Session 9 ggplot2 in Action (part 2)
9.1 Recap & Workshop
Just like the last session, I am going to present each code submission but this time I will include the image of the plot for comparison.
Together, let us try to construct a mental image of the plot from the code we are reading, but because I think they may be more complex, we will use the visualisation to assist us when needed.
9.1.1 Laura
library(tidyverse)
library(here)
library(ggplot2)
library(dplyr)
library(readr)
Panthera_raw <- read_delim("Data/Panthera_correcto.csv",
";", escape_double = FALSE, trim_ws = TRUE)
View(Panthera_raw)
Panthera_onca <- Panthera_raw %>%
rename(
lon = decimalLongitude,
lat = decimalLatitude
) %>% select(species, stateProvince, lat, lon, elevation, eventDate, day, month, year, countryCode)
#Load map packages
library(rnaturalearth)
library(rnaturalearthdata)
library(sf)
world_map <- ne_countries(scale = "small", returnclass = "sf")
class(world_map)
theme_set(theme_bw())
america <- world_map %>%
filter(continent %in% c("South America", "North America"))
ggplot(
data=america
)+
geom_sf(fill="#faf4cd")+
geom_point(data=Panthera_onca,
aes(
x = lon,
y = lat),
size=1.5,
alpha=0.1,
color="#ff9500"
)+
labs(
title="Jaguar records in America",
subtitle="Records since 1800 till 2021",
x = "Longitude",
y = "Latitude"
)+
coord_sf(ylim = c(-5, 30),
xlim = c(-50, -110))
jaguars_year <- Panthera_onca %>%
#filter(year %in% c("2000", "2001", "2002", "2003", "2004",
# "2005", "2006", "2007", "2008", "2009",
# "2010", "2011", "2012", "2013", "2014",
# "2015", "2016", "2017", "2018", "2019",
# "2020", "2021")) %>%
group_by(countryCode, year) %>%
count(name="count")
jaguars_year_count <- jaguars_year %>%
left_join(america,
by = c("countryCode" = "postal")) %>%
st_as_sf()
ggplot(data=america)+
geom_sf(fill="#faf4cd") +
geom_sf(data = jaguars_year_count,
aes(fill=count
))+
coord_sf(ylim = c(-10, 19),
xlim = c(-45, -100))+
scale_fill_gradient(low = "#f2e038", high = "#bd1b09",
breaks=c(20, 40, 60, 80, 100, 120, 140), limits=c(0, 140),
name="Jaguar count")+
labs(title="**Jaguar** records by *country* in central and northern South America",
subtitle="Mexico is excluded because it has >140 records",
x="Longitude",
y="Latitude",
caption ="Data from: GBIF.org (17 July 2021)",
)+
theme(
plot.title = element_markdown(size=10),
plot.subtitle = element_text(size=9),
legend.title = element_text(size=9),
panel.grid.major = element_line(
color = "#a2a8ab",
linetype = "dashed",
size = 0.5))+
geom_point(data=Panthera_onca,
aes(
x=lon,
y=lat),
size=1,
color="#1e2224",
alpha=0.3)
#How to group by ranges of 10 years? and then use facet_wrap(vars(year))
#How to change the middle color of the gradient scale
#How create new names for each row/value in one variable (country codes = country names)
ggplot(
data=jaguars_year,
aes(
x=year,
y=count,
color=countryCode
)
)+
geom_point()+
scale_y_continuous(breaks=c(40, 80, 120, 160, 200), limits=c(0, 200))+
theme_classic()9.1.2 Gavin
library(tidyverse)## -- Attaching packages --------------------------------------- tidyverse 1.3.1 --## v ggplot2 3.3.3 v purrr 0.3.4
## v tibble 3.1.2 v dplyr 1.0.6
## v tidyr 1.1.3 v stringr 1.4.0
## v readr 1.4.0 v forcats 0.5.1## -- Conflicts ------------------------------------------ tidyverse_conflicts() --
## x dplyr::filter() masks stats::filter()
## x dplyr::lag() masks stats::lag()library(here)## here() starts at C:/Users/gavin/Google Drive/Work/OTS - ggplot course/dvfc_deployed# Import
reptiles <- read_csv("data/reptiles.csv")##
## -- Column specification --------------------------------------------------------
## cols(
## date = col_date(format = ""),
## species = col_character(),
## array = col_character(),
## arm = col_double(),
## trap = col_character(),
## recapture = col_character(),
## size = col_character(),
## group = col_character(),
## block = col_double(),
## treatment = col_character()
## )# p1
reptiles %>%
group_by(
species,
date,
block
) %>%
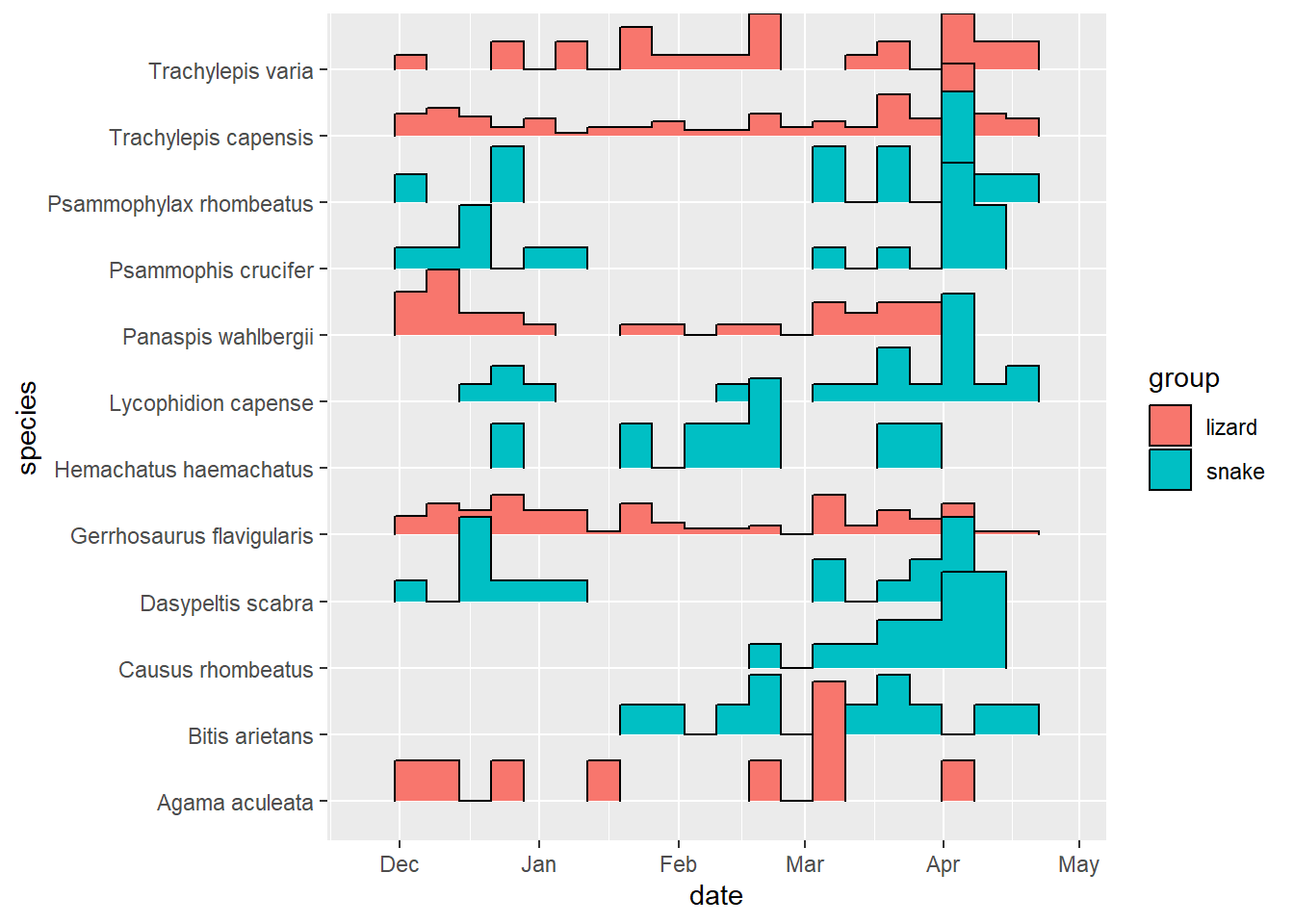
ggplot(mapping = aes(x = date, fill = group)) +
geom_density(position = "fill")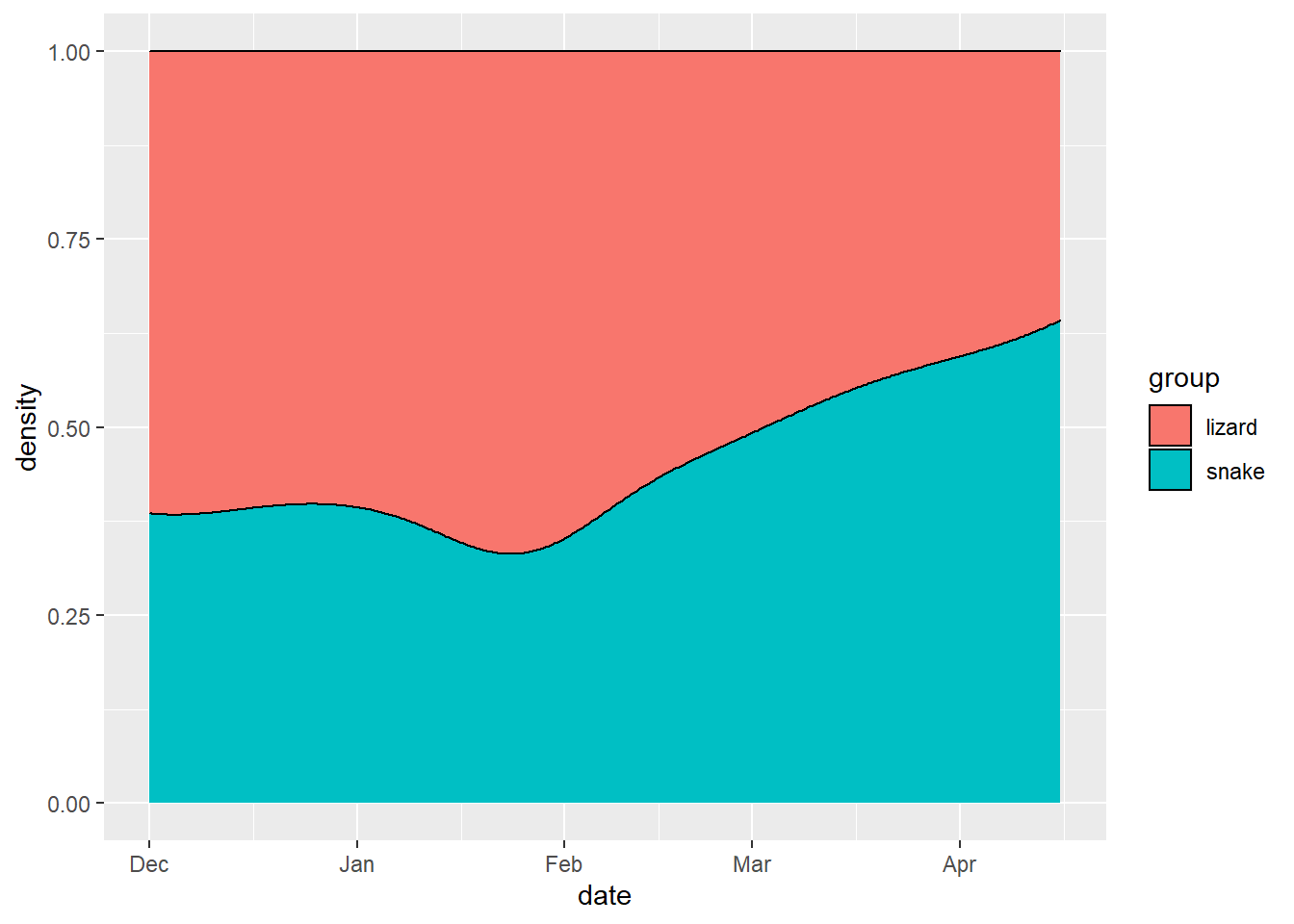
# And the same plot in base R
cdplot(x = reptiles$date, y = as.factor(reptiles$group), col = c("pink", "light blue"))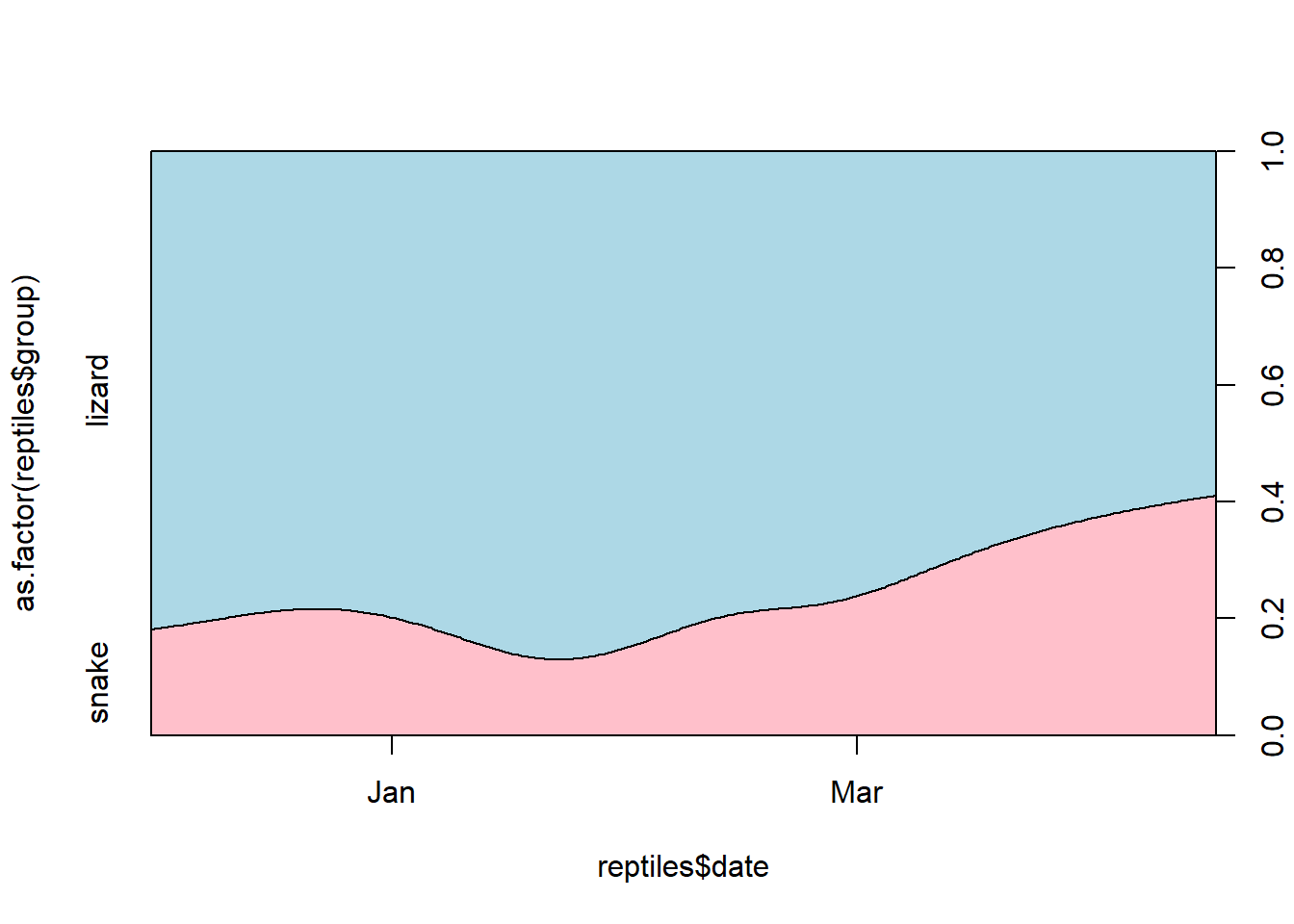
# heatmap of reptile captures through time
reptiles %>%
ggplot(aes(x = date,
y = species)) +
geom_bin2d()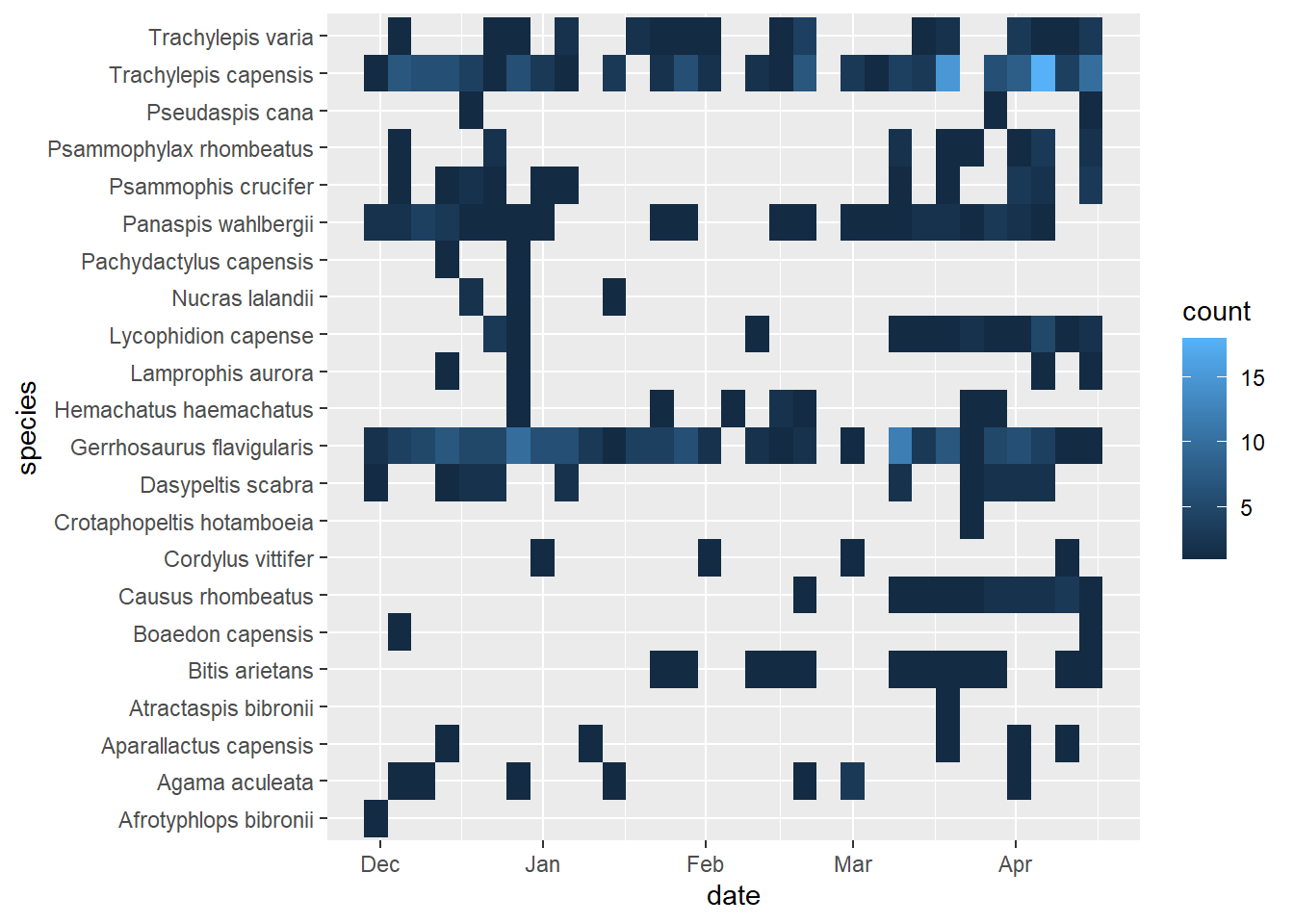
# A different vis - can discuss pros and cons
# e.g no y-axis (distorts numbers of captures across species)
# It was not in fact raining Night Adders (C. rhombeatus) in April compared to other species, 1 in Feb, 6 in March, 8 in April.
reptiles %>%
group_by(species) %>%
filter(n() > 5) %>%
ggplot(aes(x = date, y = species, fill = group)) +
ggridges::geom_density_ridges()## Picking joint bandwidth of 16.9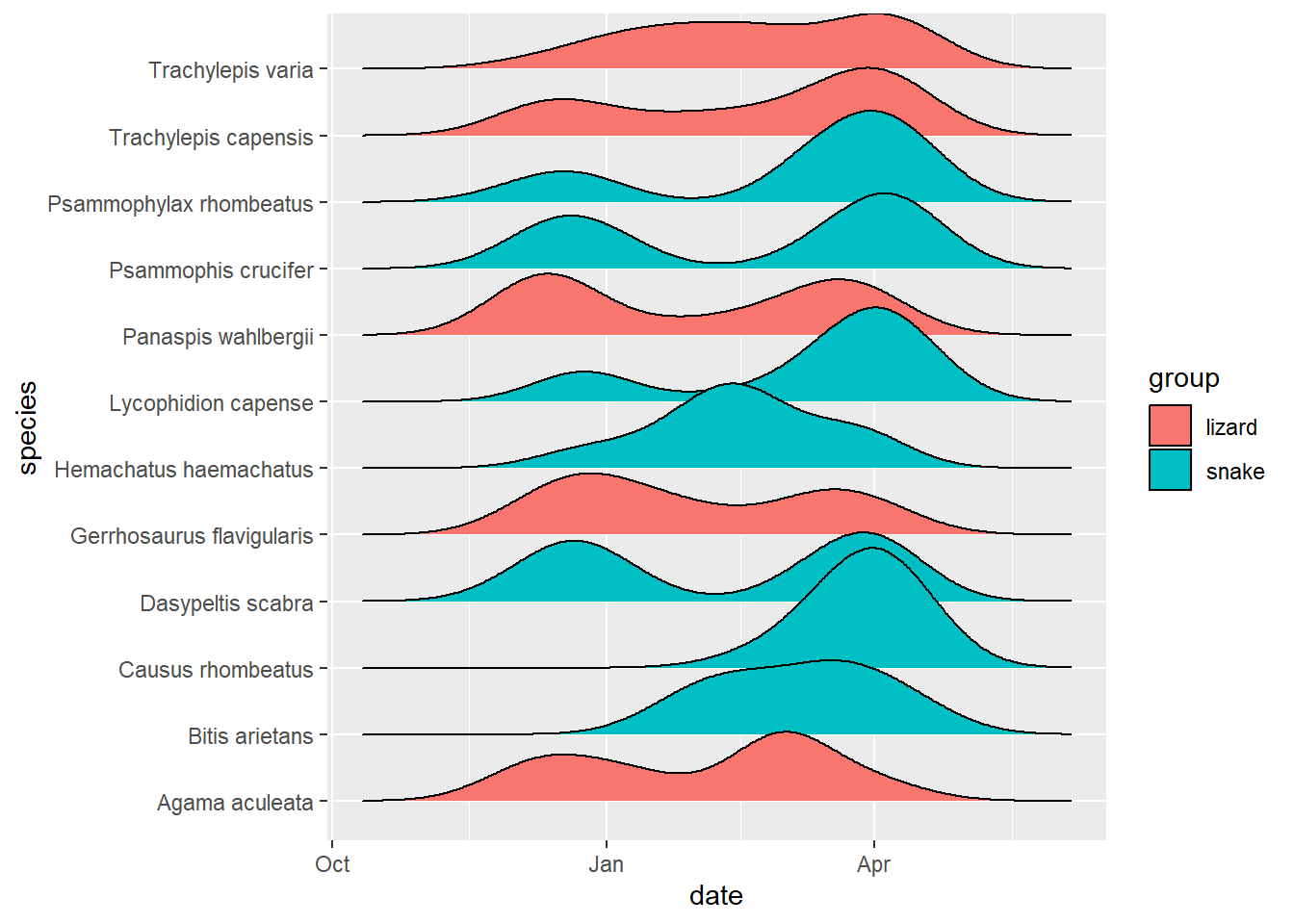
# # hist version
reptiles %>%
group_by(species) %>%
filter(n() > 5) %>%
ggplot(aes(x = date, y = species, fill = group)) +
ggridges::geom_density_ridges(
stat = "binline",
bins = 20, draw_baseline = FALSE
)