Chapter 1 Familiarization
Throughout this textbook, we will include links to a variety of video lectures developed by the original author Dr. Derek Sonderegger. The video lectures add useful additional details to the content of this textbook. At this time, the video lectures have not been updated but may be changed in future updates.
The video lecture for this chapter can be found here: Video Lecture
R is a open-source program that is commonly used in statistics and machine learning. It runs on almost every platform and is completely free and is available at www.r-project.org. Most cutting-edge statistical research is first available on R.
The basic editor that comes with R works fairly well, but but it is strongly recommended that you run R through the program RStudio which is available at rstudio.com. This is a completely free Integrated Development Environment that works on Macs, Windows and a couple of flavors of Linux. It simplifies a bunch of more annoying aspects of the standard R GUI and supports useful things like tab completion.
R is a script based language, and there isn’t a point-and-click interface for data wrangling and statistical modeling. While initially painful, writing scripts leaves a clear and reproducible description of exactly what steps were performed. This is a critical aspect of sharing your methods and results with other students, colleagues, and the world at-large.
1.1 A Basic Script
The simplest form for developing and executing code is a script file. A script file can be loaded within R studio by selecting File -> New File -> R Script. It is encouraged to develop and run code within a script file first, before placing code into an RMD file. A script file only executes R code, such as that of an R-chunk discussed below. However, the script file does not compile any of the other items an Rmarkdown file will use, and thus is a safe and easy way to check if your statistical code is working. If you are getting errors when compiling an Rmarkdown file, ensure that all of your statistical code is properly working first by executing within a basic script file. This can help you decide if the error is coming from Markdown (latex, etc…) or if you have a flaw in your solution.
1.2 Working within an Rmarkdown File
The first step in any new analysis or project is to create a new Rmarkdown file. This can be done by selecting the File -> New File -> R Markdown... drop down option and a menu will appear asking you for the document title, author, and preferred output type. In order to create a PDF, you’ll need to have LaTeX installed, but the HTML output nearly always works and I’ve had good luck with the MS Word output as well. All university computers have software prepared for compiling latex documentation. A section has been added to the end of this textbook to help setup personal computers with a lightweight package known as tinytex for compiling PDF documents.
R-Markdown (RMD) is an implementation of the Markdown syntax that makes it extremely easy to write documents and give instructions for how to do typesetting, introduce graphics, display code, and write mathematical expressions. The syntax available to embed R commands directly into the document always for transparent and easy to follow statistical programming. Perhaps the easiest way to understand the syntax is to look at the RMarkdown website or the Help -> Rmarkdown Quick Reference drop down link in RStudio.
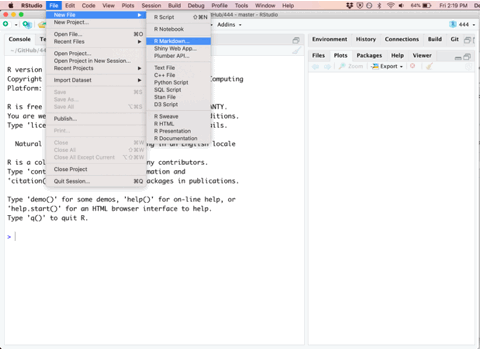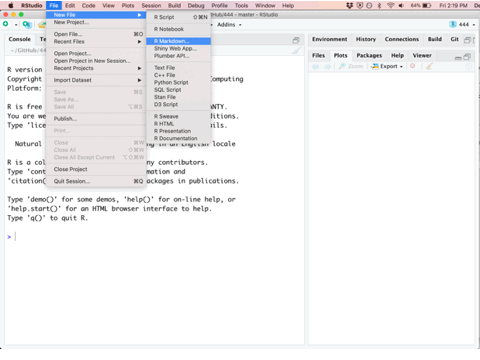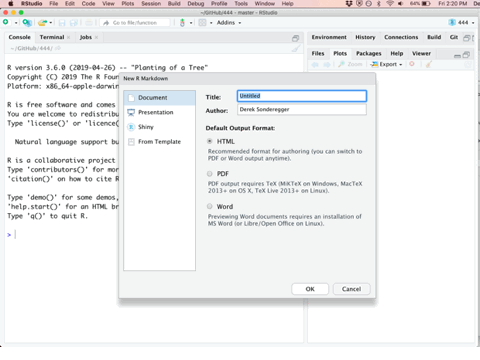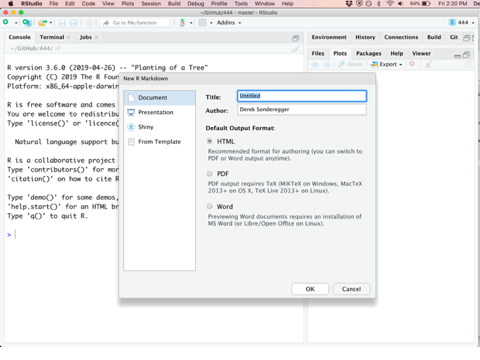
Once you’ve created a new Rmarkdown file, you’ll be presented with four different panes that you can interact with.
| Pane | Location | Description |
|---|---|---|
| Editor | Top Left | Where you edit the script. This is where you should write most all of your R code. You should write your code, then execute it from this pane. Because nobody writes code correctly the first time, you’ll inevitably make some change, and then execute the code again. This will be repeated until the code finally does what you want. |
| Console | Bottom Left | You can execute code directly in this pane, but the code you write won’t be saved. I recommend only writing stuff here if you don’t want to keep it. I only type commands in the console when using R as a calculator and I don’t want to refer to the result ever again. |
| Environment | Top Right | This displays the current objects that are available to you. I typically keep the data.frame I’m working with opened here so that I can see the column names. |
| Miscellaneous | Bottom Right | This pane gives access to the help files, the files in your current working directory, and your plots (if you have it set up to show here.) |
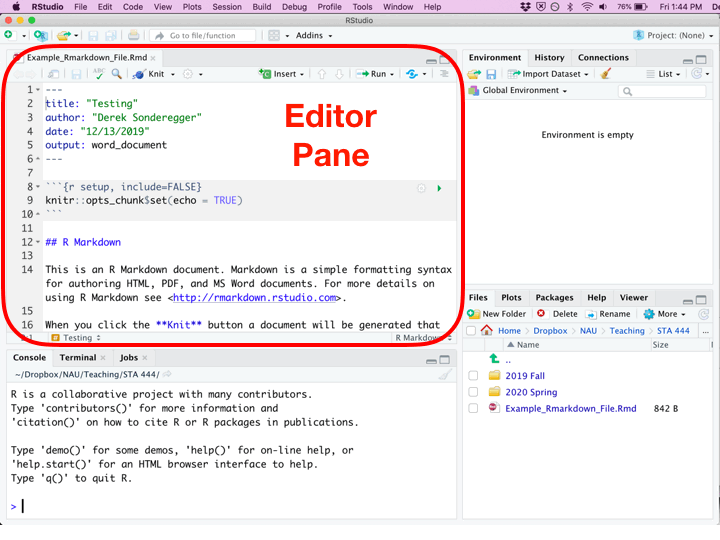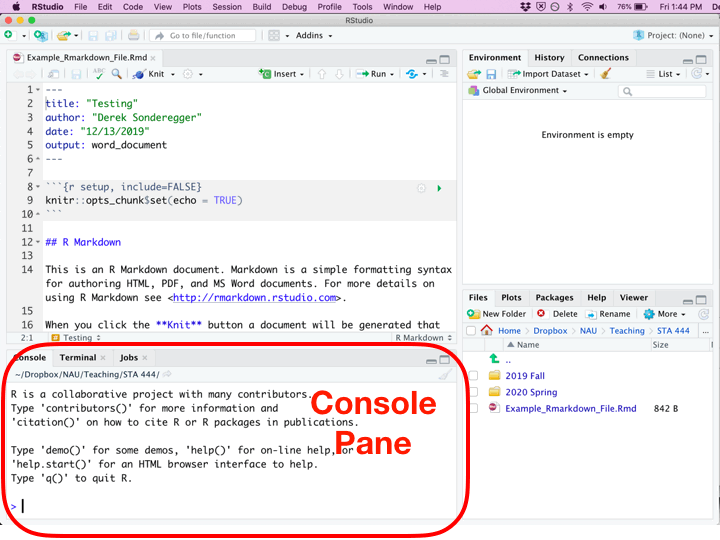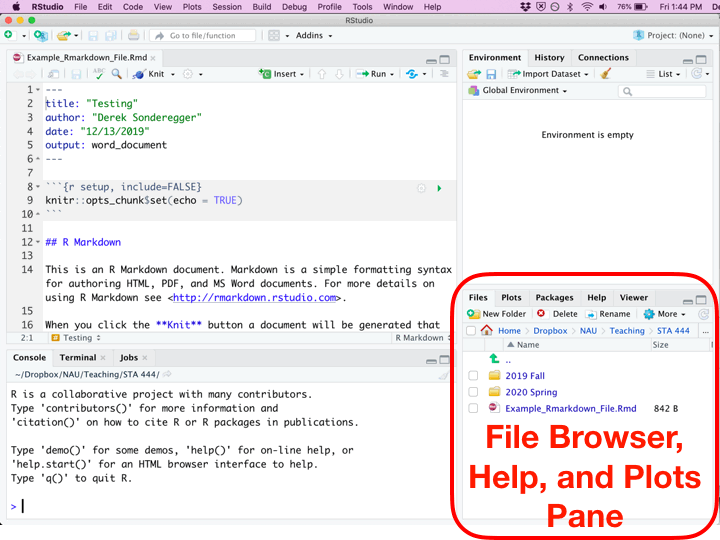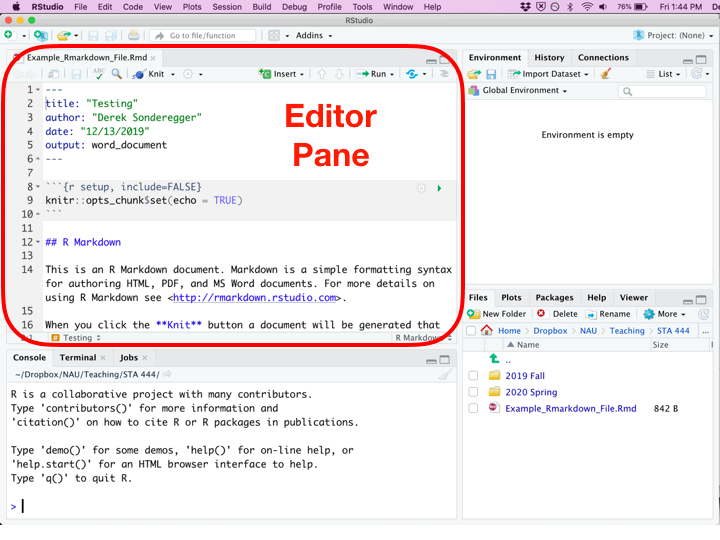
Whenever you create a new Rmarkdown document, it is populated with code and comments that attempts to teach new users how to work with Rmarkdown. Critically there are two types of regions:
| Region Type | Description |
|---|---|
| Commentary | These are the areas with a white background. You can write nearly anything here and in your final document it will be copied over. I typically use these spaces to write commentary and interpretation of my data analysis project. |
| Code Chunk | These are the grey areas. This is where your R code will go. When knitting the document, each code chunk will be run sequentially and the code in each chunk must run. |
In an R Markdown (RMD) document, R code is clearly separated from the regular text using three backticks along with an instruction indicating that it’s R code to be evaluated. You can easily insert a new R code chunk using the shortcut Ctrl+Alt+I on Windows or Cmd+Option+I on Mac, or by clicking the green insert button at the top of the RMD editor. You can also manually type the chunk delimiters. There are many chunk options available. The final output of an RMD document can be rendered in various formats, including HTML, PDF, or MS Word.
This entire book is created using R Markdown. To view the RMD file for any chapter, simply click the view source icon at the top of the online notes. This allows you to see the source code, which is especially useful if you want to copy and paste code into your own work, such as the exercises provided at the end of each chapter.
While writing an RMD file, each of the code chunks can be executed in a couple of different ways.
- Press the green arrow at the top of the code chunk to run the entire chunk.
- The run button has several options for running different amounts of code.
- There are keyboard shortcuts, on Windows use
Ctrl+Enter, on Mac useCmd+Return.
Finally, we want to produce a nice output document that combines the code, output, and commentary. To do this, you’ll knit the document which causes all of the R code to be run in a new R session, and then weave together the output into your document. This can be done using the knit button at the top of the Editor Window. Be mindful that the RMD file is a stand-alone document that must have all components necessary for the file to run. Anything that was loaded into local memory will not be seen when compiling the RMD file. For instance, if you need a particular data set available for the code to run, this must be coded into the RMD file, and not just loaded into local memory. These types of issues will become more familiar as you work through the examples and exercises available in this textbook.
When I was a graduate student, I had to tediously copy and paste tables of output from the R console and figures I had made into my Microsoft Word document. Far too often I would realize I had made a small mistake in part (b) of a problem and would have to go back, correct my mistake, and then redo all the laborious copying. I often wished that I could write both the code for my statistical analysis and the long discussion about the interpretation all in the same document so that I could just re-run the analysis with a click of a button and all the tables and figures would be updated by magic. That is what RMD does.
1.3 R as a simple calculator
Assuming that you have started R on whatever platform you like, you can use R as a simple calculator. In either your Rmarkdown file code chunk, an R script, or by running it within the console. Try running the following simple calculation in all three of the methods mentioned above:
## [1] 5In this fashion you can use R as a very capable calculator.
## [1] 48## [1] 64## [1] 2.718282R has most constants and common mathematical functions you could ever want. Functions such as sin(), cos(), are available, as are the exponential and log functions exp(), log(). The absolute value is given by abs(), and round() will round a value to the nearest integer while floor() and ceiling() will always round down or up as desired.
## [1] 3.141593## [1] 0## [1] 1.609438## [1] 0.69897Whenever I call a function, there will be some arguments that are mandatory, and some that are optional and the arguments are separated by a comma. In the above statements the function log() requires at least one argument, the number in which you want to log. However, the base argument is optional. If you do not specify what base to use, R will use a default value. You can see that R will default to using base \(e\) by looking at the help page by typing help(log) or ?log at the command prompt.
Arguments can be specified via the order in which they are passed or by naming the arguments. So for the log() function which has arguments log(x, base=exp(1)). If I specify which arguments are which using the named values, then order doesn’t matter.
## [1] 0.69897## [1] 0.69897But if we don’t specify which argument is which, R will decide that x is the first argument, and base is the second. It is suggested to use the order of options as presented in the help files. This will improve efficiency as you become more complex with your statistical programming. Notice when you use these shortcuts though, you will get different answers based on the order you enter values with provided specific option names!
## [1] 0.69897## [1] 1.430677When arguments are specified, notice the use of syntax such as name=value notation and a student might be tempted to use the <- notation here. Don’t do that as the name=value notation is making an association mapping and not a permanent assignment. Assignment of objects is one of the core ideas of using R, and is discussed more in the section below.
1.3.1 Assignment
We need to be able to assign a value to a variable to be able to use it later. R does this by using an arrow <- or an equal sign =. While R supports either, for readability, I suggest people pick one assignment operator and stick with it. I personally prefer to use the arrow and this is the most basic assignment possible. There are different levels of assignment available in R, so stick to the <- arrow as you get started. When assigning a new object, be mindful that variable names cannot start with a number, may not include spaces, and are case sensitive. Let’s practice by assigning the variable tau and my.test.var below, and using them in further computations.
tau <- 2*pi # create two variables
my.test.var = 5 # notice they show up in 'Environment' tab in RStudio!
tau## [1] 6.283185## [1] 5## [1] 31.41593As your analysis gets more complicated, you’ll want to assign the results to a variable so that you can access the results later. If you don’t assign the result to a variable, you have no way of accessing the result. Using object-oriented programming within R will have tremendous consequences when developing more complex code.
When a variable has been assigned, you will see it in the environment tab in the environment pane. This area is extremely convenient to remind ourselves how we spelled a variable name and capitalization. R is case sensitive so X and x are two different variable names, so being consistent in your capitalization scheme is quite helpful. The complexity of assignment names in R is also very useful, so that you can write out complex names for easy to recall functionality. A common theme is known as camel casing, where you write a longer name with each separate word capitalized, such as MyFirstFunction.
1.3.2 Vectors
While single values are useful, it is very important that we are able to make groups of values. The most fundamental aggregation of values is called a vector. In R, we will require vectors to always be of the same type (e.g. all integers or all character strings). To create a vector, we just need to use the concatenation function c(). This is a very common theme in R, so get used to using concatenation early! Below we create two vectors through concatenation, the first a vector of strings, the second a vector of constants.
## [1] "A" "A" "B" "C"## [1] 4 3 8 10It is very common to have to make sequences of integers, and R has a shortcut to do this. The notation A:B will produce a vector of constants starting with the value A and incrementally increasing by one until the value B. Here we quickly produce a vector of constants from 2 to 6.
## [1] 2 3 4 5 6Nearly every function in R behaves correctly when being given a vector of values. This is called vectorization and can make computations within R very fast if properly used. Vectorization is much faster than writing for-loops (learned in later chapters), so practice with vectors is very important. Notice we can feed a vector of constants to the log function, and it will calculate the log of each element of the vector.
## [1] 1.3862944 1.9459101 1.6094379 0.69314721.4 Packages
One of the greatest strengths about R is that so many people have developed add-on packages with additional functionality. For example, plant community ecologists have a large number of multivariate methods that are useful but were not part of R. So Jari Oksanen got together with some other folks and put together a package of functions that they found useful. The result is the package vegan.
To download and install the package from the Comprehensive R Archive Network (CRAN), you just need to ask RStudio it to install it. This can be done via the menu Tools -> Install Packages.... You can also use a console or script and the R function install.packages(). With either method, you just need to provide the name of the package and RStudio will download and install the package on your computer.
Many major analysis types are available via downloaded packages as well as problem sets from various books (e.g. Sleuth3 or faraway) and can be easily downloaded and installed from CRAN.
Once a package is downloaded and installed on your computer, it is available and does not need to be installed again. However, it is not loaded into your current R session by default. The reason it isn’t loaded is that there are thousands of packages, some of which are quite large and only used occasionally. So to improve overall performance only a few packages are loaded by default. When you want to use a specific package you must explicitly load them. You only need to load them once per session/script. After installing the vegan package, we can load the package into our environment using the library() function.
For a similar performance reason, many packages do not automatically load their data sets unless explicitly asked. Therefore when loading data sets from a package, you might need to do a two-step process of loading the package and then loading the data set.
##
## Attaching package: 'faraway'## The following object is masked from 'package:lattice':
##
## melanomaIf you don’t need to load any functions from a package and you just want the data, you can do it in one step.
data('butterfat', package='faraway') # just load the data set, not anything else
head(butterfat) # print out the first 6 rows of the data## Butterfat Breed Age
## 1 3.74 Ayrshire Mature
## 2 4.01 Ayrshire 2year
## 3 3.77 Ayrshire Mature
## 4 3.78 Ayrshire 2year
## 5 4.10 Ayrshire Mature
## 6 4.06 Ayrshire 2yearSimilarly, if I am not using many functions from a package, I might choose call the functions using the notation package::function(). This is particularly important when two packages both have functions with the same name and it gets confusing which function you want to use. For example the packages mosaic and dplyr both have a function tally. When such a conflict occurs, you may notice R provides a warning that certain functions have been masked. This is a not an error, instead a message informing you that more than one function of the same name is present. These messages were purposefully left in the sections above so you can observe their behavior. For example, if the dplyr package is loaded but you want to use the mosaic::tally() function, you can introduce the following syntax so that R knows which version of tally() you want to execute.
## Registered S3 method overwritten by 'mosaic':
## method from
## fortify.SpatialPolygonsDataFrame ggplot2## X
## 0 1 2
## 3 4 1Finally, many researchers and programmers host their packages on GitHub (or equivalent site) and those packages can easily downloaded using tools from the devtools package, which can be downloaded from CRAN. In later chapters of the STA 445 section, we will learn to host our own R packages through GitHub. These can then be installed directly from a github repository using the install_github() function. Below installs a demo package we will build in later Chapters. Remember, if you want to execute the code below, you’ll likely need to install the devtools package first!
1.5 Finding Help
There are many complicated details about R and nobody knows everything about how each individual package works. As a result, a robust collection of resources has been developed and you are undoubtedly not the first person to wonder how to do something.
1.5.1 How does this function work?
If you know the function you need, but just don’t know how to use it, the built-in documentation is really quite good. Suppose I am interested in how the rep function works. We could access the rep help page by searching in the help window or from the console via help(rep). The document that is displayed shows what arguments the function expects and what it will return. At the bottom of the help page is often a set of examples demonstrating different ways to use the function. As you get more proficient in R, these help files become quite handy, but initially they feel quite overwhelming.
1.5.2 How does this package work?
If a package author really wants their package to be used by a wide audience, they will provide a “vignette”. These are a set of notes that explain enough of how a package works to get a user able to utilize the package effectively. This documentation is targeted towards people the know some R, but deep technical knowledge is not expected. Whenever I encounter a new package that might be applicable to me, the first thing I do is see if it has a vignette, and if so, I start reading it. If a package doesn’t have a vignette, I’ll Google “R package XXXX” and that will lead to documentation on CRAN that gives a list of functions in the package.
1.5.3 How do I do XXX?
Often I find myself asking how to do something but I don’t know the function or package to use. In those cases, I will use the coding question and answer site stackoverflow. This is particularly effective and I encourage students to spend some time to understand the solutions presented instead of just copying working code. By digging into why a particular code chunk works, you’ll learn all sorts of neat tricks and you’ll find yourself utilizing the site less frequently.
1.6 Exercises
Create an RMarkdown file that solves the following exercises.
Exercise 1
Calculate \(\log\left(6.2\right)\) first using base \(e\) and second using base \(10\). To figure out how to do different bases, it might be helpful to look at the help page for the log function.