14 Day 14 (March 9)
14.1 Announcements
Class projects
- Proposal (submitted Feb. 17; All proposals have been retured)
- Presentations will occur between May 2 and May 9
- Peer review is due May 5
- Report and reproducible analysis is due May 10
Assignment 7 is posted and due Wednesday (March 22)
- Short in-class demonstration
Read Ch. 17 on spatial models
Question about the Bayesian view of missing data
14.2 Bayesian Prediction
Review how to do prediction using linear model
- Difference between prediction intervals and confidence intervals?
- Do we need an example using R?
Derived derived quantities
- Functions of one or more posterior distribution
Predictive/forecast distributions vs. point estimate
- Popular measures of predictive accuracy
Posterior prediction vs. prior prediction
- Remember the retirement example? (see here)
What do we mean and what do we want to show when we make predictions?
Posterior prediction
- Use data/process model to determine the full-conditional distribution of the unobserved data given the parameters and observed data
- Use composition sampling to perform integration in Eq. 12.2 to BBM2L
# Preliminary steps library(mvnfast) # Required package to sample from multivariate normal distribution url <- "ftp://aftp.cmdl.noaa.gov/products/trends/co2/co2_annmean_mlo.txt" df <- read.table(url,header=FALSE) colnames(df) <- c("year","meanCO2","unc") y <- df$meanCO2 # Response variable X <- model.matrix(~year, data=df) # Design matrix n <- length(y) # Number of observations p <- 2 # Dimensions of beta K <- 5000 #Number of MCMC samples to draw samples <- as.data.frame(matrix(,K,3)) # Samples of the parameters we will save colnames(samples) <- c(colnames(X),"sigma2") sigma2.e <- 1 # Starting values for sigma2 sigma2.beta <- 10^6 #Prior variance for beta q <- 2.01 #Inverse gamma prior with E() = 1/(r*(q-1)) and Var()= 1/(r^2*(q-1)^2*(q-2)) r <- 1 # MCMC algorithm for(k in 1:K){ # Sample beta from full-conditional distribution H <- solve(t(X)%*%X + diag(1/sigma2.beta,p)) h <- t(X)%*%y beta <- t(rmvn(1,H%*%h,sigma2.e*H)) # Sample sigma from full-conditional distribution q.temp <- q+n/2 r.temp <- (1/r+t(y-X%*%beta)%*%(y-X%*%beta)/2)^-1 sigma2.e <- 1/rgamma(1,q.temp,,r.temp) # Save samples of beta and sigma samples[k,] <- c(beta,sigma2.e) } burn.in <- 1000 # Use composition sampling to take samples from # the posterior predictive distribution year <- 2050 K <- dim(samples)[1] beta0.y <- samples[,1] beta1.y <- samples[,2] sigma2.y <-samples[,3] ppd.samples <- rnorm(K,beta0.y + beta1.y*year, sqrt(sigma2.y)) # Histogram of samples from the posterior predictive distribution hist(ppd.samples[-c(1:burn.in)],,xlab=expression(tilde(y)*"|"*bold(y)),ylab=expression("["*tilde(y)*"|"*bold(y)*"]"),freq=FALSE,col="grey",main="",breaks=30)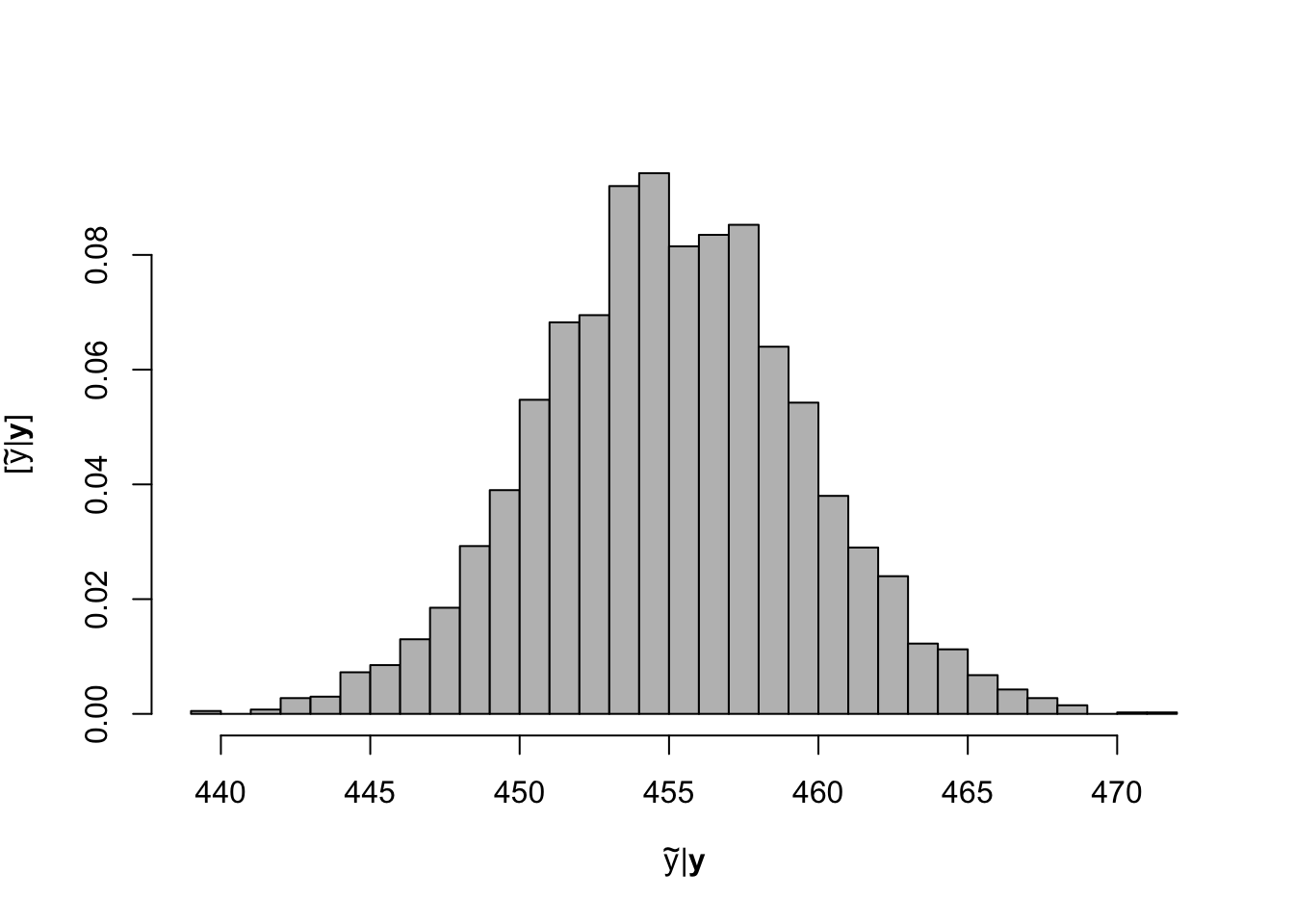
# Expected value of the posterior predictive distribution mean(ppd.samples[-c(1:burn.in)])## [1] 455.0655# 95 % equal-tailed credible interval quantile(ppd.samples[-c(1:burn.in)],prob=c(0.025,0.975))## 2.5% 97.5% ## 446.2166 464.1382# Compare to results obtained from lm function m1 <- lm(meanCO2~year,data=df) predict(m1,newdata=data.frame(year=2050),interval="prediction",level=0.95)## fit lwr upr ## 1 455.1518 446.2043 464.0992