3.4 Parsing a file
现在我们已经学会了如何用 parse_*() 函数族解析单个向量,接下来就能回到本章的最初目标,研究readr是如何解析文件的。我们将关注以下两点:
readr如何自动猜出文件每列的数据类型
- 如何修改默认设置
3.4.1 Strategies
readr 通过一种启发式过程(heuristic)来确定每列的类型:先读取文件的前1000行,然后使用(相对保守的)某种启发式算法确定每列的类型。readr 中的导入函数会先用 guess_parser() 函数返回对于所需解析函数最可信的猜测,然后尝试用可能性最大的解析函数解析该列:
guess_parser("123.45")
#> [1] "double"
guess_parser("12,352,561")
#> [1] "number"
guess_parser("1998-11-12")
#> [1] "date"
guess_parser(c("True", "False"))
#> [1] "logical"这个过程会依次尝试以下每种数据类型,直到找到匹配的类型。
逻辑值(logical):
只包括F、T、FALSE和True
整数(integer)
只包括数值型字符(以及-)
双精度浮点数(double)
只包括有效的双精度浮点数
数值(number)
只包括带有分组符号的有效双精度浮点数
时间
与默认的time_format匹配的值
日期
与默认的date_format匹配的值
日期时间
符合ISO 8601标准的任何日期
如果以上数据不符合上述要求中的任意一个,那么这一列就是一个字符串向量,readr将使用parse_character()解析它。
3.4.2 Possible challenges
这些默认设置对更大的文件并不总是有效。以下是两个可能遇到的主要问题:
readr通过前 1000 行猜测数据类型,但是前 1000 行可能只是一种特殊情况,不足以代表整列。例如,一列双精度数值的前1000行有可能都是整数
- 列中可能包含大量缺失值。如果前 1000 行都是
NA,那么readr会认为这是一个字符向量,但你其实想将这一类解析为更具体的值。
readr的安装包里包含了一份文件challenge.csv,用来说明解析过程中可能遇到的问题。这个csv文件包含两列x,y和2001行观测。x 列的前 1001 行均为整数,但之后的值均为双精度整数。y 列的前 1001 行均为NA,后面是日期型数据:
# readr_example() find path for a built-in readr file
challenge <- read_csv(readr_example("challenge.csv"))
#> Parsed with column specification:
#> cols(
#> x = col_double(),
#> y = col_logical()
#> )可以看到,read_csv() 成功解析了 x,但对于 y 则无从下手,因为使用了错误的解析函数col_logical()。使用 problems() 函数明确列出这些失败记录,以便深入探究其中的问题:
problems(challenge)
#> # A tibble: 1,000 x 5
#> row col expected actual file
#> <int> <chr> <chr> <chr> <chr>
#> 1 1001 y 1/0/T/F/TRUE/~ 2015-01~ 'C:/Users/Lenovo/Documents/R/win-library/~
#> 2 1002 y 1/0/T/F/TRUE/~ 2018-05~ 'C:/Users/Lenovo/Documents/R/win-library/~
#> 3 1003 y 1/0/T/F/TRUE/~ 2015-09~ 'C:/Users/Lenovo/Documents/R/win-library/~
#> 4 1004 y 1/0/T/F/TRUE/~ 2012-11~ 'C:/Users/Lenovo/Documents/R/win-library/~
#> 5 1005 y 1/0/T/F/TRUE/~ 2020-01~ 'C:/Users/Lenovo/Documents/R/win-library/~
#> 6 1006 y 1/0/T/F/TRUE/~ 2016-04~ 'C:/Users/Lenovo/Documents/R/win-library/~
#> # ... with 994 more rows可以使用spec_csv() 来直接查看 readr 在默认情况下用那种类型的解析函数解析数据:
为了解决这个问题,我们用read_csv()函数中的col_types指定每列的解析方法(column specification),之前我们向col_types传入一个字符串说明各列的类别,但这里是要直接指明解析函数了。这样做的指定必须通过cols()函数来创建(具体格式和spec_csv()或者read_csv()自动打印的说明是一样的):
challenge <- read_csv(readr_example("challenge.csv"),
col_types =cols(
x = col_double(),
y = col_date()
))
tail(challenge)
#> # A tibble: 6 x 2
#> x y
#> <dbl> <date>
#> 1 0.805 2019-11-21
#> 2 0.164 2018-03-29
#> 3 0.472 2014-08-04
#> 4 0.718 2015-08-16
#> 5 0.270 2020-02-04
#> 6 0.608 2019-01-06每个parse_*()函数都有一个对应的col_*()函数。如果数据已经保存在R的字符向量中,那么可以使用parse_*(),如果要告诉readr如何加载数据,则应该使用col_*()。
The available specifications are: (with string abbreviations in brackets)
col_logical()[l], containing only T, F, TRUE or FALSE.
col_integer()[i], integers.
col_double()[d], doubles.
col_character()[c], everything else.
col_factor(levels, ordered)[f], a fixed set of values.
col_date(format = "")[D]: with the locale’s date_format.
col_time(format = "")[t]: with the locale’s time_format.
col_datetime(format = "")[T]: ISO8601 date times
col_number()[n], numbers containing the grouping_mark
col_skip()[_, -], don’t import this column.
col_guess()[?], parse using the “best” type based on the input.
cols_only()代替cols()可以仅指定部分列的解析方式;.default表示未提及的所有列(read_csv()的默认设置可以表示为read_csv( col_type = cols(.default = col_guess()))
一旦我们指定了正确的解析函数,问题便迎刃而解。
3.4.3 Other tips
我们再介绍其他几种有注意解析文件的通用技巧:
- 在前面的示例中,如果比默认方式再多检查一行,就可以解析成功:
type_convert()re-convert character columns in existing data frame. This is useful if you need to do some manual munging - you can read the columns in as character, clean it up with (e.g.) regular expressions and then let readr take another stab at parsing it.
df <- tibble(
x = as.character(runif(10)),
y = as.character(sample(10)),
z = FALSE
)
df %>% glimpse()
#> Rows: 10
#> Columns: 3
#> $ x <chr> "0.78455981053412", "0.525363321648911", "0.336503465892747", "0....
#> $ y <chr> "2", "10", "1", "6", "8", "7", "4", "5", "3", "9"
#> $ z <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE
# note changes in column types
df %>% type_convert() %>% glimpse()
#> Parsed with column specification:
#> cols(
#> x = col_double(),
#> y = col_double()
#> )
#> Rows: 10
#> Columns: 3
#> $ x <dbl> 0.7846, 0.5254, 0.3365, 0.5136, 0.5227, 0.5568, 0.7613, 0.5780, 0...
#> $ y <dbl> 2, 10, 1, 6, 8, 7, 4, 5, 3, 9
#> $ z <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE- 如果正在读取一个非常大的文件,那么应该将
n_max设置为一个较小的数,比如10,000 或者 100,000,这可以让加速重复试验的过程。
3.4.4 Example: Dealing with metadata
https://alison.rbind.io/post/2018-02-23-read-multiple-header-rows/
This dataset is from an article published in PLOS ONE called “Being Sticker Rich: Numerical Context Influences Children’s Sharing Behavior”. In this study, children (ages 3–11) received a small (12, “sticker poor”) or large (30, “sticker rich”) number of stickers, and were then given the opportunity to share their windfall with either one or multiple anonymous recipients.
Data in a plain text editor:
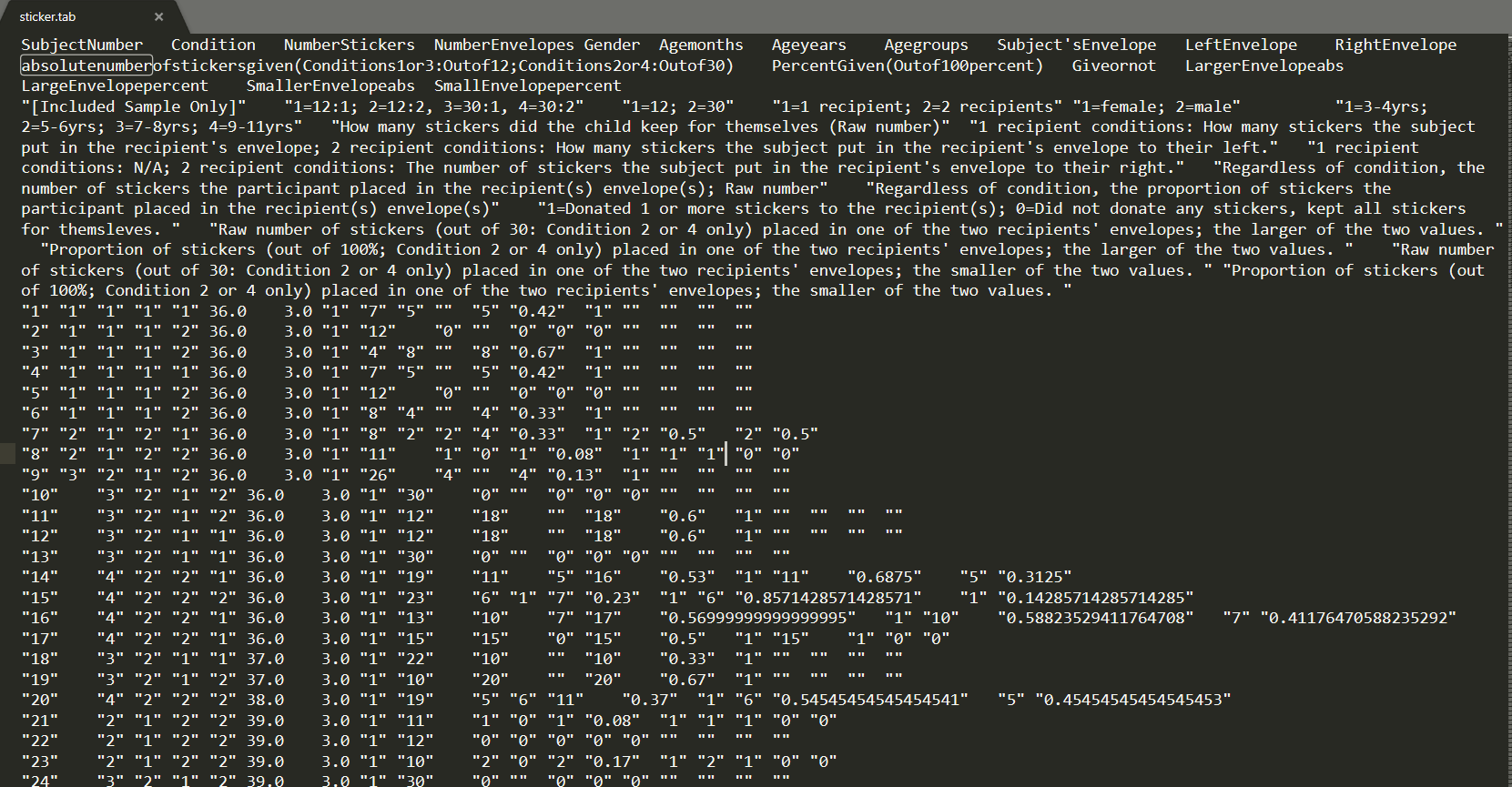
To read in a .tab file, use read_tsv()
# https://dataverse.harvard.edu/api/access/datafile/2712105
stickers <- read_tsv("data/sticker.tab")
spec(stickers)
#> cols(
#> SubjectNumber = col_character(),
#> Condition = col_character(),
#> NumberStickers = col_character(),
#> NumberEnvelopes = col_character(),
#> Gender = col_character(),
#> Agemonths = col_double(),
#> Ageyears = col_double(),
#> Agegroups = col_character(),
#> `Subject'sEnvelope` = col_character(),
#> LeftEnvelope = col_character(),
#> RightEnvelope = col_character(),
#> `absolutenumberofstickersgiven(Conditions1or3:Outof12;Conditions2or4:Outof30)` = col_character(),
#> `PercentGiven(Outof100percent)` = col_character(),
#> Giveornot = col_character(),
#> LargerEnvelopeabs = col_character(),
#> LargeEnvelopepercent = col_character(),
#> SmallerEnvelopeabs = col_character(),
#> SmallEnvelopepercent = col_character()
#> )The problem here is that the second row is actually metadata or descriptions about each column header. But this make read_tsv() recognize as character type many of our columns. To verify this, see the first and last 6 rows:
head(stickers)
#> # A tibble: 6 x 18
#> SubjectNumber Condition NumberStickers NumberEnvelopes Gender Agemonths
#> <chr> <chr> <chr> <chr> <chr> <dbl>
#> 1 [Included Sa~ 1=12:1; ~ 1=12; 2=30 1=1 recipient;~ 1=fem~ NA
#> 2 1 1 1 1 1 36
#> 3 2 1 1 1 2 36
#> 4 3 1 1 1 2 36
#> 5 4 1 1 1 1 36
#> 6 5 1 1 1 2 36
#> # ... with 12 more variables: Ageyears <dbl>, Agegroups <chr>,
#> # `Subject'sEnvelope` <chr>, LeftEnvelope <chr>, RightEnvelope <chr>,
#> # `absolutenumberofstickersgiven(Conditions1or3:Outof12;Conditions2or4:Outof30)` <chr>,
#> # `PercentGiven(Outof100percent)` <chr>, Giveornot <chr>,
#> # LargerEnvelopeabs <chr>, LargeEnvelopepercent <chr>,
#> # SmallerEnvelopeabs <chr>, SmallEnvelopepercent <chr>
tail(stickers)
#> # A tibble: 6 x 18
#> SubjectNumber Condition NumberStickers NumberEnvelopes Gender Agemonths
#> <chr> <chr> <chr> <chr> <chr> <dbl>
#> 1 396 1 1 1 2 136
#> 2 397 4 2 2 1 136
#> 3 398 1 1 1 1 137
#> 4 399 1 1 1 2 137
#> 5 400 4 2 2 2 139
#> 6 401 3 2 1 1 143
#> # ... with 12 more variables: Ageyears <dbl>, Agegroups <chr>,
#> # `Subject'sEnvelope` <chr>, LeftEnvelope <chr>, RightEnvelope <chr>,
#> # `absolutenumberofstickersgiven(Conditions1or3:Outof12;Conditions2or4:Outof30)` <chr>,
#> # `PercentGiven(Outof100percent)` <chr>, Giveornot <chr>,
#> # LargerEnvelopeabs <chr>, LargeEnvelopepercent <chr>,
#> # SmallerEnvelopeabs <chr>, SmallEnvelopepercent <chr>To solve this, we will first create a (tidier) character vector of the column names only. Then we’ll read in the actual data and skip the multiple header rows at the top. When we do this, we lose the column names, so we use the character vector of column names we created in the first place instead.
When we set n_max = 0 and col_names = TRUE(the default), ony column headers will be read. Then we could ask janitor::clean_names() to produce a tidier set of names:
sticker_names <- read_tsv("data/sticker.tab",
n_max = 0) %>% # default: col_names = TRUE
rename("stickers_give" = 'absolutenumberofstickersgiven(Conditions1or3:Outof12;Conditions2or4:Outof30)') %>%
janitor::clean_names() %>%
names()
sticker_names
#> [1] "subject_number" "condition"
#> [3] "number_stickers" "number_envelopes"
#> [5] "gender" "agemonths"
#> [7] "ageyears" "agegroups"
#> [9] "subjects_envelope" "left_envelope"
#> [11] "right_envelope" "stickers_give"
#> [13] "percent_given_outof100percent" "giveornot"
#> [15] "larger_envelopeabs" "large_envelopepercent"
#> [17] "smaller_envelopeabs" "small_envelopepercent"sticker_tidy <- read_tsv("data/sticker.tab",
col_names = sticker_names,
skip = 2)
# or
read_tsv("data/sticker.tab",
col_names = FALSE,
skip = 2) %>%
set_names(sticker_names)
#> # A tibble: 401 x 18
#> subject_number condition number_stickers number_envelopes gender agemonths
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 1 1 1 1 1 36
#> 2 2 1 1 1 2 36
#> 3 3 1 1 1 2 36
#> 4 4 1 1 1 1 36
#> 5 5 1 1 1 2 36
#> 6 6 1 1 1 2 36
#> # ... with 395 more rows, and 12 more variables: ageyears <dbl>,
#> # agegroups <dbl>, subjects_envelope <dbl>, left_envelope <dbl>,
#> # right_envelope <dbl>, stickers_give <dbl>,
#> # percent_given_outof100percent <dbl>, giveornot <dbl>,
#> # larger_envelopeabs <dbl>, large_envelopepercent <dbl>,
#> # smaller_envelopeabs <dbl>, small_envelopepercent <dbl>What if we want to include that meta data? Create a variable description column use pivot_longer()
sticker_dict <- read_tsv("data/sticker.tab", n_max = 1) %>%
rename(stickersgiven = 'absolutenumberofstickersgiven(Conditions1or3:Outof12;Conditions2or4:Outof30)') %>%
janitor::clean_names() %>%
pivot_longer(everything(),
names_to = "variable_name",
values_to = "variable_description")
sticker_dict
#> # A tibble: 18 x 2
#> variable_name variable_description
#> <chr> <chr>
#> 1 subject_number [Included Sample Only]
#> 2 condition 1=12:1; 2=12:2, 3=30:1, 4=30:2
#> 3 number_stickers 1=12; 2=30
#> 4 number_envelopes 1=1 recipient; 2=2 recipients
#> 5 gender 1=female; 2=male
#> 6 agemonths <NA>
#> # ... with 12 more rows3.4.5 Example: multi-row headers
https://debruine.github.io/posts/multi-row-headers/
In most cases, a header will only take up one row. And when multi-row headers present itself, readr fails to recognize all these rows as header
# create a small demo csv
demo_csv <- "SUB1, SUB1, SUB1, SUB1, SUB2, SUB2, SUB2, SUB2
COND1, COND1, COND2, COND2, COND1, COND1, COND2, COND2
X, Y, X, Y, X, Y, X, Y
10, 15, 6, 2, 42, 4, 32, 5
4, 43, 7, 34, 56, 43, 2, 33
77, 12, 14, 75, 36, 85, 3, 2"
read_csv(demo_csv)
#> Warning: Duplicated column names deduplicated: 'SUB1' => 'SUB1_1' [2], 'SUB1'
#> => 'SUB1_2' [3], 'SUB1' => 'SUB1_3' [4], 'SUB2' => 'SUB2_1' [6], 'SUB2' =>
#> 'SUB2_2' [7], 'SUB2' => 'SUB2_3' [8]
#> # A tibble: 5 x 8
#> SUB1 SUB1_1 SUB1_2 SUB1_3 SUB2 SUB2_1 SUB2_2 SUB2_3
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 COND1 COND1 COND2 COND2 COND1 COND1 COND2 COND2
#> 2 X Y X Y X Y X Y
#> 3 10 15 6 2 42 4 32 5
#> 4 4 43 7 34 56 43 2 33
#> 5 77 12 14 75 36 85 3 2In demo_csv, the first three rows are intended to be an “overall” header. read_csv() outputs a message and modifes the first row for a unqiue set of column names, since it assumes this is a one-row header.
Based on what we have learned in the previous example, the solution should be easy. First set n_max to gain rows of interest, then compose a one-row header to set column names.
demo_names <- read_csv(demo_csv,
n_max = 3,
col_names = FALSE) %>%
map_chr(~ str_c(.x, collapse = "")) %>%
unname()
demo_names
#> [1] "SUB1COND1X" "SUB1COND1Y" "SUB1COND2X" "SUB1COND2Y" "SUB2COND1X"
#> [6] "SUB2COND1Y" "SUB2COND2X" "SUB2COND2Y"
read_csv(demo_csv, col_names = demo_names, skip = 3)
#> # A tibble: 3 x 8
#> SUB1COND1X SUB1COND1Y SUB1COND2X SUB1COND2Y SUB2COND1X SUB2COND1Y SUB2COND2X
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 10 15 6 2 42 4 32
#> 2 4 43 7 34 56 43 2
#> 3 77 12 14 75 36 85 3
#> # ... with 1 more variable: SUB2COND2Y <dbl>For reading multi-row headers in Excel, check section 3.5.1