6 Assembling the ecological puzzle
Jannice Friedman ![]() and Aly Van Natto
and Aly Van Natto ![]()
How to cite this chapter
Friedman J. and van Natto A. C. (2023) Assembling the ecological puzzle. In: Cousens R. D. (ed.) Effective ecology: seeking success in a hard science. CRC Press, Taylor & Francis, Milton Park, United Kingdom, pp. 81-96. Doi:10.1201/9781003314332-6
Abstract Ecological research is often concerned with complex and tangled systems. Sometimes by necessity and sometimes by choice, researchers make simplifying assumptions and focus only on one part of the ecological puzzle. That is not to say that a single question then leads to a direct, simple answer. It is still a puzzle, but at a finer resolution. If we are to be confident that our answers are probable rather than merely plausible explanations, we need to integrate a diversity of methods and approaches, in an iterative process that eventually leads to a deep understanding. Here we review a range of empirical methods that can be integrated to make the most effective progress in studies of evolutionary aspects of ecology. We use three case studies of research programs – on threespine stickleback, deer mouse, and seep monkeyflower – to highlight successful approaches to assembling the pieces of an evolutionary puzzle. To implement a diversity of approaches, collaboration with scientists with complementary skills can be beneficial.
6.1 From the picture on the outside to the pieces in the box
Ecologists often choose to focus on particular aspects of an organism, community or ecosystem. Despite reducing the scope of their questions, they are still dealing with the outcome of a myriad of components, processes, and interactions. Examining any one aspect in isolation may fail to lead to an adequate understanding of the entire (sub)system.
The steps necessary to fully understand any scientific issue ideally involve a series of studies, at appropriate scales, addressing the interactions between the components and their effects on each other. Surveys can generate hypotheses about what is going on; theoretical modelling can suggest what is liable to occur under particular scenarios, while experiments can test hypotheses by showing how the system changes as variables are altered in a controlled manner. Researchers learn from each of these steps, propose further questions that will unravel the system and develop appropriate follow-up studies: an iterative process that leads to ever-greater knowledge. Perhaps they model it or simulate its workings in greater detail; they test whether some explanations are better supported by others; and they modify their ideas accordingly. And, step by step, they get closer to a true comprehension of how the system works (see Figure 1.1).
Ecology is rooted in natural history, and indeed natural exploration and observation form the foundation of almost every discipline in biology. Natural history surprises us, amazes us, and often leaves us wanting more explanation. It is intuitive to go directly from an interesting observation to the inference of a process that caused it – whether it be explanations for a giraffe’s long neck, why bipedalism evolved in humans or how the leopard got its spots. These ‘just-so stories’ reflect a desire to explain form and function by imagining and reconstructing the processes that gave rise to them. The phrase ‘just-so stories’ is often used pejoratively and critically in the scientific academic literature; however, these types of observations can form the basis of hypotheses and be an important part of the scientific process (Smith et al. 2016). The crux of science is that we do not accept claims without evidence and substantiation. Plausible hypotheses need to be challenged and tested until we are as certain as we can be that we have found the probable explanation. The early speculations need to be just that, the beginning step that will lead towards rigorous explorations. Follow-up research that tests hypotheses, modifies explanations, interprets data and builds upon knowledge is essential.
Research in the area of evolutionary ecology provides outstanding examples of how to move through the steps described above. The goal of research in this area is to understand the processes generating and maintaining variation in nature. Having observed patterns of phenotypic and genetic variation in time and space, we use a wide array of techniques to best determine how, and over what timescales, those patterns arose. We can start by speculating. We can look at survey data for evidence of particular processes, and use theory and modelling to understand what to expect under specific circumstances. We can conduct experiments to show that these explanations are plausible because we can replicate them; we can further elucidate those mechanisms. And we can end up identifying the genes or mutations encoding differences in phenotypes.
In this chapter, we break down the foundational concepts that underlie the integration of some of the main techniques used in evolutionary ecology, and then use three case studies that exemplify the kind of traits and species that have been used to successfully connect the multitude of factors relating to the environment, phenotype and genotype that underlie adaptive variation.
6.2 Difficulties in drawing inferences
Despite a deep history and rich tradition of research in evolutionary ecology, there remain fundamental issues that we need to grapple with in every study, complications at the core of how we define the concepts that ultimately constrain the inferences that we can make. These complications include things like how we specify the traits on which selections acts, how we decide on the fitness ‘currency’ through which evolution proceeds, how we cope with correlations among traits and interactions among processes, and how we recognise adaptation when we are confronted with it. Thorny issues such as these are part of the reason why iterative and integrative methods are necessary to link together the pieces of the research puzzle, so that a complete picture of the system can emerge. While the issues we discuss in this section are specific to a particular type of research, we hope they act to exemplify the value of being fully cognisant of our assumptions and obstacles in all ecological research. Each of us makes many assumptions, some small, some large. It is only by critical deliberation of them that we can determine what to measure and what to do with the data that we collect (see also Chapter 7).
6.2.1 What are the traits on which selection has acted?
Identifying a trait of ecological significance often begins with natural history observations of variation within or among populations. The very existence of this variation often hints at its evolutionary potential. While these traits may not be causally related to fitness, they may be related to components of fitness, either mechanistically through genetic, developmental, morphological or physiological connections, or statistically through their correlational impact on survival and reproduction. While identifying the existence of trait variation can be a starting point in a research program, careful experiments and modelling are needed to demonstrate the role of natural selection and adaptation in determining the extent and maintenance of that variation (Stinchcombe et al. 2002). Crucially, the relative fitness of any particular trait depends on the environment the organism experiences. Thus, traits that confer high fitness now may become less fit in the future if the environment changes (and vice versa). On the other hand, currently beneficial traits may have been present long before the current environmental conditions, without having conferred any previous advantage.
6.2.2 How do we recognise adaptation?
Living organisms have some amazing complex adaptations that, it is tempting to assume, are the result of natural selection. But not all characteristics or seemingly adaptive traits are adaptations at all. Evolutionary biologists (and natural history documentaries) are sometimes inclined to invoke ‘just-so stories’ or speculate that selection could have produced a particular trait for a particular function.
While it is tempting to look for adaptive explanations to describe the extraordinary diversity around us, alternative explanations must be considered. For example, it is possible that a particular trait of interest reflects genetic drift; or that it is constrained through physiological, developmental or genetic mechanisms; or that it emerged as a by-product of selection on other traits; or that it was once adaptive but due to environmental changes it is no longer. Carefully designed experiments or robust comparisons between appropriate groups are necessary to demonstrate adaptation (Olson-Manning et al. 2012).
6.2.3 How do we measure fitness?
The word ‘fitness’ is meant to capture the total relative performance of individuals that will predict their success and their long-term contribution to population dynamics. But the definition of fitness often depends on the question of interest and the biological system under consideration (Hendry et al. 2018). A useful metric of fitness comprises both survival and reproduction, measured as lifetime reproductive success. For species that persist for multiple years and with multiple bouts of reproduction (i.e. iteroparous species), determining lifetime fitness can be challenging or near impossible. Thus, we usually measure just components of fitness, or make measurements in a single year or season. Additionally, for many organisms it can be difficult to ascertain male reproductive success, and co-sexual organisms can have different fitness through female- and male- function. Molecular approaches to assigning paternity have facilitated the ability to quantify male fitness empirically, but quantifying male fitness continues to pose technical and logistical challenges.
6.2.4 What if there are trade-offs and correlations between traits?
A trade-off exists when an evolutionary change that increases fitness in one phenotypic trait or context causes a decrease in fitness in another trait or context. Correlations between traits can constrain or facilitate evolutionary responses to selection (e.g. Etterson and Shaw 2001). Various analytical approaches can help to parse out the tangle of correlations between traits (Lande and Arnold 1983). Using multiple regression, it is possible to determine how much of the covariance between a trait and fitness is independent of other correlated traits. This approach has changed the way that selection is studied in the wild, and it ameliorates the problem of confounding trait correlations. Furthermore, the genetic architecture underlying trait correlations can help us understand both their evolutionary causes and consequences. For example, when one gene affects two or more traits – known as pleiotropy – the two traits are unable to evolve independently. A similar outcome can occur when the genes for traits are in close physical proximity on a chromosome and experience ‘linkage disequilibrium’ (see Slatkin 2008). Alternatively, one trait may be influenced by more than one gene – known as epistasis – and depending on the nature of the interactions between the genes can mean that the phenotypic outcomes are constrained. Well-designed experiments using genetic mapping populations, or lines where genotypes are known and fixed, can allow researchers to gain an understanding of the independent effects of single traits or genes.
6.2.5 What are the timescales?
One of the challenges of linking ecology and environmental differences to evolutionary processes that generate phenotypic variation is the timescale that underlies these processes. Research in the past few decades has demonstrated that selection and evolution in natural populations is often much faster, more dynamic and on shorter timescales than originally conceived (Hairston et al. 2005). In many recent studies, rapid evolutionary change has been shown to occur on the timescale of 1 - 100 years (Messer et al. 2016). To understand the processes generating and maintaining trait variation, it is useful to look at the evolution of novel traits when they first arise. This approach often relies on comparisons between populations that differ in their environment, in which natural selection has acted on genetic variation to produce phenotypic differences between the populations. A challenge in studying variation at short timescales is that the differences (genetic or phenotypic) between individuals may not be great. Nonetheless, studying recent evolutionary changes is informative because we know or can reliably infer the ecological context in which selection is acting.
6.3 The methods available
How do we go about dealing with the conceptual or methodological issues above and assess the validity of how the different parts of a system work and contribute to the overall outcome? The specific methods employed will depend on the research field and the type of information required. The important aspect that we wish to emphasise here is that relying on any one method, or only one approach, is unlikely to lead to a complete picture. To demonstrate this, we use examples from evolutionary biology highlighted by the complications from the previous section.
In many cases, there is a clear logical flow beginning with natural history observations, to identifying the mechanisms and processes that could give rise to the phenotypic and genotypic variation and then demonstrating which have (probably) given rise to that variation (Figure 6.1). One of the strengths of evolutionary ecology is that it has a rich tradition of formal theoretical modelling, which often provides the foundation for how we tackle empirical research. Here we outline the key research themes and the types of methods available to first drill down and understand mechanisms and processes, and then to integrate and connect between the various pieces of research.
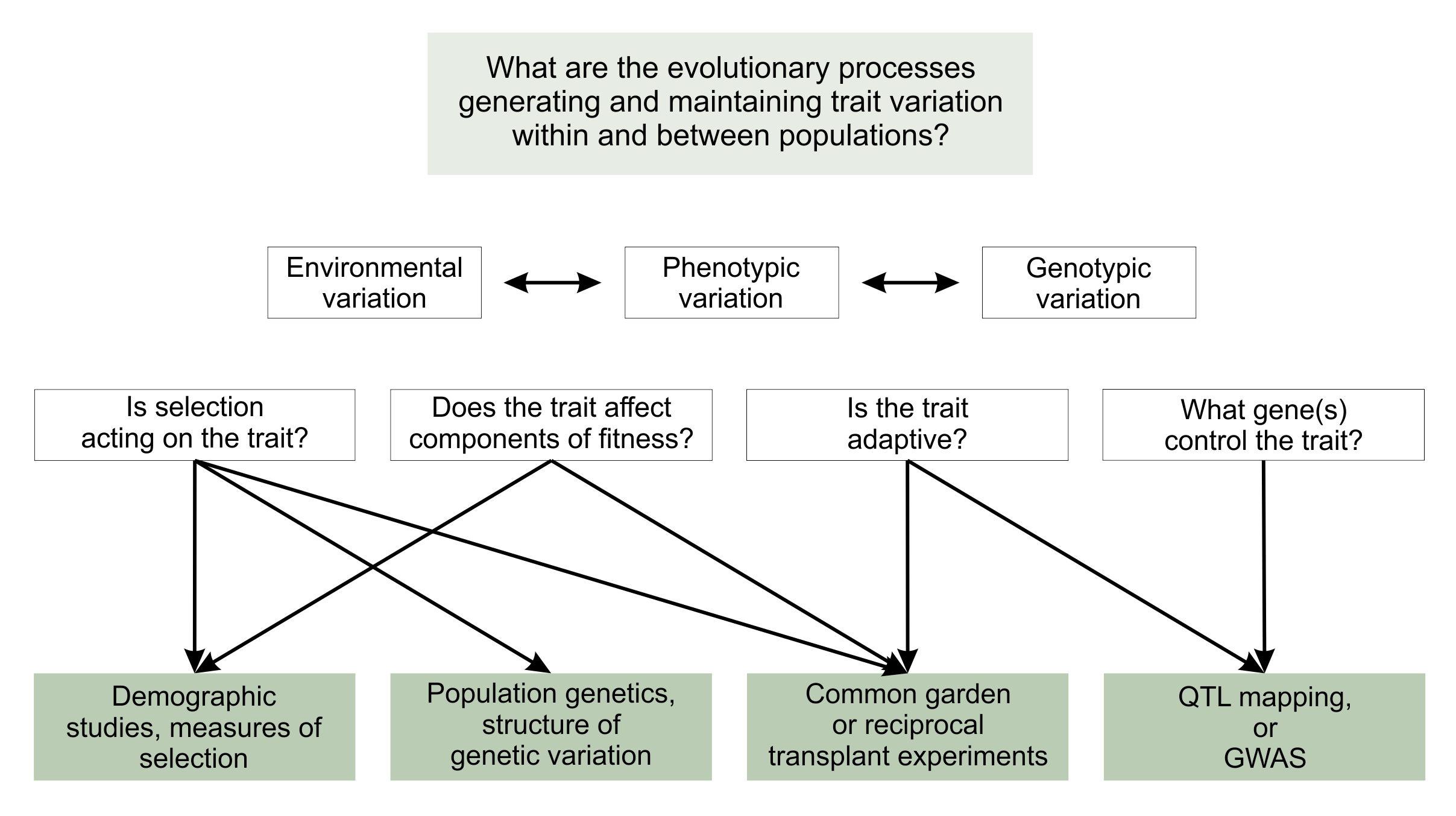
Figure 6.1. A schematic of the ideas discussed in this chapter. Beginning with the broad question aimed at understanding the evolutionary processes generating and maintaining trait variation in nature, we illustrate the component questions that can be tackled with specific methods, and then integrated to return to the larger question.
An understanding of natural selection is at the core of evolutionary ecology, and yet identifying and studying natural selection is notoriously difficult (Endler 1986). Natural selection is generally ubiquitous in nature and a characteristic of populations everywhere. Breaking down the process of evolution by natural selection reveals the nature of its parts.
Whenever trait values are associated with differential reproductive success (fitness) within a generation, phenotypic selection is at play. The effect of phenotypic selection is then mediated through the heritability of the trait, and its underlying genetic architecture, to determine evolutionary change. The first step then for understanding evolution by natural selection is to quantify the nature and strength of selection and the patterns of phenotypic selection observed in nature.
To determine whether selection is likely to be acting on a particular trait in a population, we first estimate the fitness associated with the various trait values (Box 6.1, Figure 6.2a). This allows us to establish that there exists an average, non-random variation in fitness, and that the difference in fitness between individuals is caused by, or correlated with, their phenotypes (trait values). Typically, this is done by measuring trait values for a sample of individuals within a population that are of similar age (e.g. juvenile animals or annual plants). Individuals are marked and then followed over a period of time to determine their survival and/or reproductive success. In an ideal scenario, we would attain total lifetime fitness. In reality, this is seldom achieved, and most researchers measure just some components of fitness: survival, mating success or fecundity. Statistical approaches are then used to relate the trait values to fitness: a regression analysis allows us to determine the strength and mode of selection acting on the trait of interest.
We provide a brief description of some of the methods used as part of an iterative process to understand the factors generating and maintaining trait variation within a species. Although these methods can be used to answer a myriad of questions, this example focuses on the process of natural selection and how it affects phenotypes in different environments.
Many studies of adaptation by natural selection begin with an observation of variation in a trait of interest within or among populations. The trait of interest may range from the molecular level (e.g. expression of an immunity-related gene) to easily identifiable traits (e.g. flower colour), and usually precedes a prediction of adaptive significance. In Figure 6.2a we illustrate a trait that is under both directional and stabilising selection, and the pattern suggests that the trait influences fitness directly in that environment. However, the fitness function may not be consistent between populations, years or environments.
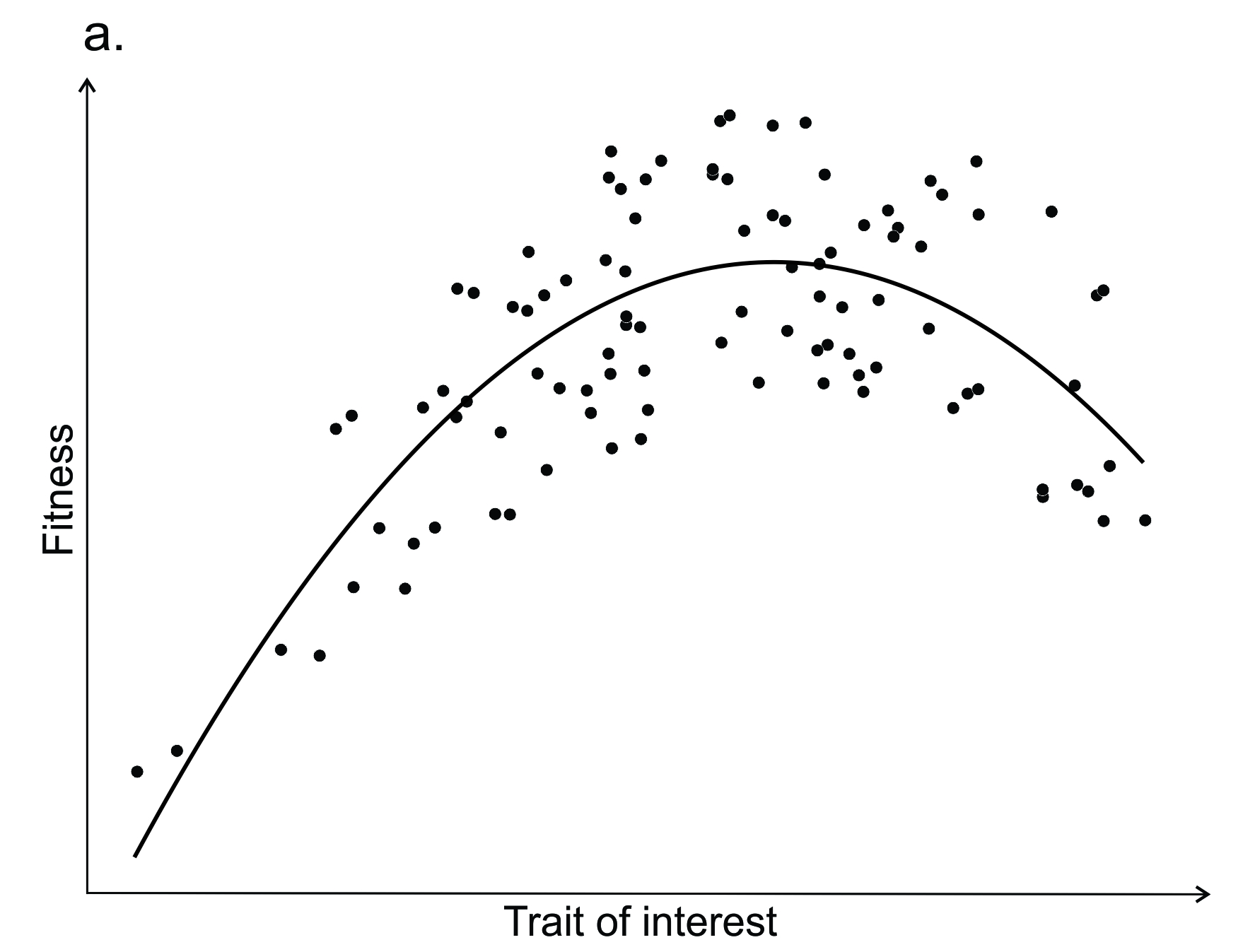
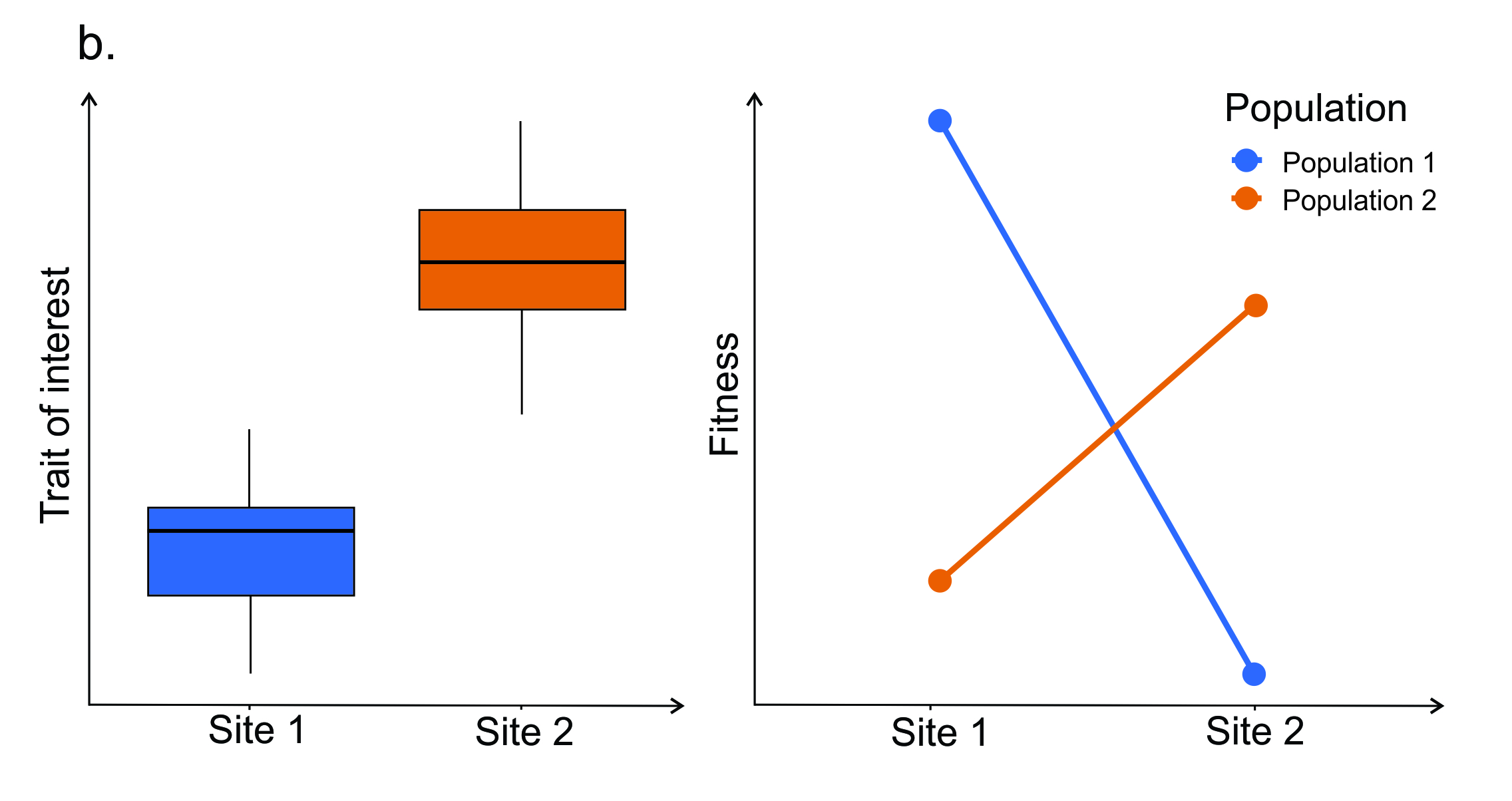
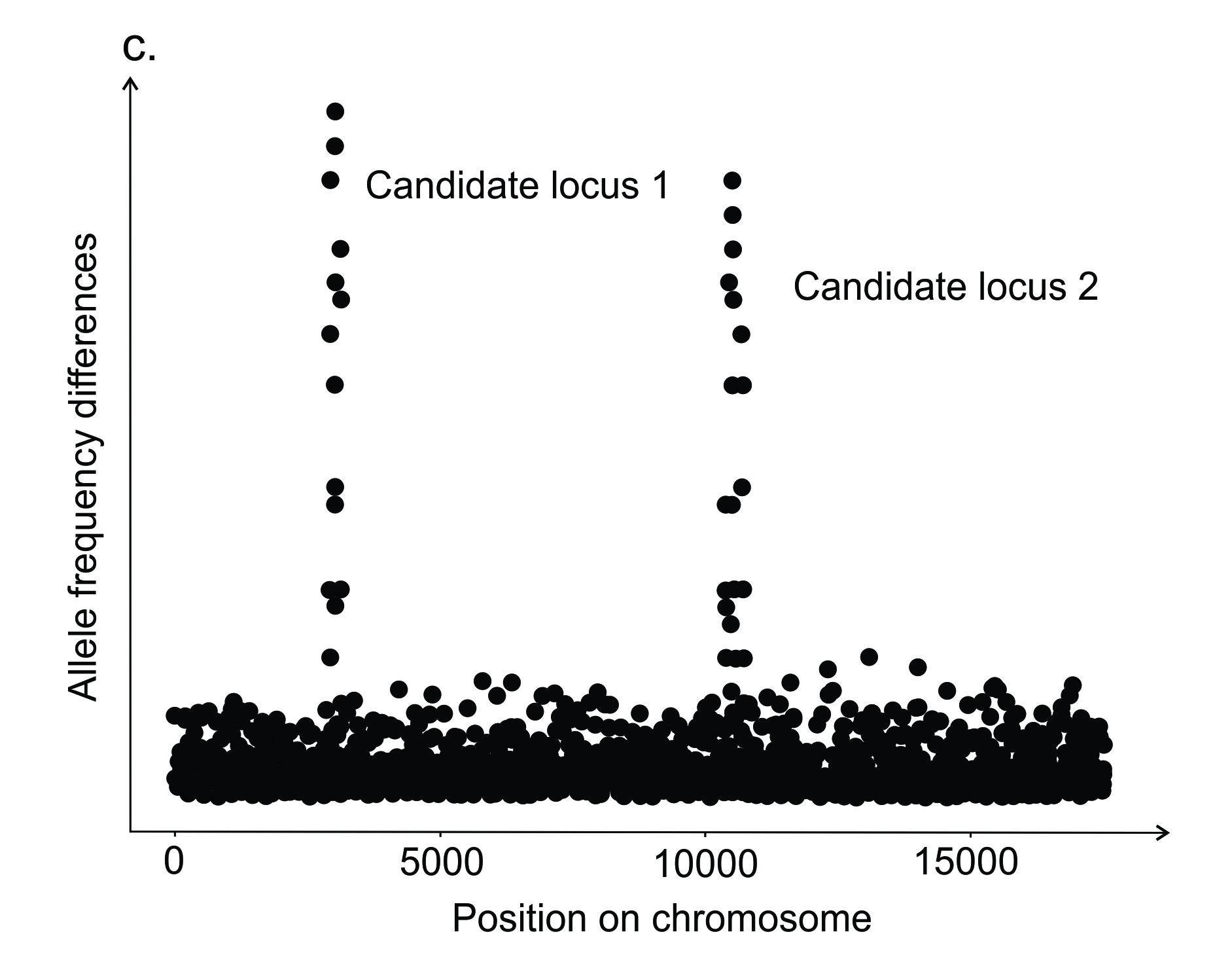
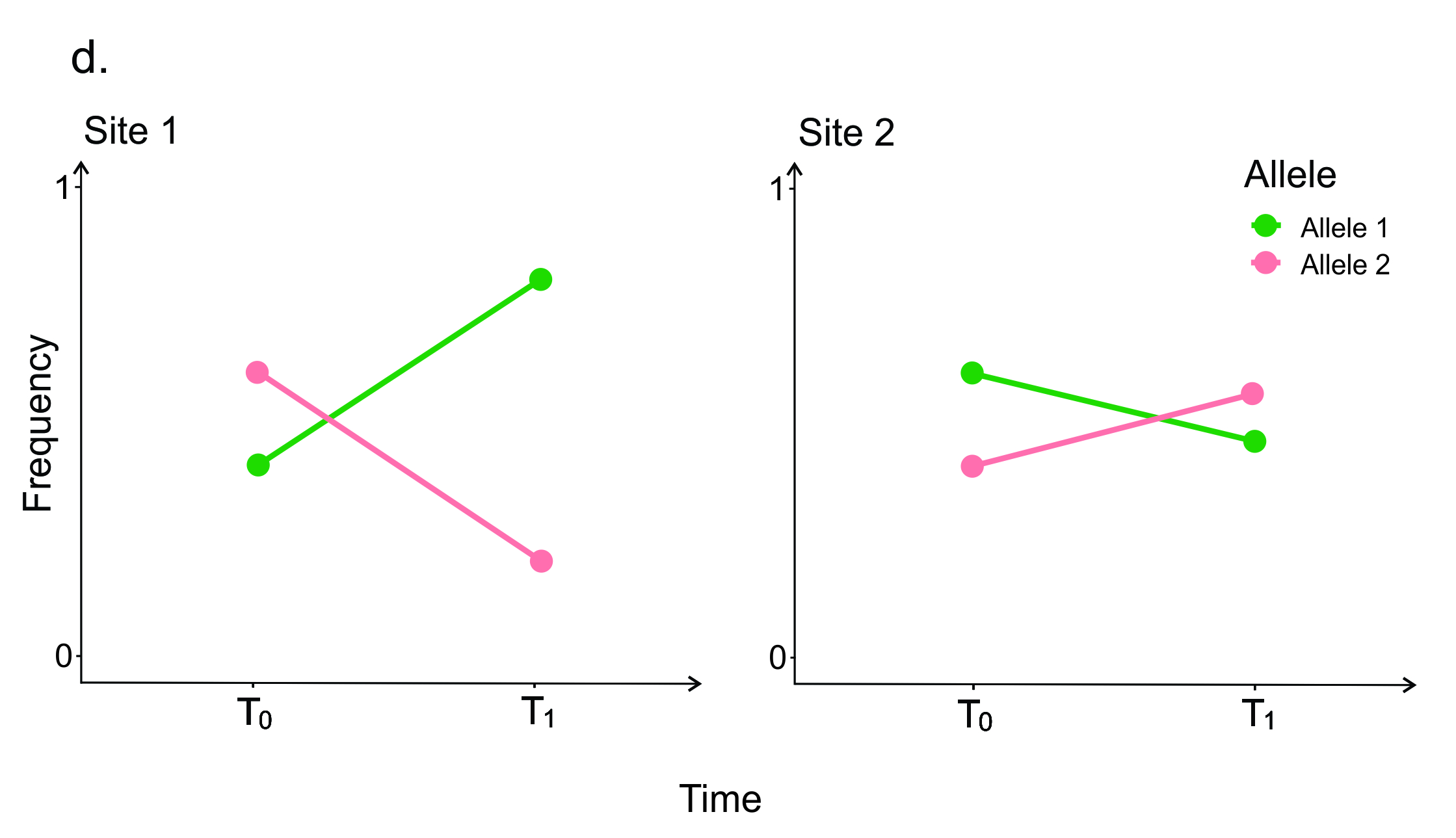
Figure 6.2. Some of the methods used as part of an iterative process to understand the factors generating and maintaining trait variation within a species. See text for details.
One approach to testing for adaptive significance of a trait is to determine if populations are locally adapted to their home environment. To compare populations, common garden and reciprocal transplant experiments are useful methods. These traditional approaches have stood the test of time because of their effectiveness. Figure 6.2b (left) shows differences between traits in two populations. Figure 6.2b (right) shows hypothetical results from a reciprocal transplant study where individuals from ‘Population 1’ are transplanted into the environment of ‘Population 2’ and vice versa. The change of phenotypes expressed by a genotype in different environments indicates local adaptation resulting from divergent selection because fitness is higher in the home environment (see Anderson et al. 2011 for further details). Researchers may conclude that the trait has diverged due to selection in opposing directions in these populations.
The next question may be to understand the evolutionary history of the causal genes underlying the trait differences. One approach to this is to use population genomic sequencing to compare allele frequency differences across the genome between populations (Figure 6.2c). Regions of the genome that have higher differences than expected by neutral processes (e.g. Candidate loci 1 and 2 in Figure 6.2c) can identify putative candidate genes that have evolved under spatially divergent selection and indicate adaptive loci. Further investigations may assess possible function of candidate genes by comparing them to an annotated genome. Genome-wide studies can tell us about the evolutionary history of adaptive loci (Barrett and Schluter 2008). For example, if selection favours a beneficial allele from the moment it enters the population as a single copy mutation (hard selective sweep) there is a different pattern of genetic variation around the locus compared to when multiple mutations at the same gene or existing variation becomes beneficial (soft selective sweep).
Once researchers have identified the genes controlling adaptive traits, they can connect the genotypic differences back to the ecological mechanisms causing selection. One way to do this is to combine a common garden experiment with genetic sequencing. For example, in Figure 6.2d, individuals were genotyped at ‘Candidate gene 1’ and then put into a reciprocal transplant experiment and the frequency of the allele is then sampled through time. In this example, ‘Allele 1’ originates from ‘Population 1’, and ‘Allele 2’ originates from ‘Population 2’. If allele frequencies change more drastically at one site, then it suggests that selection is stronger in that environment: linking evolution back to ecology (see Savolainen et al. 2013).
The process of adaptation occurs as populations of organisms evolve to become better suited to their environments, through natural selection increasing the prevalence of advantageous traits. A curious feature about the concept of natural selection leading to adaptation is that the chief action of natural selection is to eliminate or reduce the frequency of low-fitness traits. In fact, most forms of natural selection will deplete genetic variation within populations. Nonetheless, adaptation has given rise to extraordinarily elaborate and complex characters. The key generative process occurs as new genetic variation is introduced by mutation and recombination (in sexually reproducing organisms), or through gene flow via migration or hybridisation with other species. Beneficial traits increase through time, and the direction of adaptive change depends on the environment. Implicit in this (and sometimes underappreciated) is that adaptations are always to the conditions experienced by past generations.
Ecologists and evolutionary biologists often focus on differences between populations, and these differences are frequently interpreted in the context of selection and adaptation. But not all differences among populations or species are adaptive, an idea eloquently described by Gould and Lewontin (1979). Differences between populations or species could be caused by genetic drift and reflect neutral processes (e.g. Husband and Barrett 1992). It is also possible that the traits of interest that differ between populations did not evolve for those purposes at all or evolved as a by-product of selection on other traits (e.g. Bourdeau 2010). Particularly when considering differences between species, it is possible that trait differences are due to historical contingency and that they are simply the features that happened to be inherited from ancestors (Blount et al. 2018). For all these reasons, defining and studying adaptation can be challenging.
Experiments are among the most powerful tools to isolate the effect of a single, well-defined factor. A powerful and long-established experimental approach for identifying adaptation, and particularly local adaptation, involves the use of common garden experiments and reciprocal transplant experiments (Blanquart et al. 2013). Because local adaptation occurs when organisms have higher average fitness in their local environment compared to individuals from elsewhere, by using an approach that explicitly measures fitness of individuals from different environments, researchers can compare their performance and aim to identify the traits that contribute to those fitness differences (Box 6.1, Figure 6.2b). The history of transplant experiments goes back to studies of spatial phenotypic variation in plants (von Marilaun and Oliver 1895). Turesson (1922) first used transplant experiments to reveal that spatial variation across a species range is genetically based and could be associated with performance differences and adaptive traits between populations.
The mid-20th century reciprocal transplant experiments of Clausen et al. (1940), demonstrating adaptation by plants along an altitudinal climatic gradient, are arguably the most famous, and following these classic experiments, a large number of studies have since estimated local adaptation in a wide variety of species (Hereford 2009; Briscoe Runquist et al. 2020). These studies indicate that habitat-mediated natural selection plays a pervasive role in the maintenance of genetic variation among locations with contrasting environments.
Natural variation within populations has long been the fodder of ecologists and evolutionary biologists alike. The largely phenotypic approaches discussed above allow us to predict and interpret changes in traits over time or space. However, not all approaches require an understanding of the traits under selection – researchers can use DNA sequence variation to examine population genetic processes, like the effects of gene flow, population structure and inbreeding. Further, many long-standing questions about the genetic basis of phenotypic evolution and adaptation can be addressed by combining an understanding of trait variation with the genetic mechanisms. Identifying the genetic changes underlying adaptation allows researchers to address whether phenotypic convergence involves repeatable genetic changes (Stern 2013), whether adaptive mutations are more likely to occur in coding or regulatory regions (Wittkopp and Kalay 2012), whether adaptive alleles are more often of large or small effect (Yeaman 2015), and assess the relative contributions of adaptive evolution, balancing selection, deleterious variation and genetic drift in maintaining variation (Mitchell-Olds et al. 2007).
Before modern molecular biology, studying genetic variation in natural populations was limited to visible phenotypes with a known genetic basis, and early theoretical population geneticists used this type of variation to study evolutionary processes (e.g. Fisher 1930; Wright 1931; Haldane 1932). Many of the studies on major gene polymorphisms come from highly visible trait variation, with examples including male horn type in sheep (Johnston et al. 2013), wing spot variation in butterflies (Reed et al. 2011) and flower colour variation (Brown and Clegg 1984). However, organisms vary in complex ways in their anatomy, physiology and behaviour. For most traits, this variation reflects the combination of multiple environmental and genetic factors. Later we will discuss three case studies of selection - coat colour in deer mouse, freshwater adaptation in stickleback, and copper tolerance in monkeyflowers – all of which identify the cause of natural selection, connect this with changes in allele frequencies, and use genomic and bioinformatic approaches to identify the loci responsible for phenotypic variation.
Advances in technology enabled experimental approaches to identify the genetic architecture of more complex characters, using a combination of genomics and bioinformatics. First, quantitative trait locus (QTL) mapping was used to identify the genomic location, number and effect sizes of genetic changes that underlie phenotypic differences among natural populations of plants and animals. Approaches that use whole genome sequencing, such as genome-wide association studies (GWAS), can allow researchers to address whether the genetic changes that underlie phenotypic evolution and adaptation are found together in particular regions of the genome or whether the genes are distributed across the genome (Box 6.1, Figure 6.2c,d). These questions are important if we wish to understand the process of an organism adapting to a new environment, because adaptation usually requires multiple phenotypic changes. For example, consider an ant species that is adapting to a warmer, urban environment from a cooler, rural environment (Martin et al. 2021). Adaptation to the new environment will require multiple phenotypic differences, including morphology, life history and physiology. The process of adaptation may be facilitated if the same genetic changes give rise to multiple phenotypic differences (pleiotropy), or if multiple phenotypes are inherited together due to linkage of genes. Filling in these details allows us to broadly understand the evolutionary process without making simplifying assumptions.
6.4 Case studies that exemplify integrative methods
Here we use three case studies to highlight study systems where researchers have combined some of the methods outlined above to make significant progress in assembling the pieces of the evolutionary ecological puzzle. The foundation for this research was natural history observations on populations that varied in key traits or experienced different ecological conditions. From these observations, researchers proceeded to design successive careful experiments to test the evolutionary and genetic mechanisms responsible for the variation, through an iterative process where discoveries feed back to refine and reformulate future research.
6.4.1 Case study 1: threespine stickleback
An understanding of the genetics of adaptive evolution has emerged from extensive studies on the evolution of threespine stickleback fish as they adapted from living in the ocean to living exclusively in freshwater environments. Threespine sticklebacks (Gasterosteus aculeatus) are fish that inhabit saltwater and freshwater habitats in the Northern Hemisphere.
Most populations of sticklebacks are either exclusively marine, or live in coastal waters and enter streams and lakes to spawn and then move back out to sea, but some populations have adapted to live their entire lives in lakes. We know that ancestors of the species were trapped in lakes that formed at the end of the glacial maximum approximately 12,000 years ago (McPhail 1993). Fish in freshwater populations have fewer spines and drastically reduced lateral plating compared to their marine relatives, and this phenotypic variation suggested the differences were adaptive. The research on freshwater stickleback populations highlights the benefit of combining rigorous studies of natural selection, with research on developmental and genetic mechanisms underlying phenotypic differences, to develop a holistic picture of the adaptation to freshwater environments.
One of the earliest studies aimed at identifying whether lateral armour in threespine stickleback was associated with environmental variation demonstrated a phenotypic correlation, where fish from saline environments had more plates and fish from freshwater had fewer plates (Bertin 1925). To investigate whether the morphological differences were due to genetic or environmental variation, Heuts (1947) used a reciprocal transplant experiment to compare the development of eggs parented by fish of varying armour in aquaria that differed in temperatures and salinities. The results indicated that survival and hatching success existed in narrow ranges of temperature and salinity for each plate morph and were thus the result of natural selection. Later studies have investigated other possible mechanisms influencing body armour in threespine stickleback including predation (Reimchen 2000; Vamosi and Schluter 2004; Marchinko 2009), buoyancy (Myhre and Klepaker 2009), swimming performance (Bergstrom 2002; Blake 2004; Hendry et al. 2011), growth rate (Marchinko and Schluter 2007; Barrett et al. 2009) and ionic concentrations (Giles 1983; Bell et al. 1993). Contemporary studies are still aiming to determine the key ecological factors driving the evolution of body armour in threespine stickleback.
The next question in the research program was to connect the phenotypic variation to causal genetic variants, and to ask whether there were consistent differences between oceanic and freshwater populations. First, phylogenetic relations among populations of fish were examined with mitochondrial and microsatellite markers to show independent introductions to freshwater environments (Taylor and McPhail 1999). Next, genetic crosses between freshwater and marine fish revealed the genetic loci controlling the phenotypic differences in body armour, initially using a linkage map with about 200 microsatellite markers (Peichel et al. 2001). The introduction of population genomics expanded threespine stickleback research dramatically. Shortly after, researchers used QTL mapping to identify the genetic loci related to differences in plate morph number (Colosimo et al. 2005), and genome-wide patterns of differentiation to identify novel regions of the genome responsible for adaptation (Hohenlohe et al. 2010). One of the genes involved is known as Eda, which codes for a signalling protein involved in the growth of lateral armour plates in fish. By comparing the genetic differences in the low-Eda allele in different freshwater populations, researchers found that it likely arose at least two million years ago in the marine ancestors of freshwater sticklebacks. Because the allele is recessive, it would have lingered at very low frequencies in ancestral marine populations, and this standing genetic variation provided a selective advantage when populations colonised freshwaters. These new environments lacked predators and contained different ionic compounds, so that elaborate defences no longer resulted in higher fitness. The low-Eda allele now increased fitness and rapidly increased in frequency across replicate freshwater populations.
The genetic patterns of variation in the Eda gene strongly suggested that these alleles were associated with adaptation, but formal experiments are necessary to confirm the connection between genotypic variation and phenotypic performance in natural populations. Experiments using living populations of fish directly tested how natural selection caused the increase in the low-Eda allele in freshwater populations. Researchers used a common garden experiment, where they populated ponds with marine and freshwater fish, tracked individual survival and growth, and genotyped fish at the Eda locus. The results revealed that individuals with less body armour, and low-Eda alleles, had an advantage in predator-free lakes (Barrett et al. 2008). For example, the reduction in armour gives insect predators less area to grab onto juvenile fish in freshwater environments. The energy investment to grow armour is also costly, and in freshwater environments this is compounded with low concentrations of the ions necessary for bone growth. Juvenile fish with low armour can grow faster and increase lipid storage, which result in higher overwinter survival and earlier reproduction the following season, both key components of fitness.
6.4.2 Case study 2: deer mouse
Variation in coat colour pigmentation in deer mice (genus Peromyscus) has provided a rich backdrop for understanding natural selection and the genetic basis of colour adaptation. Deer mice are ubiquitous in North America with ‘not one square mile not inhabited by Peromyscus’ (Osgoode 1909). Early research documented variation in coat colour across populations in different environments. Based on measurements of thousands of deer mice in field observations and experiments in laboratory conditions, Sumner (1929) showed a strong relation between coat colour and soil colour among field populations and demonstrated that coat colour was genetically inherited. A plausible inference was that coat colour evolved for camouflage against visual predators. These extensive natural history observations form the basis of more recent research on Peromyscus, which has become one of the best model systems for connecting ecology with genetics to understand evolutionary processes.
One of the species of Peromyscus that varies dramatically in coat colour and associated soil colour is P. maniculatus. Populations of this species live on the Sand Hills of Nebraska, a dune field with light quartz grain soil that is lighter in colour than the surrounding soils. Mice living on these soils have evolved a dorsal coat that matches the soil, plausibly due to selection imposed by visual avian predators against dark coats that would be conspicuous on light soils. Because the Sand Hills are geologically young, dating back to the receding of the Wisconsin glacial period 8000 to 15,000 years ago (Loope and Swinehart 2000), the light-coloured mice are evidence of recent adaptation. Research for the past 75 years has aimed to answer what ecological factors drive this evolution and what genes cause the divergent phenotypes.
While it seems intuitive that light mice should have a survival advantage on light soils, empirical evidence is necessary to demonstrate that colour differences matter and to what extent they matter. Using experiments to show that coat colour influences predation risk, Dice (1947) employed a common garden approach in which mice with varying dorsal coat colours were released in two indoor enclosures with either dark and light soil colours and subjected to predation by two owls. They found that mice that matched the soil colour were 70% less likely to be captured by a predator, providing direct support for the hypothesis that coat colour does indeed aid in predation avoidance.
In a more recent experiment, this time in the natural Sand Hills environment, Linnen et al. (2013) measured attack rates on light and dark clay models and similarly found that conspicuous dark models were attacked significantly more than light models. These experiments demonstrate that natural selection is almost certainly occurring, due to survival differences in mice with different coat colours.
The next question was to understand how colour varies. To begin, researchers drew on knowledge from the extensively studied mouse, Mus musculus, where knockouts of the Agouti gene resulted in dark-coloured mice, while overexpression of Agouti resulted in light-coloured mice. Using a crossing experiment between light and dark mice, and mRNA expression levels, Linnen et al. (2009) found that Agouti is responsible for the light-coloured phenotype in P. maniculatus. Now that the phenotypic effect of Agouti was understood, the ecological consequences of changes in Agouti could be investigated.
To test whether selection favours locally adapted colour phenotypes, Barrett et al. (2019) conducted a reciprocal transplant experiment similar to experiments done by Dice (1947) and Linnen et al. (2013) but with live mice in natural habitats. They collected dark- and light-coloured mice and released them in enclosures that either had dark- or light-coloured soil. All enclosures were open to predators. They tracked the survival of all mice regularly using a mark-recapture method until mortality reached almost 100%. They found that mice introduced to enclosures that matched their original habitat type had greater survival than mice from a different habitat, demonstrating local adaptation in each environment. Additionally, they demonstrated selection on pigmentation: in light enclosures, mice were 1.44 times lighter than mice from the founding populations; and in dark enclosures, mice were 1.98 times darker. Finally, they fully connected the research on natural selection with the genetics of adaptation by determining how allele frequencies at the Agouti locus changed over the course of the experiment. They found significant allele frequency changes consistent with selection in the dark enclosures, and similarly in the light enclosures, although low survival reduced their statistical power.
6.4.3 Case study 3: seep monkeyflower
Much like the research on stickleback and deer mice, natural history observations of extensive variation in nature underpin current research on plants in the genus Mimulus (sensu lato; monkeyflowers). Several species in this genus have become model systems in evolutionary ecology and evolutionary genetics because of their variation in reproduction, life history and physiology. Mimulus guttatus (syn. Erythranthe guttata) is a widespread species with a native range from Mexico to Alaska and from the Pacific coast to the Rocky Mountains. It is part of a species complex with about a dozen other species that have evolved in response to a wide range of abiotic and biotic conditions. Populations can be found in grasslands, forests, desert streams, peat bogs, alpine meadows and seeps, coastal cliffs and sand dunes and even in toxic soils of serpentine barrens and copper mine tailings (Wu et al. 2008).
Here we use the research on copper tolerance in M. guttatus to demonstrate an integrative research program. Common garden greenhouse experiments were first used by Allen and Sheppard in the early 1970s to confirm genetic differences among plants from copper-contaminated soils compared to nearby plants on unpolluted land. They demonstrated that populations from non-contaminated soil had less root growth (a proxy for tolerance) than populations from contaminated soils when grown in contaminated aqueous solutions or soil, suggesting that plants from contaminated populations had adapted to high metal concentrations. Further, they used progeny from tolerant and intolerant parents to demonstrate that the trait was heritable. This study (Allen and Sheppard 1971) set the stage for future efforts to determine the exact genetic mechanism underlying copper tolerance.
Early experiments to identify genetic mechanisms underpinning copper tolerance used classic Mendelian crossing experiments. MacNair (1977) crossed putative tolerant individuals with non-tolerant individuals, backcrossing them and measuring root growth in copper-contaminated solution. The ratio of back-crossed plants that were copper tolerant was consistent with a hypothesis that copper tolerance is controlled by two major genes. However, upon continued breeding experiments with selfed and back-crossed F1 hybrids, research found ratios more aligned with a single gene controlling copper tolerance (MacNair 1983). These crosses also indicated that the locus controlling copper tolerance may also cause hybrid lethality in crosses with non-tolerant populations, perhaps due to pleiotropic effects or a tightly linked locus that has hitchhiked with the copper tolerance locus.
Thirty years later, Wright et al. (2013) took advantage of advances in molecular technologies by using QTL genetic mapping and bioinformatic approaches to identify the loci controlling copper tolerance. They discovered the gene Tol1 was responsible for copper tolerance. Using additional crosses between tolerant and non-tolerant plants, they found that copper tolerance and hybrid inviability between tolerant and intolerant plants are controlled by two distinct but tightly linked genes rather than a pleiotropic by-product of copper tolerance. The allele for hybrid inviability causes necrosis (and consequently named Nec1), and exists in surprisingly high frequencies in copper-tolerant populations.
Hypotheses as to why Nec1 exists at high frequency include: (1) genetic drift caused the allele to increase in frequency, (2) the hybrid lethality allele confers a fitness advantage in the mine habitat and has been directly selected and (3) the allele may have hitchhiked to high frequency due to tight linkage with Tol1 which is under positive selection.
To distinguish between these hypotheses, researchers used population genomics and bioinformatics to compare genetic variation and divergence between copper mine and non-mine populations at the genomic region of Tol1 and Nec1. They found mine populations have low genetic variation and high genetic differentiation compared to non-mine populations, indicating that the alleles increased in frequency due to selection. The results could not distinguish whether there was direct selection on both loci independently. However, the Tol1 allele has been discovered at multiple mine sites, while the Nec1 allele has only been found at one recently derived population, suggesting that direct selection is much more likely on the Tol1 allele (Wright et al. 2013).
The presence of Tol1 alleles in multiple mine populations provides an opportunity to investigate the potential for repeated evolution and local adaptation. A reciprocal transplant study coupled with genomic sequencing and coalescent analyses provided a powerful approach to identify the evolutionary processes generating and maintaining trait variation. First, to determine if mine and off-mine populations were locally adapted, researchers reciprocally planted individuals from mine habitats and non-mine habitats into both environments (Wright et al. 2015). They found that populations from mine populations performed better in mine habitats and vice versa, demonstrating strong local adaptation to mine conditions. Interestingly, they did not find a cost (or trade-off) to copper tolerance in off-mine environments. Next, they tested whether the fitness advantage in mine environments is directly related to Tol1 alleles, by creating introgression lines with individuals from mine and non-mine sites, with or without the tolerance allele. They found that individuals with tolerant alleles perform equivalently in non-mine sites and have increased survival in mine sites. Finally, to understand how copper tolerance arose at multiple mine sites, they used population genomics and bioinformatics to compare genetic variation around the candidate gene across populations to determine whether the allele arose from standing variation or new mutations. They found low genetic differentiation among mine populations, indicating that tolerance alleles at Tol1 likely arose from standing genetic variation rather than independent mutations. This hypothesis is plausible because mine sites where M. guttatus exist are only about 150 years old and thus time for multiple mutations to arise is unlikely.
6.5 So what?
In this chapter we have explored the process of combining and integrating research methods to understand the generation and maintenance of trait variation in wild populations. Although we have focused on a specific area of study – evolutionary ecology – the approach is generalisable to all of ecology. We usually begin with speculation based on experience and common sense, but to understand the process and how the system works, we need to use an iterative and integrative approach that employs a variety of methods. Research that aims to mechanistically link the ecological and environmental differences among populations to the evolutionary processes that generate diversity is no easy task.
One of the strengths of evolutionary biology is that it is deeply rooted in a robust theoretical framework. This provides the foundation for making predictions about population genetic changes and the way selection operates. Although we may begin with highly plausible speculations about the selection pressures that are involved, or the traits that are responding to these selection agents, those factors may not necessarily be the most important influences. Even a carefully designed experiment may fail to control or account for the environmental variation that really matters to fitness. We may identify genetic differences among populations, or identify the genetic architecture of traits, but these may not be general across populations even within a single species. These challenges are often what make evolutionary ecology dynamic and stimulating, and why there remain many fundamental questions to address and hypotheses to test.
Regardless of the specific research question or field of study, there will always be merits to applying a diversity of methods (see Chapter 3). Ecology is, by nature, a field with complex, interacting pieces. By focusing solely on only one piece of the puzzle, or employing only one type of method, we are prone to making simplified (wrong) assumptions. We advocate for using a diversity of interlocking techniques and approaches, and encourage researchers to borrow across fields. Applying evolutionary perspectives to traditional ecological questions has proved fruitful and provided novel insight, and vice versa (e.g. Urban et al. 2020). The case studies discussed here exemplify this strategy.
Nonetheless, not all research requires this level of detail, and indeed intensive research on a large number of ‘model systems’ may be overkill. There will always be a place for a spectrum of research that ranges from broad-based observations to projects that go into incredible detail using the most advanced methodologies. It is an issue of balance, as well as a matter of scientific credibility and using suitable methods for the question. Natural history observations and straightforward experiments can tell us what is plausible. The field of Ecology can be developed further with studies that go deeper to demonstrate that the initial ideas are valid (Figure 1.1c).
A common concern among new researchers is that all the interesting studies have already been conducted, or that there are few new research avenues. As highlighted in the case studies discussed in this chapter, insightful new research is conducted by using different approaches, borrowing techniques from other fields, and testing old, fundamental ideas with new methods. New molecular technologies and analytical tools are facilitating new research directions, enabling us to answer questions that seemed impossible to achieve only a few decades ago.
Few researchers will have the expertise or time to independently apply a diversity of methods. Actively seeking out collaborators with varied skills can be rewarding, productive or fun – and perhaps essential for progress. But it is not always easy to identify appropriate collaborators. How does an early-career researcher attract collaborators with the skills, knowledge and experience that they need to further their research or apply different methods? Collaborators need not be leaders in the field; peers at the same career stage working on different research questions can make excellent collaborators for integrative work. Regardless of who you are approaching, being well-informed and well-read on the person’s work is respectful and necessary. Most researchers are excited to hear about other people’s ideas and to share their own. It’s okay if you find it intimidating at first - it becomes easier with experience. It’s also okay to admit that you don’t know everything and that there are gaps in your research expertise – that is the very point of seeking out collaborations! When you identify researchers with specific expertise, offer them a seminar on your intriguing fledgling research questions, this can help communicate the excitement of your investigations and the need for your work. Such offers are seldom rejected and can sow the seeds for collaboration. Small, intimate workshops (see Chapter 10) may provide better rewards for your attendance, even at the cost of missing out on big international conferences. With hope, your persistence will eventually be rewarded.
6.6 References
Allen, W. R., and P. M. Sheppard. 1971. Copper tolerance in some Californian populations of the monkey flower, Mimulus guttatus. Proceedings of the Royal Society B: Biological Sciences 177:177–196. doi:10.1098/rspb.1971.0022
Anderson J. T., J. H. Willis, and T. Mitchell-Olds. 2011. Evolutionary genetics of plant adaptation. Trends in Genetics 27:258-266. doi:10.1016/j.tig.2011.04.001
Barrett, R. D. H., S. Laurent, R. Mallarino, et al. 2019. Linking a mutation to survival in wild mice. Science 363:499–504. doi:10.1126/science.aav3824
Barrett, R. D. H., S. M. Rogers, and D. Schluter. 2008. Natural selection on a major armor gene in threespine stickleback. Science 322:255–257. doi:10.1126/science.1159978
Barrett, R. D. H., S. M. Rogers, and D. Schluter. 2009. Environment specific pleiotropy facilitates divergence at the Ectodysplasin locus in threespine stickleback. Evolution 63:2831–2837. doi:10.1111/j.1558-5646.2009.00762.x
Barrett, R. D. H., and D. Schluter. 2008. Adaptation from standing genetic variation. Trends in Ecology and Evolution 23:38–44. doi:10.1016/j.tree.2007.09.008
Bell, M. A., G. Ortí, J. A. Walker, and J. P. Koenings. 1993. Evolution of pelvic reduction in threespine stickleback fish: a test of competing hypotheses. Evolution 47:906–914. doi:10.1111/j.1558-5646.1993.tb01243.x
Bergstrom, C. A. 2002. Fast-start swimming performance and reduction in lateral plate number in threespine stickleback. Canadian Journal of Zoology 80:207–213. doi:10.1139/z01-226
Bertin, L. 1925. Recherches bionomiques, biométriques et systématiques sur les épinoches (Gastérostéidés). Paris: Éd. Blondel La Rougery.
Blake, R. W. 2004. Fish functional design and swimming performance. Journal of Fish Biology 65:1193–1222. doi:10.1111/j.0022-1112.2004.00568.x
Blanquart, F., O. Kaltz, S. L. Nuismer, and S. Gandon. 2013. A practical guide to measuring local adaptation. Ecology Letters 16:1195–1205. doi:10.1111/ele.12150
Blount, Z. D., R. E. Lenski, and J. B. Losos. 2018. Contingency and determinism in evolution: Replaying life’s tape. Science 362:eaam5979. doi:10.1126/science.aam5979
Bourdeau, P. E. 2010. An inducible morphological defence is a passive by-product of behaviour in a marine snail. Proceedings of the Royal Society B: Biological Sciences 277:455–462. doi:10.1098/rspb.2009.1295
Briscoe Runquist, R. D., A. J. Gorton, J. B. Yoder, et al. 2020. Context dependence of local adaptation to abiotic and biotic environments: a quantitative and qualitative synthesis. American Naturalist 195:412–431. doi:10.1086/707322
Brown, B. A., and M. T. Clegg. 1984. Influence of flower color polymorphism on genetic transmission in a natural population of the common morning glory, Ipomoea purpurea. Evolution 38:796–803. doi:10.2307/2408391
Clausen J., D. D. Keck, and W. M. Hiesey. 1940. Experimental studies on the nature of species. I. Effect of varied environment on Western North American plants. Washington, DC: Carnegie Institution of Washington. Publications No. 520.
Colosimo, P. F., K. E. Hosemann, S. Balabhadra, et al. 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307:1928–1933. doi:10.1126/science.1107239
Darwin, C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray. doi:10.4324/9780203509104
Dice, L. R. 1947. Effectiveness of selection by owls of deer-mice (Peromyscus maniculatus) which contrast in color with their background. Contributions of the Laboratory of Vertebrate Biology, University of Michigan 34:1–20
Etterson, J. R., and R. G. Shaw. 2001. Constraint to adaptive evolution in response to global warming. Science 294:151–154. doi:10.1126/science.1063656
Endler, J. A. 1986. Natural selection in the wild. Princeton: Princeton University Press. doi:10.2307/j.ctvx5w9v9
Fisher, R. A. 1930. The genetical theory of natural selection. Oxford: Clarendon.
Giles, N. 1983. The possible role of environmental calcium levels during the evolution of phenotypic diversity in Outer Hebridean populations of the three‐spined stickleback, Gasterosteus aculeatus. Journal of Zoology 199:535–544. doi:10.1111/j.1469-7998.1983.tb05104.x
Gould, S. J., and R. C. Lewontin. 1979. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proceedings of the Royal Society B: Biological Sciences 205:581–598. doi:10.1098/rspb.1979.0086
Haldane, J. B. S. 1932. The causes of evolution. Princeton: Princeton University Press.
Hairston, N. G., S. P. Ellner, M. A. Geber, et al. 2005. Rapid evolution and the convergence of ecological and evolutionary time. Ecology Letters 8:1114–1127. doi:10.1111/j.1461-0248.2005.00812.x
Hendry, A. P., K. Hudson, J. A. Walker, et al. 2011. Genetic divergence in morphology-performance mapping between Misty Lake and inlet stickleback. Journal of Evolutionary Biology 24:23–35. doi:10.1111/j.1420-9101.2010.02155.x
Hendry, A.P., D. J. Schoen, M. E. Wolak, and J. M. Reid. 2018. The contemporary evolution of fitness. Annual Review of Ecology Evolution and Systematics 49:457–476. doi:10.1146/annurev-ecolsys-110617-062358
Hereford, J. 2009. A quantitative survey of local adaptation and fitness trade-offs. American Naturalist 173:579–588. doi:10.1086/597611
Heuts, M. J. 1947. Experimental studies on adaptive evolution in Gasterosteus aculeatus L. Evolution 1:89-102. doi:10.2307/2405407
Hohenlohe, P. A., S. Bassham, P. D. Etter, et al.. 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genetics 6:e1000862. doi:10.1371/journal.pgen.1000862
Husband, B. C., and S. C. H. Barrett. 1992. Genetic drift and the maintenance of the style length polymorphism in tristylous populations of Eichhornia paniculata (Pontederiaceae). Heredity 69:440–449. doi:10.1038/hdy.1992.148
Johnston, S. E., J. Gratten, C. Berenos, et al. 2013. Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature 502:93–95. doi:10.1038/nature12489
Lande, R., and S. J. Arnold. 1983. The measurement of selection on correlated characters. Evolution 37:1210–1226. doi:10.2307/2408842
Linnen, C. R., E. P. Kingsley, J. D. Jensen, and H. E. Hoekstra. 2009. On the origin and spread of an adaptive allele in deer mice. Science 325:1095–1098. doi:10.1126/science.1175826
Linnen, C. R., Y.-P. Poh, B. K. Peterson, et al. 2013. Adaptive evolution of multiple traits through multiple mutations at a single gene. Science 339:1312–1316. https://doi.org/10.1126/science.1233213
Loope, D. B., and J. B. Swinehart. 2000. Thinking like a dune field. Great Plains Research 10:5–35.
Macnair, M. R. 1977. Major genes for copper tolerance in Mimulus guttatus. Nature 268:428–430. doi:10.1038/268428a0
MacNair, M. R. 1983. The genetic control of copper tolerance in the yellow monkey flower, Mimulus guttatus. Heredity 50:283–293. doi:10.1038/hdy.1983.30
Marchinko, K. B. 2009. Predation’s role in repeated phenotypic and genetic divergence of armor in threespine stickleback. Evolution 63:127–138. doi:10.1111/j.1558-5646.2008.00529.x
Marchinko, K. B., and Schluter, D. 2007. Parallel evolution by correlated response: lateral plate reduction in threespine stickleback. Evolution 61:1084–1090. doi:10.1111/j.1558-5646.2007.00103.x
Martin, R. A., L. D. Chick, M. L. Garvin, and S. E. Diamond. 2021. In a nutshell, a reciprocal transplant experiment reveals local adaptation and fitness trade‐offs in response to urban evolution in an acorn‐dwelling ant. Evolution 75:876–887. doi:10.1111/evo.14191
McPhail, J. D. 1993. Speciation and the evolution of reproductive isolation in the sticklebacks (Gasterosteus) of southwestern British Columbia. In: The Evolutionary Biology of the Threespine Stickleback, eds M. A. Bell, and S. A. Foster, 399–437. Oxford: Oxford Science Publications.
Messer P. W., S. P. Ellner, and N. G. Hairston. 2016. Can population genetics adapt to rapid evolution? Trends in Genetics 32:408–418. doi:10.1016/j.tig.2016.04.005
Myhre, F., and T. Klepaker. 2009. Body armour and lateral-plate reduction in freshwater three-spined stickleback Gasterosteus aculeatus: adaptations to a different buoyancy regime? Journal of Fish Biology 75:2062–2074. doi:10.1111/j.1095-8649.2009.02404.x
Mitchell-Olds, T., J. H. Willis, and D. B. Goldstein. 2007. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nature Reviews Genetics 8:845–856. doi:10.1038/nrg2207
Moran, E. V., F. Hartig, and D. M. Bell. 2016. Intraspecific trait variation across scales: implications for understanding global change responses. Global Change Biology 22:137–150. doi:10.1111/gcb.13000
Olson-Manning, C. F., M. R. Wagner, and T. Mitchell-Olds. 2012. Adaptive evolution: evaluating empirical support for theoretical predictions. Nature Reviews Genetics 13:867–877. doi:10.1038/nrg3322
Osgood, W. H. 1909. Revision of the mice of the American genus Peromyscus. North American Fauna 28:1–285
Peichel, C. L., K. S. Nereng, K. A. Ohgi, et al. 2001. The genetic architecture of divergence between threespine stickleback species. Nature 414:901–905. doi:10.1038/414901a
Reed, R. D., R. Papa, A. Martin, et al. 2011. Optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333:1137–1141. doi:10.1126/science.1208227
Reimchen, T. E. 2000. Predator handling failures of lateral plate morphs in Gasterosteus aculeatus: functional implications for the ancestral plate condition. Behaviour 137:1081–1096. doi:10.1163/156853900502448
Savolainen, O., M. Lascoux, and J. Merilä. 2013. Ecological genomics of local adaptation. Nature Reviews Genetics 14:807–820. doi:10.1038/nrg3522
Slatkin, M. 2008. Linkage disequilibrium — understanding the evolutionary past and mapping the medical future. Nature Reviews Genetics 9:477–485. doi: 10.1038/nrg2361
Smith, R. J. 2016, Explanations for adaptations, just-so stories, and limitations on evidence in evolutionary biology. Evolutionary Anthropology 25:276–287. doi:10.1002/evan.21495
Stern, D. L. 2013. The genetic causes of convergent evolution. Nature Reviews Genetics 14:751–764. doi:10.1038/nrg3483
Stinchcombe, J. R, M. T. Rutter, D. S. Burdick, et al. 2002. Testing for environmentally induced bias in phenotypic estimates of natural selection: theory and practice. American Naturalist 160:511–523. doi:10.1086/342069
Sumner, F. B. 1929. The analysis of a concrete case of intergradation between two subspecies. Proceedings of the National Academy of Science of the United States of America 15:110–120. doi:10.1073/pnas.15.6.481
Taylor, E. B., and J. D. McPhail. 1999. Evolutionary history of an adaptive radiation in species pairs of threespine sticklebacks (Gasterosteus): insights from mitochondrial DNA. Biological Journal of the Linnean Society 66:271–291. doi:10.1111/j.1095-8312.1999.tb01891.x
Turesson, G. 1922. The species and the variety as ecological units. Hereditas 3:110–113. doi:10.1111/j.1601-5223.1922.tb02727.x
Urban, M. C., S. Y. Strauss, F. Pelletier, et al. 2020. Evolutionary origins for ecological patterns in space. Proceedings of the National Academy of Sciences of the United States of America 117:17482-17490. doi:10.1073/pnas.1918960117
Vamosi, S. M., and D. Schluter. 2004. Character shifts in the defensive armor of sympatric sticklebacks. Evolution 58:376–385. doi:10.1111/j.0014-3820.2004.tb01653.x
von Marilaun, A. K., and F. W. Oliver. 1895. The natural history of plants, their forms, growth, reproduction, and distribution. New York: Henry Holt. doi:10.5962/bhl.title.54631
Wittkopp, P. J., and G. Kalay. 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nature Reviews Genetics 13:59–69. doi:10.1038/nrg3095
Wright, K. M., U. Hellsten, C. Xu, et al. 2015. Adaptation to heavy-metal contaminated environments proceeds via selection on pre-existing genetic variation. bioRxiv 029900. doi:10.1101/029900
Wright, K. M., D. Lloyd, D. B. Lowry, M. R. Macnair, and J. H. Willis. 2013. Indirect evolution of hybrid lethality due to linkage with selected locus in Mimulus guttatus. PLoS Biology 11:e1001497. doi:10.1371/journal.pbio.1001497
Wright, S. 1931. Evolution in Mendelian populations. Genetics 16:97–159. doi:10.1093/genetics/16.2.97
Wu, C. A., D. B. Lowry, A. M. Cooley, et al. 2008. Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity 100:220–230. doi:10.1038/sj.hdy.6801018
Yeaman, S. 2015. Local adaptation by alleles of small effect. American Naturalist 186:S74-S89. doi:10.1086/682405
Young A., T. Boyle, and T. Brown. 1996. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution 11:413–418. doi:10.1016/0169-5347(96)10045-8