Chapter 14 Customizing Graphics
library(tidyverse) # loading ggplot2 and dplyr
library(viridis) # The viridis color schemes
library(latex2exp) # For plotting math notation
library(plotly) # for interactive hover-textDr. Sonderegger’s Video Companion: Video Lecture.
This chapter will address additional tasks that occur while polishing graphics from the ggplot2 package. This chapter builds upon the topics presented in Chapter 3.
14.1 Multi-plots
There are several cases where it is reasonable to need to take several possibly unrelated graphs and put them together into a single larger graph. This is not possible using facet_wrap or facet_grid as they are intended to make multiple highly related graphs that can be formulated in a single data frame. Instead we have to turn to other packages that enhance the ggplot2 package and allow for graphics to be combined from multiple ggplot objects, possibly built from many different data frames..
14.1.1 plot_grid() from package cowplot
Claus O. Wilke wrote a lovely book about data visualization and also wrote an R package to help him tweak his plots. One of the functions in his cowplot package is called plot_grid() and it takes in any number of plots and lays them out on a grid. Below we use the iris and trees data sets to create several visualizations of different types. Notice that the graphs are stored to objects for further use. Once we have all information prepared in the manner we would like to display it, we arrange these graphs together as a single output using the plot_grid() function.
P1 <- ggplot(iris, aes(x=Sepal.Length, y=Sepal.Width, color=Species)) +
geom_point(size=3) + theme(legend.position='bottom')
P2 <- ggplot(trees, aes(x=Height, y=Volume, color=Girth)) +
geom_point(size=3) + theme(legend.position='bottom')
P3 <- ggplot(iris, aes(x=Sepal.Length)) +
geom_histogram(bins=30, fill='grey50', color='black')
P4 <- ggplot(iris, aes(x=Species, y=Sepal.Length, fill=Species)) +
geom_boxplot() + theme(legend.position='bottom')
### place graphics into a grid
cowplot::plot_grid(P1, P2, P3, P4)
Notice that the graphs are by default are arranged in a 2x2 grid. We could adjust the number or rows/columns using the nrow and ncol arguments. Furthermore, alphabetic labels can be added to each graph so that the figure caption can refer to “Panel A” or “Panel B” as appropriate using the labels option. Below creates a single row of three graphics, which shows we can place the graph objects in any order desired, and then adds labels for easy discussion of the graphic.
14.1.2 multiplot() from package Rmisc
In his book, R Graphics Cookbook Winston Chang produced a really nice function to address this issue, but just showed the code. The folks that maintain the R miscellaneous package Rmisc kindly included his function. The benefit of using this function is that you can control the layout to not be on a grid. For example we might want two graphs side by side, and then the third be short and wide underneath both. By specifying different numbers and rows and columns in the layout matrix (below my.layout), we can highly customize how the plot looks.
# Define where the first plot goes, etc.
my.layout <- matrix(
c(1, 2, 2,
1, 2, 2,
3, 3, 3),
byrow = TRUE, nrow=3)
my.layout # show the matrix...## [,1] [,2] [,3]
## [1,] 1 2 2
## [2,] 1 2 2
## [3,] 3 3 3
The layout matrix controls the dimension of each graphics output. Based on the layout matrix above, you notice that the first graphic is one unit wide and two units long, the second graphic is square with 2 units wide and long, and finally the third graph is only one unit long but three units wide. Any number of units can be added to achieve the layout you desire. Unfortunately, Rmisc::multiplot doesn’t have a label option, so if you want to refer to “Panel A”, you need to insert the label into each plot separately.
14.2 Color Scale Customization
While ggplot typically produces reasonable choices for the values of each axis scale and color choices for the color and fill type options, we often want to tweak them. One common objective in modern visualization tasks is making graphics accessible to all readers, so we may want to consider schemes such as colorblind or grey-scale. Below gives a first review of how to manipulate color schemes, change color palettes using different R packages, and how to customize and axis scales and fonts.
14.2.1 Manually Select Colors
For an individual graph, we might want to set the color manually. Within ggplot there are a number of functions of the type scale_XXX_ where the XXX is either color or fill, and can be followed by many arguments such as manual, continuous, gradient, etc. Below we change the color of the P1 and P2 objects created above using the manual and gradient schemes.
cowplot::plot_grid(
P1 + scale_color_manual( values=c('red','navy','forestgreen') ),
P2 + scale_color_gradient(low = 'blue', high='red')
)
Notice that when we used scale_color_manual, we were required to provide a color argument for each species of iris. Similarly, when we used scale_color_gradient, which works with a continuous number scale, we provide a low scale value and high scale value, where a blue to red gradient was then produced. For continuous scales for fill and color, there is also a scale_XXX_gradient2() function which results in a divergent scale where you set the low and high values as well as the midpoint color and value. There is also a scale_XXX_grandientn() function that allows you to set as many colors as you like to move between. Here is an example of gradient2 and gradient.
cowplot::plot_grid(
P2 + scale_color_gradient2(low = 'black', mid='white', midpoint=14, high='red' ),
P2 + scale_color_gradientn(colors = c('red','orange','yellow','green','blue','violet') )
)
Generally I find that I make poor choices when picking colors manually, and luckily R provides a large number of different color palettes for those of us who are not so color-inclined.
14.2.2 Color Palettes
In choosing color schemes, a good approach is to use a color palette that has already been created by folks that know about how colors are displayed and what sort of color blindness is possible. There are two palette options that we’ll discuss, but there are a variety of other palettes available by downloading a package. The palettes given below I have found to be most useful when developing high caliber presentations for publication, large public presentations, or working with different companies.
14.2.2.1 RColorBrewer palettes
Using the ggplot::scale_XXX_brewer() functions, we can easily work with the package RColorBrewer which provides a nice set of color palettes. These palettes are separated by purpose.
Qualitative palettes employ different hues to create visual differences between classes. These palettes are suggested for nominal or categorical data sets.

Sequential palettes progress from light to dark. When used with interval data, light colors represent low data values and dark colors represent high data values.

Diverging palettes are composed of darker colors of contrasting hues on the high and low extremes and lighter colors in the middle.

To use one of these palettes, we just need to pass the palette name to scale_color_brewer or scale_fill_brewer
cowplot::plot_grid(
P1 + scale_color_brewer(palette='Dark2'),
P4 + scale_fill_brewer(palette='BrBG')
)
14.2.2.2 viridis palettes
The package viridis sets up a few different color palettes that have been well thought out and maintain contrast for people with a variety of color-blindness types as well as being converted to grey-scale. These can be excellent accessibility options when working with large diverse groups.

Similar to the RColorBrewer functions, these color palettes can be added to a graph by using a synonymous color_viridis_ functions, which can have either discrete (_d) or continuous (_c) scaling.
cowplot::plot_grid(
P1 + scale_color_viridis_d(option='plasma'), # _d for discrete
P2 + scale_color_viridis_c(option='viridis')) # _c for continuous
There are many other packages that manage color palettes, a few other options include: paletteer, ggsci and wesanderson. Studying color schemes and accessibility options is an important area for data science.
14.3 Axis Scale & Customization
14.3.1 Major & minor ticks
For continuous variables, we need to be able to control what tick and grid lines are displayed. In ggplot, there are major and minor breaks. Major breaks are typically labeled and show as large tick marks on the axis line. Minor breaks appear as either smaller axis ticks or opaque lines on the graph and are found in-between the major breaks. All axis information can be customized. Below provides a method for setting major (breaks) and minor_breaks on a ggplot graph. Minor and major ticks are shown in the default ggplot theme as white background lines. We can also introduce our own axis labels within the _continuous functions, where the y-axis has been labeled either “big” or “small”.
ggplot(trees, aes(x=Height, y=Volume)) + geom_point(size = 3) +
scale_x_continuous( breaks=seq(65,90, by=5), minor_breaks=65:90) +
scale_y_continuous( breaks=c(30,50), labels=c('small','big') ) 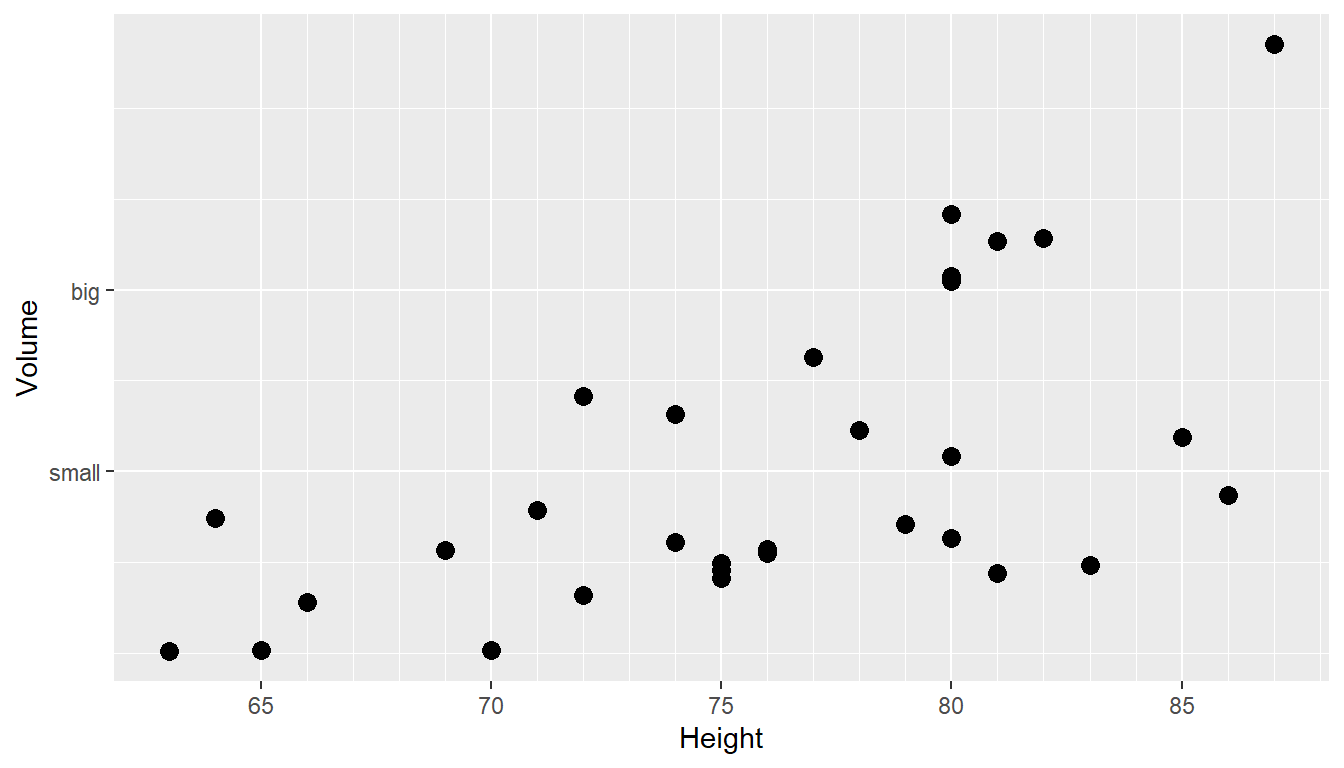
14.3.2 Axis Font, Size, and Style
A very common task to improving graphs is the readability of our axes, which can be modified by changing the text size, font, and style. These can be added within the theme() command to add axis.text and axis.title arguments, which take on the element_text command for augmenting the text output. Each element text can take on their own size, face, and family (font), allowing for customization of text output, most importantly ensuring readability and accessibility. Axis text refers to the labels of the tick marks, while axis title refers to the labels given to each axes (below, Height and Volume).
ggplot(trees, aes(x=Height, y=Volume)) + geom_point(size = 3) +
scale_x_continuous( breaks=seq(65,90, by=5), minor_breaks=65:90) +
scale_y_continuous( breaks=c(30,50), labels=c('small','big') ) +
theme(
# Change x-axis font type
axis.text.x = element_text(size = 12, face = "bold", family = "serif"),
# Change y-axis font type
axis.text.y = element_text(size = 12, face = "italic", family = "mono"),
# Change x-axis title font type
axis.title.x = element_text(size = 14, face = "bold.italic"),
# Change y-axis title font type
axis.title.y = element_text(size = 14, face = "bold.italic")
)
Although the code block above may seem to be getting out of hand and too large for just making a graph, saving the code within a script allows for easy changes and further manipulation from a user. Commonly, someone else working on a project may have suggested changes for a graphic. By saving your output as a nice code block (preferably a script), when changes are requested, only small updates to the code are needed, and then the graphic can be recompiled quickly with the requested changes. This is far preferable to a GUI interface, where you may have to click you way through many options to reformat the graph, whereas by having a large code block, you need only alter the requested lines. At first I recall being unsure about this style. As my work and research has grown, I now handle a large number of very intricate and highly customized graphics for publications, books, and lecture materials with ease.
14.3.3 Log Scales
For this example, lets create a ggplot graphic from the the ACS data of the Lock5Data package. The plot looks at information about Income (in thousands of dollars) against an individuals Age. A simple ggplot scatter graph of the data might be prepared using the following.
# Import the data and drop any zeros
data('ACS', package='Lock5Data')
ACS <- ACS %>%
drop_na() %>% filter(Income > 0)
ggplot(ACS, aes(x=Age, y=Income)) +
geom_point()
Plotting the raw data results in an ugly graph because six observations dominate the graph and the bulk of the data (income < \(\$100,000\)) is squished together. One solution is to plot income on the \(\log_{10}\) scale. Lets plot the log10 data next to the original graph using a plot_grid() for easy comparison.
cowplot::plot_grid(
ggplot(ACS, aes(x=Age, y=Income)) +
geom_point(),
ggplot(ACS, aes(x=Age, y=log10(Income))) +
geom_point()
)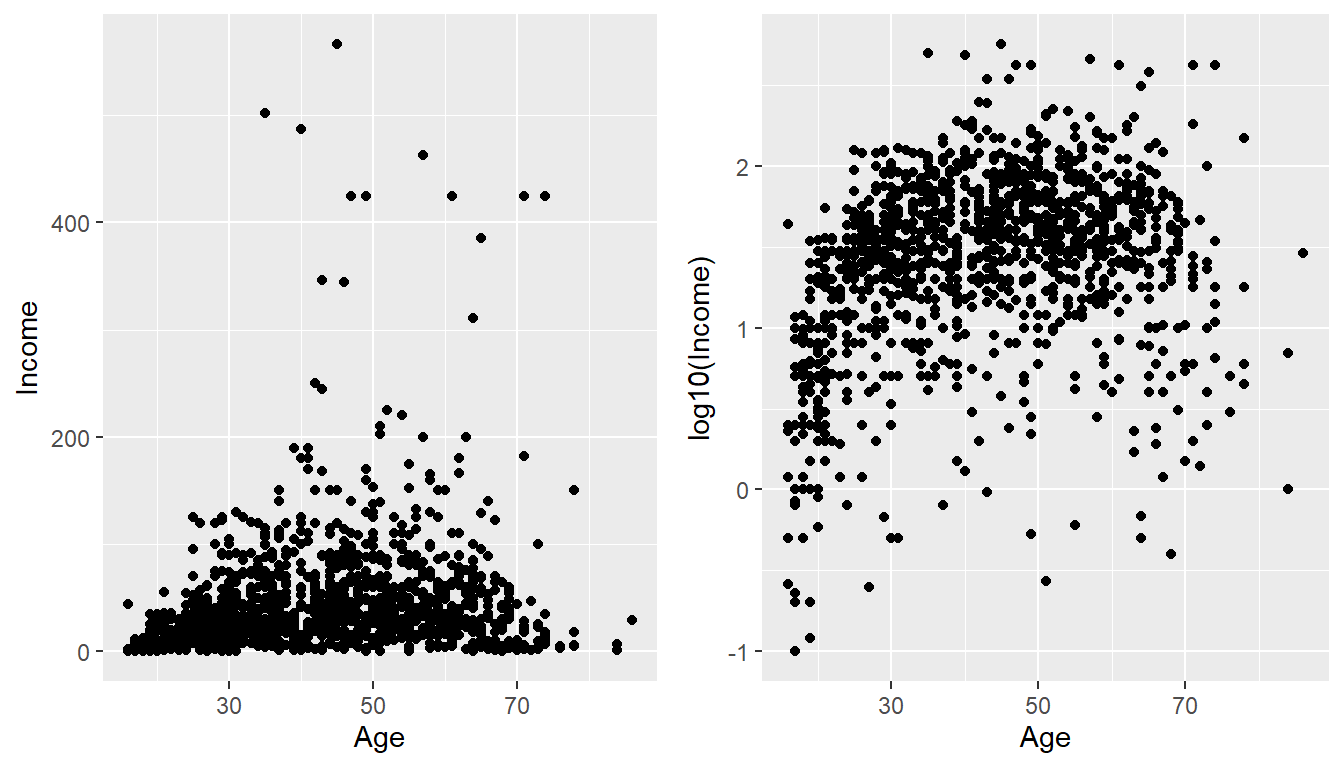
Although our changes to the log-scale worked well to see the trend of peak earning happening in a persons 40s and 50s, the scale is difficult for me to understand (what does \(\log_{10}\left(X\right)=1\) mean here? Oh right, that is \(10^{1}=X\) so that is the \(\$10,000\) line). It would be really nice if we could do the transformation but have the labels on the original scale. Rather than changing the data, which was done in the above graph, we can use the scale_y_log10() function within our ggplot structure. This will create a graph on the log-scale, but retain the original data values. The theme() command and its options will be discussed in more detail below, but to accentuate how the log scaling works, darker minor tick colors were added.
ggplot(ACS, aes(x=Age, y=Income)) +
geom_point() +
scale_y_log10(breaks=c(1,10,100),
minor=c(
seq(0,1,by=0.1),
1:10,
seq(10,100,by=10 ),
seq(100,1000,by=100))) +
ylab('Income (1000s of dollars)') +
theme(
panel.grid.minor = element_line(color = "grey65", linewidth = 0.5))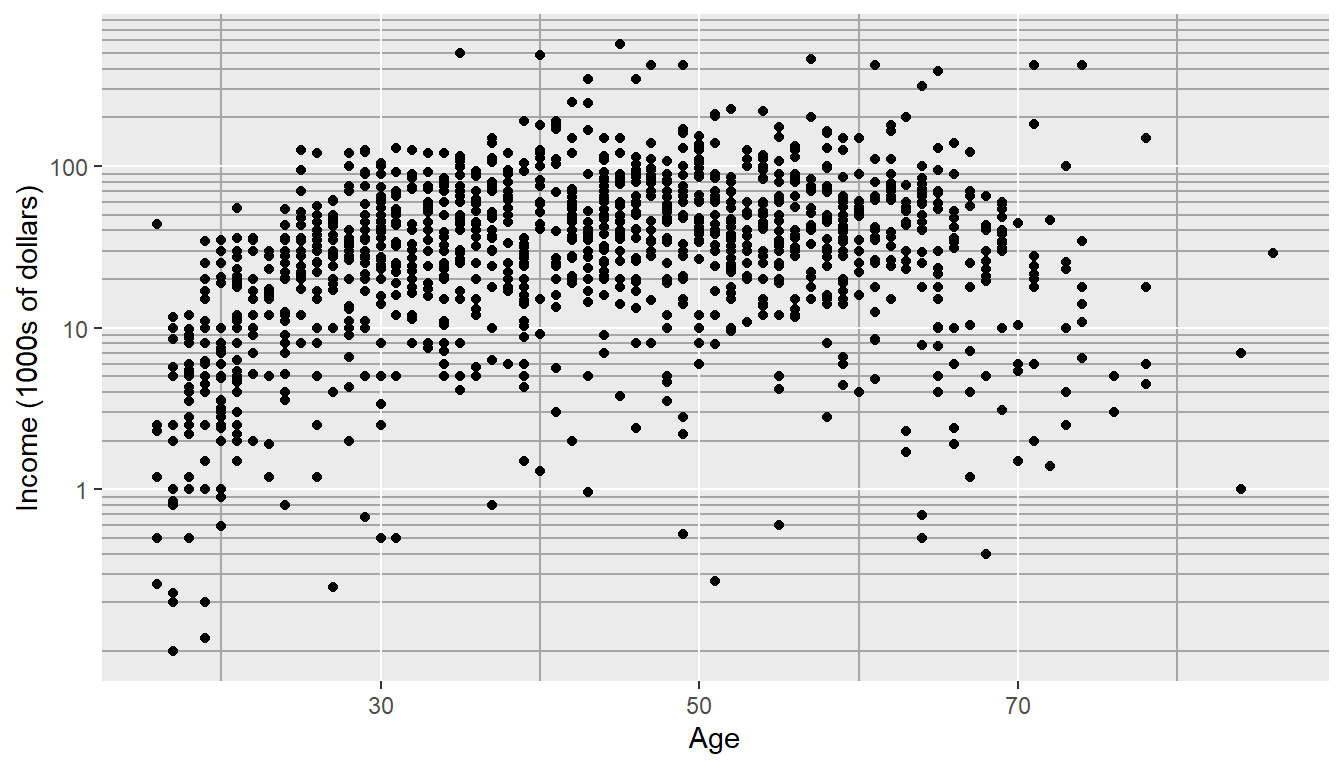
14.4 Mathematical Notation (LaTex)
Now that we have full control over colors and axes styling, it would be good to consider how to include items like mathematical formulas and notation. Commonly mathematical expressions need to be added to plot axes, titles, and on-graph text annotations. R plotting has a notation scheme which it calls expressions. You can learn more about how R expressions are defined by looking at the plotmath help help page. They are similar to LaTeX but different enough that it can be frustrating to use. It is particularly difficult to mix character strings and math symbols. I recommend not bothering to learn R expressions, but instead learn LaTeX and use the R package latex2exp that converts character strings written in LaTeX to be converted into R’s expressions.
LaTeX is an extremely common typesetting program for mathematics and is widely used. The key idea to include LaTeX expression on our graphs is that $ will open/close the LaTeX mode. With LaTeX mode opened using the backslash represents that something special should be done. For example, just typing $alpha$ produces \(alpha\), but putting a backslash in front means that we should interpret it as the Greek letter alpha. So in LaTeX, $\alpha$ is rendered as \(\alpha\). An short introduction to using LaTeX within your RMarkdown documents was given in the Rmarkdown Advanced transition chapter.
This is another good time to remind you about how the escape character works in R. To write character strings with LaTeX syntax inside of an R chunk, recall that the backslash is used to represent special characters. To get the backslash into the character string, we actually need to do the same double backslash trick introduced in the String Manipulation chapter, which was particularly important for Regular Expressions.
Lets start with an example of labeling all aspects of a graph: title, x-axis, y-axis, and on-graph text. The chunk below creates some random data and a best-fit line from the lm() command. A common task is to want to add the results of the model to the graph, which we have studied in the previous Statistical Modeling chapter. Once we have a nice graph with our best-fit line present, we will want to annotate the graph carefully. We have spent a significant amount of time above learning how to augment these annotations, but what if we have very specific units, need to include Greek letters, or generally use LaTeX expressions? The code below now includes four lines of code that allow for inclusion of LaTeX on our graphic.
seed <- 7867
N <- 20
data <- data.frame(x=runif(N, 1, 10)) %>% # create a data set to work with
mutate(y = 12 - 1*x + rnorm(N, sd=1)) # with columns x, y
model <- lm( y ~ x, data=data) # Fit a regression model
data <- data %>% # save the regression line yhat points
mutate(fit=fitted(model))
ggplot(data, aes(x=x)) +
geom_point(aes(y=y)) +
geom_line(aes(y=fit), color='blue') +
annotate('text', x=9, y=9.5,
# latex on the graph
label=latex2exp::TeX('$\\hat{\\rho}$ = 0.916') ) +
# x-axis latex
labs( x=latex2exp::TeX('X-axis $\\alpha$'),
# y-axis latex
y=latex2exp::TeX('Y: $\\log_{10}$(Income)'),
# latex in our title!
title=latex2exp::TeX('Linear Models use: $\\hat{\\beta} = (X^TX)^{-1}X^Ty$'))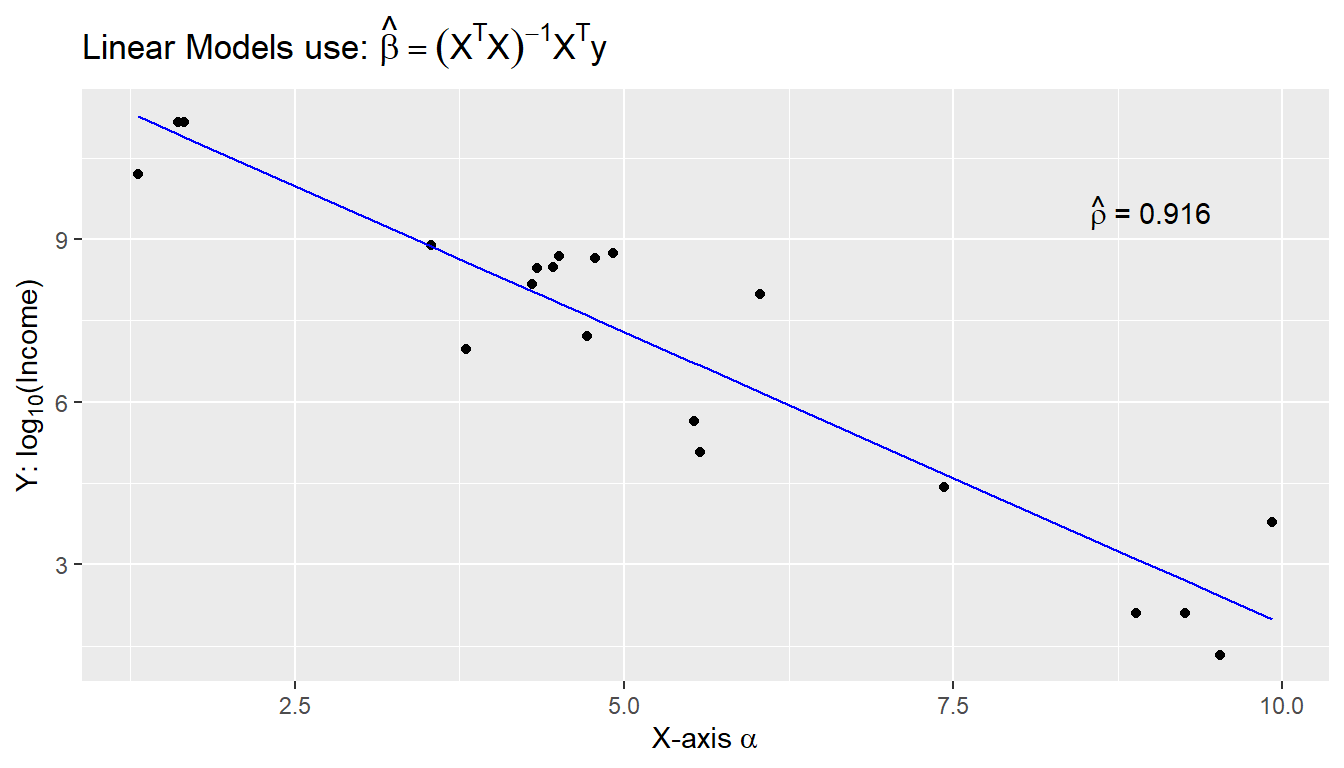
First, notice the use of the double backslash operator within the character strings! As a second note, sometimes R produces a warning message when adding LaTeX to the on-graph environment, but there does not seem to be anything wrong with executing this code. I have not found a reason or fix for this problem, so if you notice it, but the graphic is output correctly, ignore it for now.
Another issue is how to add expression to a data frame. Unfortunately, neither data.frame nor tibble will allow a column of expressions, so instead we have store it as a character string and then introduce the conversion to LaTeX. Below creates three character strings using standard LaTeX syntax, and then converts the character string that represents the R expression. Finally, in ggplot, we tell the geom_text layer to parse the label and interpret it as an R expression. This is a bit of an advanced topic, but can be very useful if you want to add many LaTeX expressions to a graph.
foo <- data.frame( x=c(1,2,3), y=c(2,2,2) ) %>%
# label is a TeX character string
mutate( label1 = paste('$\\alpha$ = ', x) ) %>%
# label2 is an expression character string
mutate( label2 = latex2exp::TeX(label1, output = 'character') )
ggplot(foo, aes(x=x, y=y) ) +
# parse=TRUE forces an expression interpretation
geom_label( aes(label=label2), parse=TRUE ) 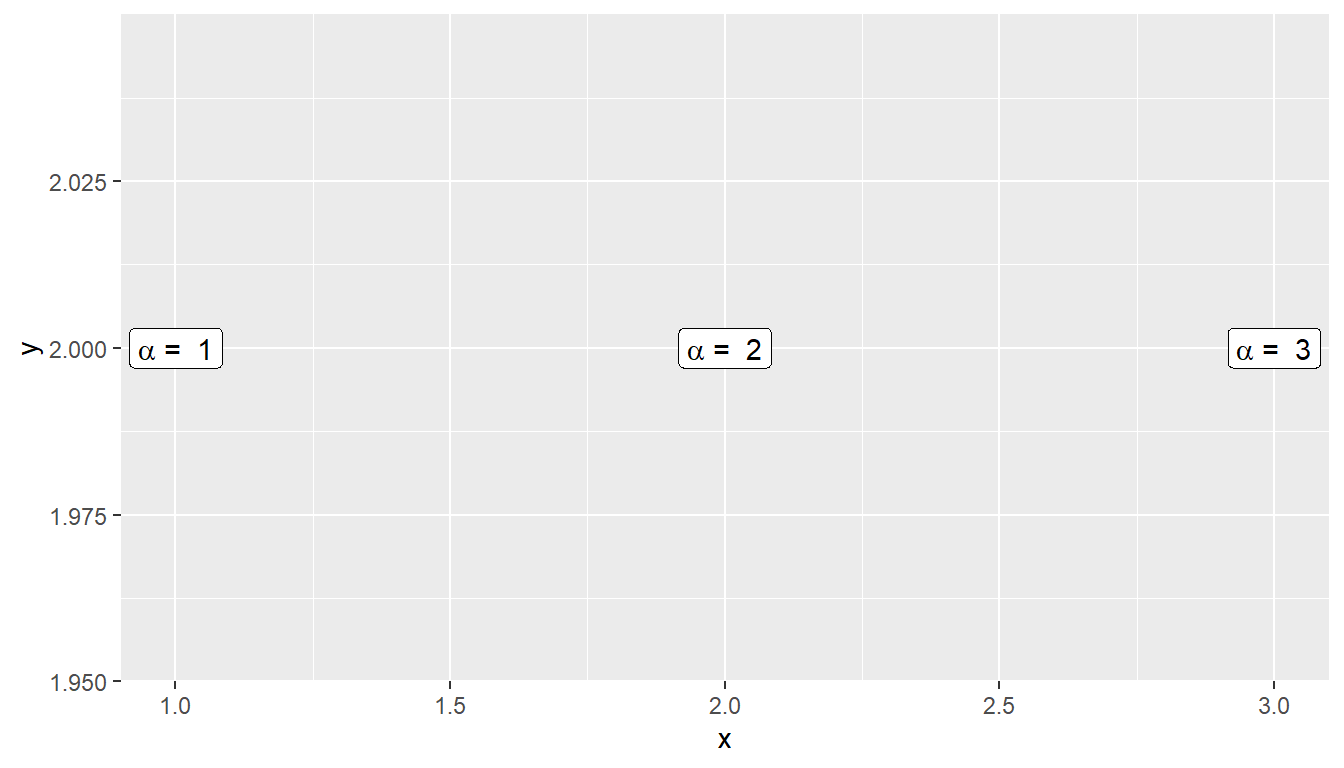
14.5 Themes
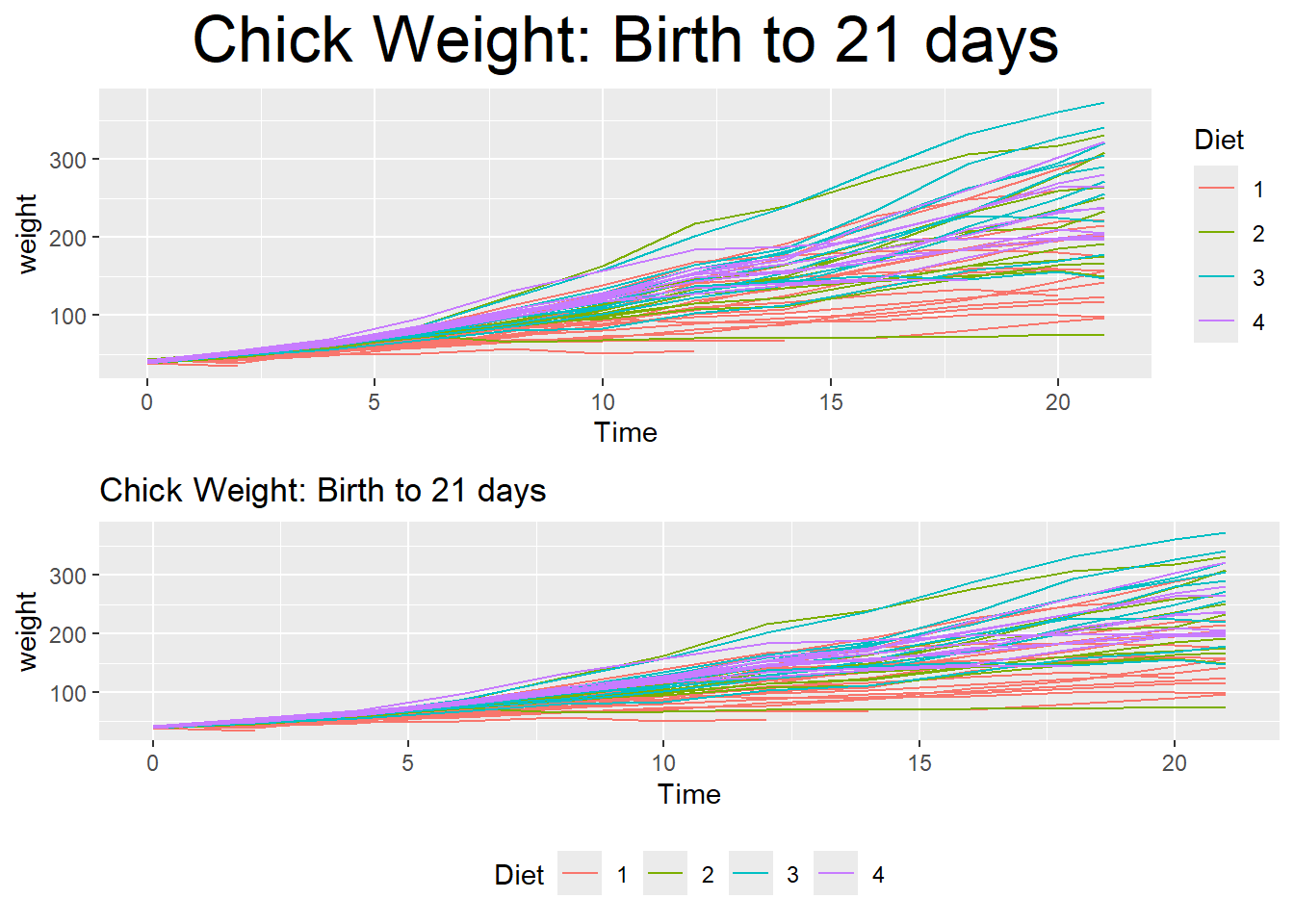
Many fine-tuning settings in ggplot2 can be manipulated using the theme() function. In the log-scale example above, a theme was used to color the minor tick lines. There are a wide variety of theme options available, allowing a user to control nearly all aspects of the graphic that is output. This link will help you understand why I am not introducing them all in this section. One common task with a graph is creating a legend and a graph title. Below uses theme options to change the position and size of the graph title, and in the second panel the legend is shifted to below the graph instead of to the side. It is likely you might want to use these two options in the future, so without showing many different theme possibilities, the two examples below give the generic syntax and concept.
# Create line graphs from the ChickWeight data
p1 <- ggplot(ChickWeight, aes(x=Time, y=weight, colour=Diet, group=Chick)) +
geom_line() + labs(title='Chick Weight: Birth to 21 days')
# Two common examples theme changes
cowplot::plot_grid(nrow=2,
### center and resize the plot title
p1 + theme(plot.title = element_text(hjust = 0.5, size=25)),
### shift the legend to below the graph
### using 'none' will remove the legend!
p1 + theme(legend.position = 'bottom')
)
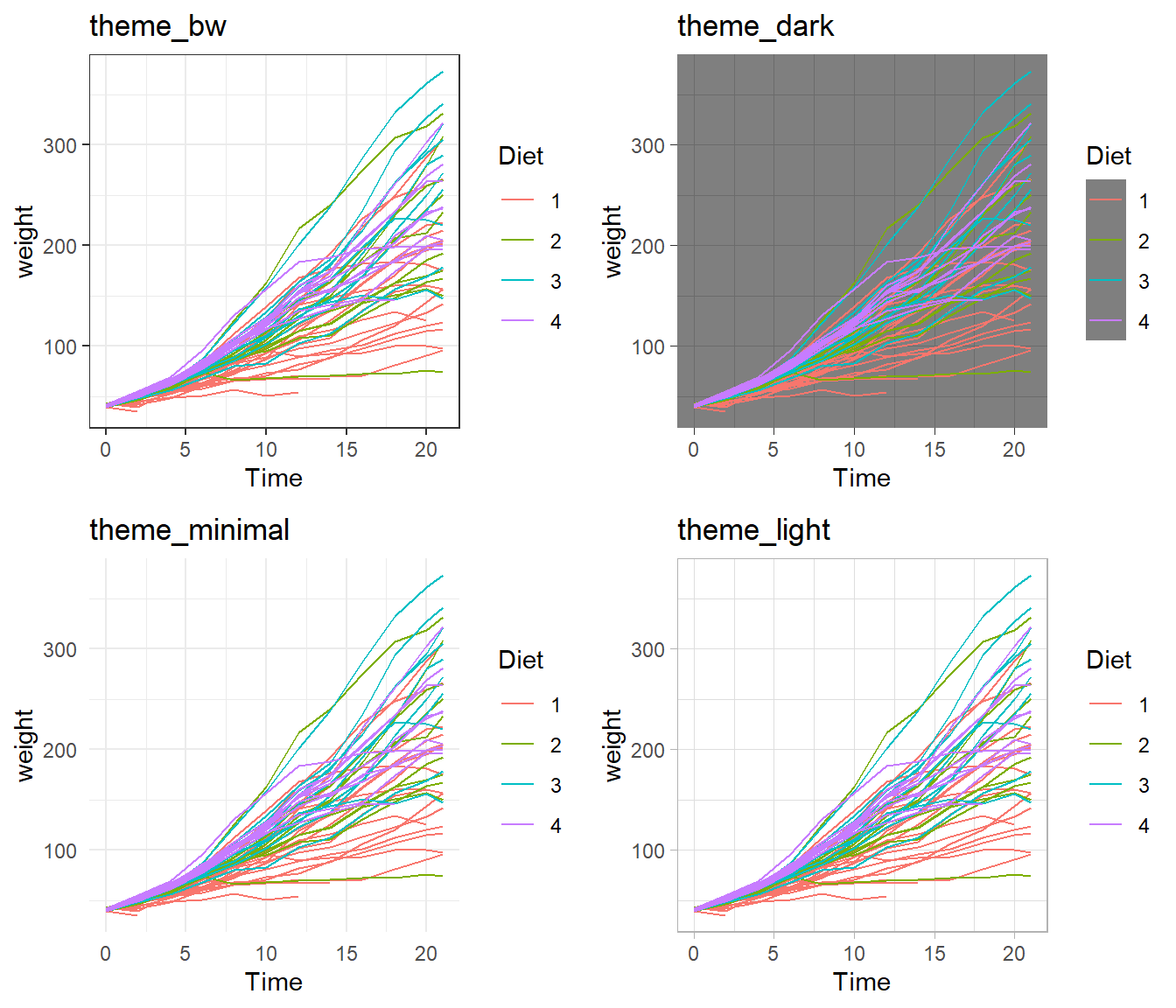
A great deal of thought went into the default settings of ggplot2 to maximize the visual clarity of the graphs. However, it happens frequently that you may need to change the defaults for many of the tiny graphical settings. You can modify each of these as shown in the syntax above, but it is often easier to modify them all at once by selecting a different pre-built theme. The ggplot2 package includes several, with a nice reference page found here. Other packages, such as cowplot and ggthemes, have a bunch of other themes that you can select from. Here is a gallery of ggthemes. It is best to see how these alter a particular graphic, so below creates some nice grids of different themes.
Default Themes.
Rmisc::multiplot( cols = 2,
p1 + theme_bw() + labs(title='theme_bw'), # Black and white
p1 + theme_minimal() + labs(title='theme_minimal'),
p1 + theme_dark() + labs(title='theme_dark'),
p1 + theme_light() + labs(title='theme_light')
)
cowplot.
Rmisc::multiplot( cols = 2,
p1 + cowplot::theme_cowplot() + labs(title='theme_cowplot()'),
p1 + cowplot::theme_minimal_grid() + labs(title='theme_minimial_grid')
)
ggthemes.
Rmisc::multiplot( cols = 2,
p1 + ggthemes::theme_stata() + labs(title='theme_stata()'),
p1 + ggthemes::theme_tufte() + labs(title='theme_tufte()'),
p1 + ggthemes::theme_economist() + labs(title='theme_economist()'),
p1 + ggthemes::theme_fivethirtyeight() + labs(title='theme_fivethirtyeight()'),
p1 + ggthemes::theme_excel_new() + labs(title='theme_excel_new()'),
p1 + ggthemes::theme_gdocs() + labs(title='theme_gdocs()')
)
It is possible to change the theme for all plots within a document or modify specific aspect of the theme. Two of the common options are provided below, which can be added to the start of your Rmarkdown/R-script document and have the settings applied throughout. Some caution should be given in that these will change all graphs within a document, so only change these settings if you are sure you want to apply those changes throughout.
| Command | Result |
|---|---|
theme_set( theme_bw() ) |
Set the default theme to be the theme_bw() theme. |
theme_update( ... ) |
Update the current default them. |
14.6 plotly - Interactive Plots
Plotly is a technical computing company that specializes in data visualization. They have created an open source JavaScript library to produce graphs, which is confusingly referred to as plotly. Because plotly is JavaScript, and RStudio’s Viewer pane is actually a web browser, it easily provides interactive abilities in RStudios Viewer pane. A good tutorial book about using plotly was written by Carson Sievert.
The simple version is that you can take a ggplot graph and pipe it into the ggplotly and it will be rendered into an interactive version of the graph.
P1 <- iris %>%
ggplot(aes(y=Sepal.Length, x=Sepal.Width, color=Species)) +
geom_point()
P1 %>% plotly::ggplotly()We can use the widgets to zoom in and out of the graph. Notice that as we hover over the point, it tells us the x/y coordinates. To add information to the hover-text, we just need to add a text aesthetic mapping to the plot.
P1 <- iris %>%
mutate(Obs_ID = row_number()) %>%
ggplot(aes(y=Sepal.Length, x=Sepal.Width, color=Species,
text=paste('Petal.Length: ', Petal.Length, '\n',
'Petal.Width: ', Petal.Width, '\n',
'Obs #: ', Obs_ID, sep=""))) +
geom_point()
P1 %>% plotly::ggplotly()Interactive plots can be extremely useful when developing web-based platforms. They are not very useful for your static PDFs rendered from RMD. This short introduction should help you see their power, and maybe in the future you will find useful ways to incorporate interactive graphics.
14.7 Exercises
Exercise 1
The infmort data set from the package faraway gives the infant mortality rate for a variety of countries. The information is relatively out of date (\(\approx 1970s\)), but is a great data set for making graphs. Visualize the data by creating scatter plots of mortality vs income while faceting using region and setting color by oil export status. Introduce a \(\log_{10}\) transformation for both mortality and income axes. This can be done either by doing the transformation inside the aes() command or by utilizing the scale_x_log10() or scale_y_log10() layers. The questions below
a) The rownames() of the table gives the country names. Create a new column that contains the country names.
b) Create scatter plots with the log10() transformation inside the aes() command.
c) Create the scatter plots using the scale_x_log10() and scale_y_log10(). Set the major and minor breaks to be useful and aesthetically pleasing.
d) Comment on which version you find easier to read.
e) Learn on your own! The package ggrepel contains functions geom_text_repel() and geom_label_repel() that mimic the basic geom_text() and geom_label() functions in ggplot2, but work to make sure the labels don’t overlap. Select 10-15 countries to label and do so using the geom_text_repel() function.
Exercise 2
Using the datasets::trees data, complete the following. This question refreshes create a linear model, graphing the linear model, and introduces using some LaTeX expressions on the graph.
a) Create a regression model for \(y=\) Volume as a function of \(x=\) Height.
b) Display the summary of the model to view the y-intercept and slope of the regression line.
c) Using ggplot2, create a scatter plot of Volume vs Height.
d) Color the scatter using the Girth variable.
e) Modify the color scheme using a RColorBrewer palette.
f) Create a nice white filled rectangle to add text information. The following might be useful.
g) Use the broom package to extract the coefficients of the best-fit line. Add this information as an annotation to the graph, which should follow a form that looks like \(\hat{y}_i = (INTERCEPT) + (SLOPE)* x_i\). Place the annotation within the white text box.
h) Use the broom package to extract the coefficient of determination \(r^2\) from the model. Add the annotation to your graph, which should look something like \(R^2 = (VALUE)\)
i) Add the regression line in red. There are several ways to do this.
j) Properly label the axes of the graph.
k) Add a descriptive title to the graph.
Exercise 3
The datasets::Titanic table summarizes the survival of passengers aboard the ocean liner Titanic. It includes information about passenger class, sex, and age (adult or child). Create a bar graph showing the number of individuals that survived based on the passenger Class, Sex, and Age variable information. You’ll need to use faceting and/or color to get all four variables on the same graph. Make sure that differences in survival among different classes of children are perceivable. Unfortunately, the data is stored as a table and to expand it to a data frame, the following code can be used.
a) Make this graph using the default theme. If you use color to denote survivors, modify the color scheme so that a cold color denotes death.
b) Make this graph using the theme_bw() theme.
c) Make this graph using the cowplot::theme_minimal_hgrid() theme.
d) Why would it be beneficial to drop the vertical grid lines?