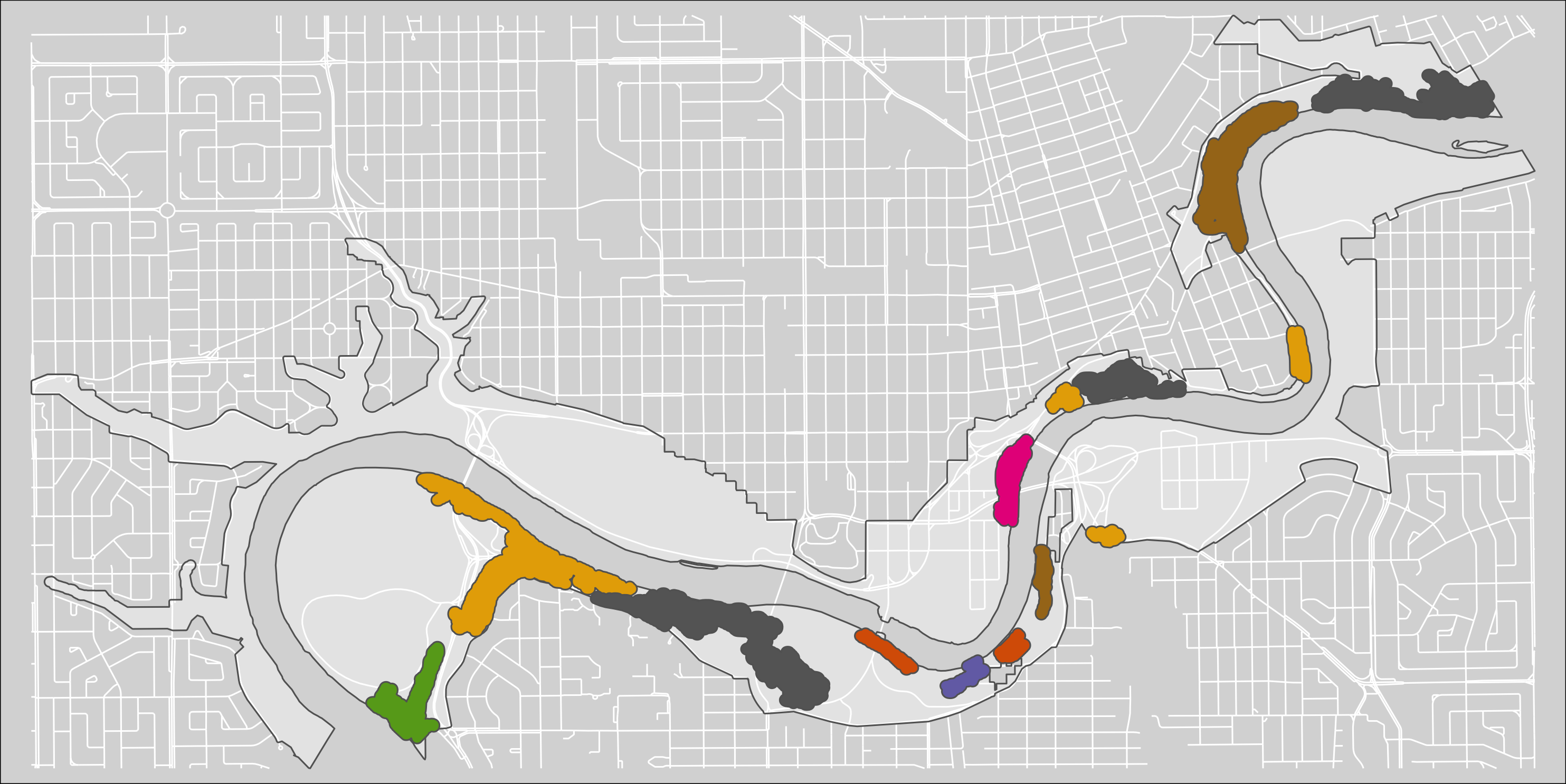
Figure 2.6: Clusters of pinch-points with a 30m buffer.
Exterior only buffer
pp_s_clip_clus_buf_ext<-st_buffer(st_boundary(pp_s_clip_clus), dist= 30, singleSide = TRUE, endCapStyle = "ROUND",
joinStyle = "ROUND")
#create a unique ID column
pp_s_clip_clus_buf_ext<-cbind(ID=1:nrow(pp_s_clip_clus_buf_ext), pp_s_clip_clus_buf_ext)
save(pp_s_clip_clus_buf_ext, file="3_pipeline/store/pp_s_clip_clus_buf_ext.rData")
## plot buffered clusters
m7<-
tm_shape(v_road_3)+tm_lines(col="white")+
tm_layout(bg.col="#d9d9d9")+
tm_shape(nscr)+
tm_fill(col="white", alpha=.4)+
tm_borders(col = "#636363")+
tm_shape(pp_s_clip_clus_buf_ext)+
tm_fill(col="MAP_COLORS", palette = "Dark2", contrast = c(0.26, 1),alpha = 1, n= nrow(pp_s_clip_clus), legend.show = FALSE)+
tm_borders(col = "#636363")
m7
tmap_save(m7, "4_output/maps/pp_s_clus_buf_ext_map.png", asp=0, outer.margins=c(0,0,0,0))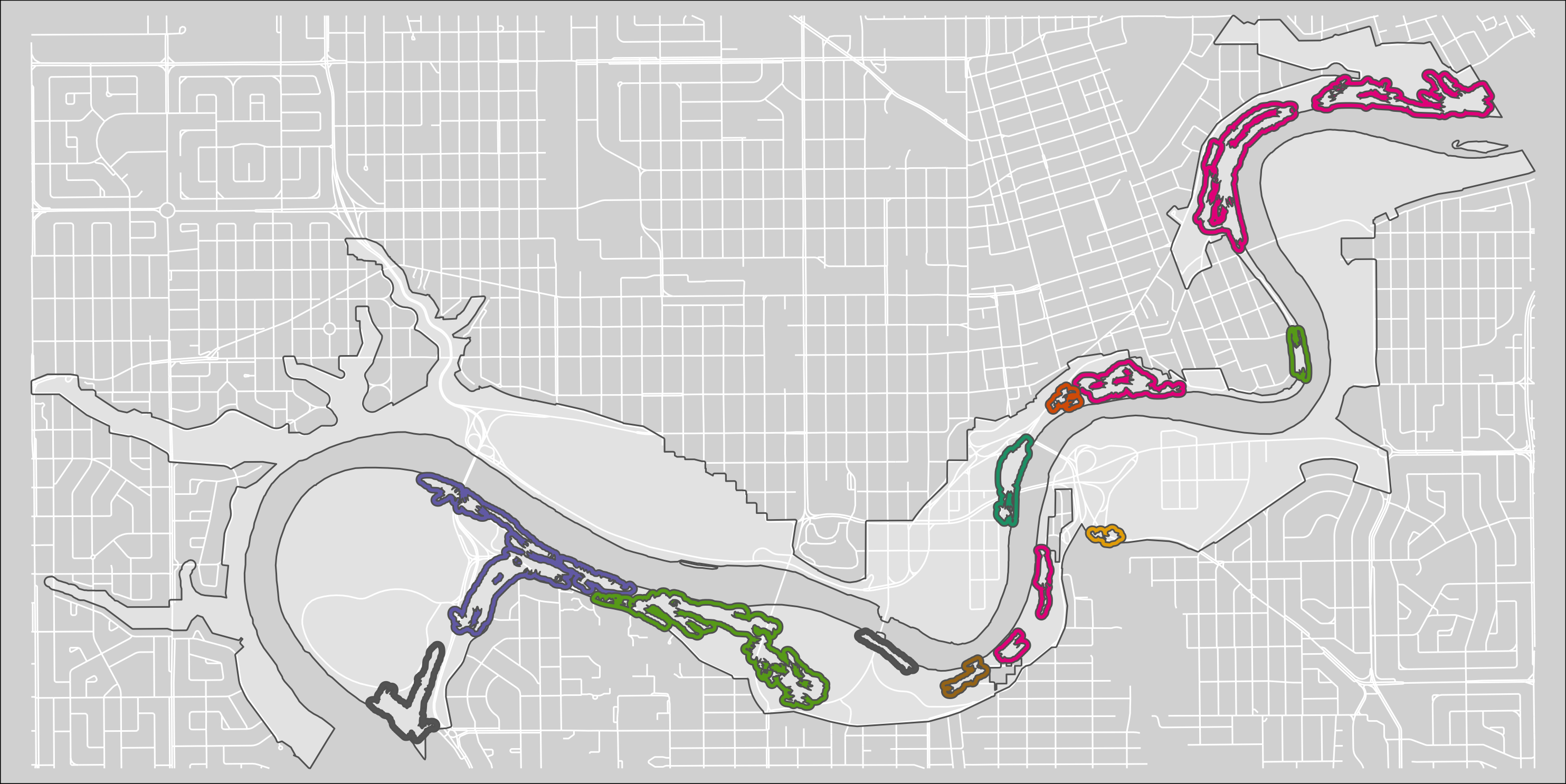
Figure 2.7: Clusters of pinch-points with a 30m exterior buffer.
Current map
Load and clip the terrestrial circuitscape current map.
Summer coyote
cur_map_sum<-raster("1_data/external/Circuitscape/Map/Coyote_Summer_All_curmap.tif")
projectRaster(cur_map_sum, crs=st_crs(study_area))
cur_map_sum_clip<-crop(cur_map_sum, study_area)
save(cur_map_sum_clip, file="1_data/manual/cur_map_sum_clip.rData")
m8<-
tm_shape(cur_map_sum_clip)+
tm_raster(palette="-RdBu",
title = "Current",
style="cont",
drop.levels = TRUE,
contrast = c(0,1)
)+
tm_layout(legend.show = FALSE,
legend.outside = FALSE,
legend.text.size = .8,
legend.bg.color="white",
legend.frame=TRUE,
legend.title.size=1,
legend.frame.lwd=.8,
)
m8
tmap_save(m8, "4_output/maps/cur_map_sum_map.png", outer.margins=c(0,0,0,0)) 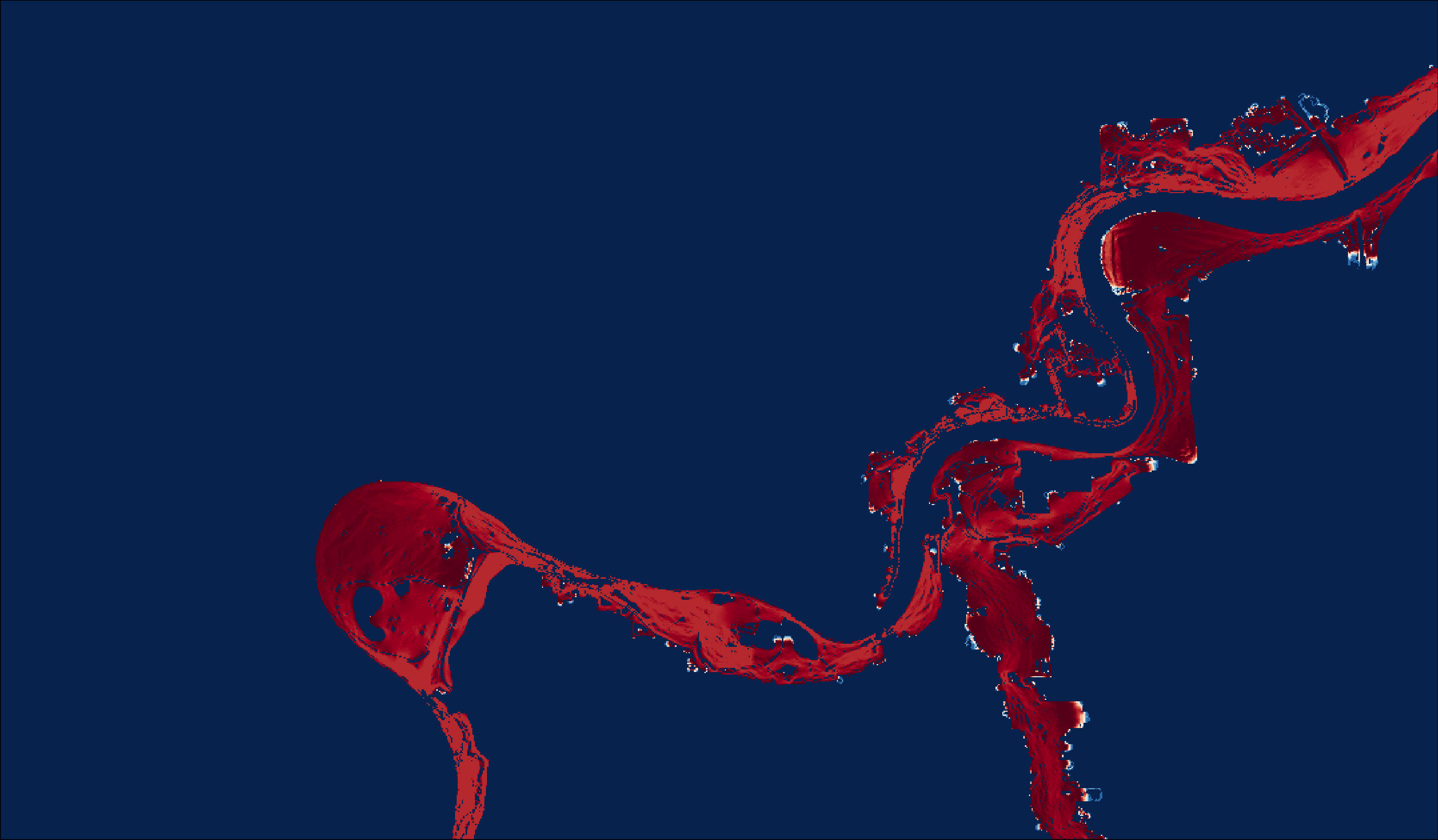
Figure 2.8: Summer coyote current map.
Winter coyote
cur_map_win<-raster("1_data/external/Circuitscape/Map/Coyote_Winter_All_curmap.tif")
projectRaster(cur_map_win, crs=crs(study_area))
cur_map_win_clip<-crop(cur_map_win, study_area)
save(cur_map_win_clip, file="1_data/manual/cur_map_win_clip.rData")
m9<-
tm_shape(cur_map_win_clip)+
tm_raster(palette="-RdBu",
title = "Current",
style="cont",
contrast = c(0,1),
)+
tm_layout(legend.show = FALSE,
legend.outside = FALSE,
legend.text.size = .8,
legend.bg.color="white",
legend.frame=TRUE,
legend.title.size=1,
legend.frame.lwd=.8,
)
m9
tmap_save(m9, "4_output/maps/cur_map_win_map.png", outer.margins=c(0,0,0,0)) 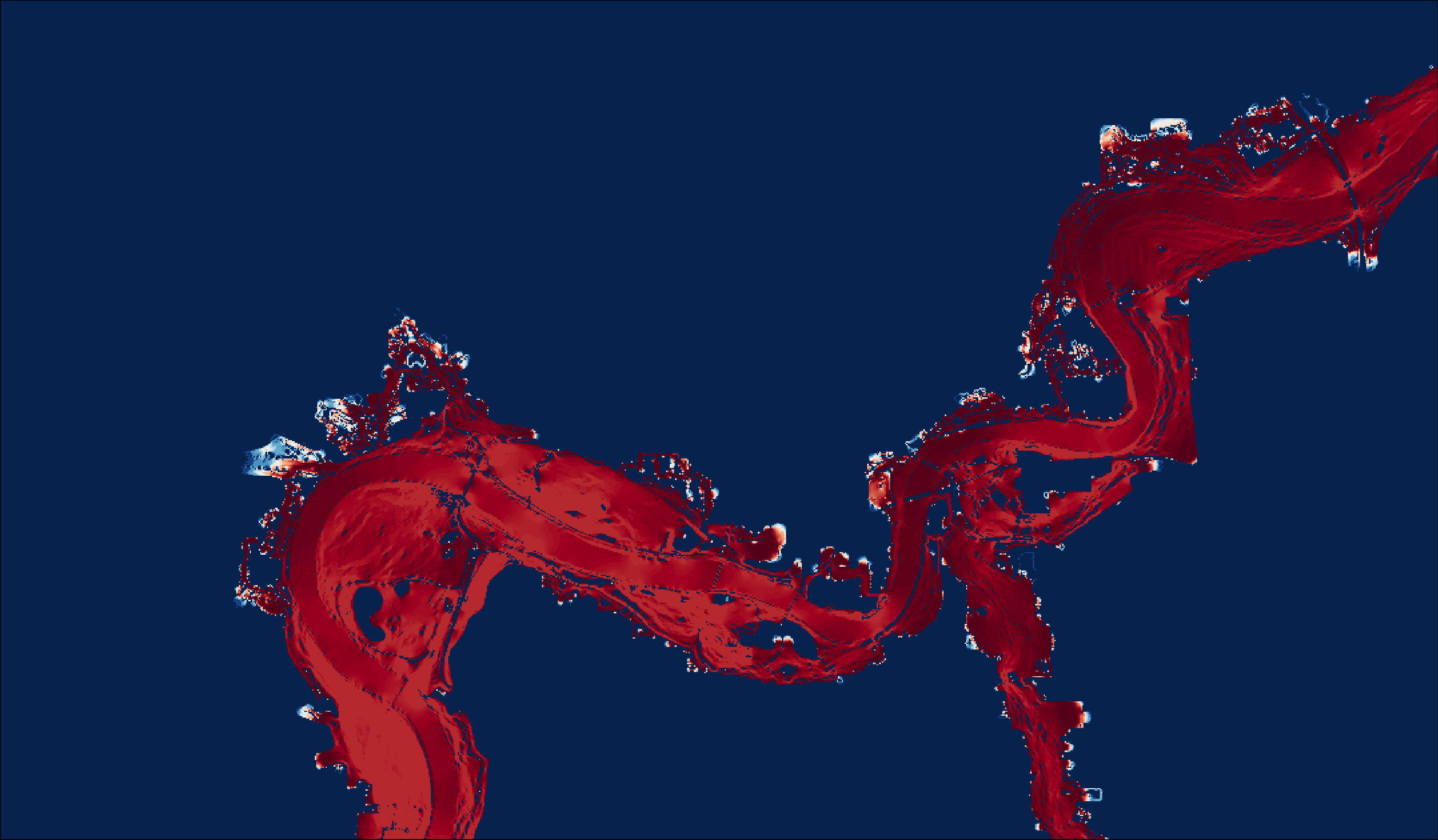
Figure 2.9: Winter coyote current map.