10 Multipanel figure for MS
10.1 Goals:
Pull code for the following models together into one place:
- Leaf area in 1990
- Front survival in 1991
- Front leaf area in 1991
- Back shoot production in 1991
- Back leaf area in 1991
Then, using that code, make the following figure panels:
| Response | Sever | Time | Interaction |
|---|---|---|---|
| Leaf 1990 | Yes | Yes | Yes |
| Front survival 1991 | Yes | Yes | |
| Front leaf area 1991 | Yes | Yes | |
| Back shoot production 1991 | Yes | Yes | |
| Back leaf area 1991 | Yes | Yes | Yes |
Then, put all of those panels into one 5 x 3 grid!
10.2 Leaf area in 1990
may<-read.csv("Datasets/mayapple_1990_updated.csv")
##need to make the variables we are working with numeric:
may$Lf_90<-as.numeric(as.character(may$Lf_90))
may<-may %>%
filter(!is.na(Lf_90))10.2.1 Best model
model.bm<-lmer(Lf_90~Sex_90*Time+Sever*Time+(1|COLONY), data=may, na.action = "na.fail", REML=FALSE)The best model includes fixed effects of Sever, Sex_90, Time, the Sever x Time interaction, and the Sex_90 x Time interaction.
10.2.2 Estimated marginal means
The main effect of Sever:
#emmeans(model.bm, pairwise~Sever, type="response")
library(mdthemes)
Sever_1 <- emmip(model.bm, ~ Sever ,CIs = TRUE)+xlab("")+mdthemes::md_theme_classic() +ggtitle("**Sever**
*Leaf area in 1990*")The main effect of Time:
#emmeans(model.bm, pairwise~Time, type="response")
Time_1 <- emmip(model.bm, ~ Time ,CIs = TRUE)+xlab("")+ylab("")+mdthemes::md_theme_classic() +ggtitle("**Time**") The Sever x Time interaction:
10.3 Front survival 1991
# set na.strings="." to recode '.' to NA
mayappleDatasetFull<-readxl::read_excel("Datasets/mayapple_1990_updated.xlsx", na=".")# select variables for analysis and filter out rows that don't have a leaf area recorded in 1990
mayappleData <- mayappleDatasetFull %>%
dplyr::select(COLONY,Lf_90,brnoF_91,Sex_90,Sever,Time,SexF1_91,SexF2_91,SexF3_91,LfFtot_91) %>%
filter(!is.na(Lf_90))
# create binary variable for plants that are dead/have no branches (0) or have 1 or more branches (1)
# append new variable to the dataframe
mayappleData<- mayappleData %>%
dplyr::mutate(deadAliveBinary=ifelse(brnoF_91>0,1,0))10.3.1 Best model
10.3.2 Estimated marginal means
The main effect of severing
#emmeans(sur_91f_bm, pairwise~Sever, type="response" )
Sever_2 <- emmip(sur_91f_bm, ~ Sever , type="response",CIs = TRUE)+xlab("")+ylab("Predicted probability")+mdthemes::md_theme_classic()+ggtitle("*Front survival in 1991*")The main effect of time
10.4 Front conditional leaf area 1991
mayappleData$LfFtot_91<-as.numeric(mayappleData$LfFtot_91)
mayappleData <- mayappleData %>%
filter(!is.na(LfFtot_91)) %>%
filter(LfFtot_91>0)10.4.1 Best model
10.4.2 Estimated marginal means
the main effect of sever
10.5 Front leaf area 1991
mayappleData$LfFtot_91<-as.numeric(mayappleData$LfFtot_91)
mayappleData <- mayappleData %>%
filter(!is.na(LfFtot_91))10.5.1 Best model
10.5.2 Estimated marginal means
the main effect of sever
#emmeans(mod91laf.bm, pairwise~Sever, type="response")
Sever_3 <- emmip(mod91laf.bm, ~ Sever ,CIs = TRUE)+xlab("")+mdthemes::md_theme_classic()+ggtitle("*Front leaf area in 1991*")Control systems have the most leaf area. S1 systems have the least leaf area. S2 and S4 systems are intermediate and not different from each other.
the main effect of time
10.6 Back shoot production 1991
# set na.strings="." to recode '.' to NA
mayappleDatasetFull<-readxl::read_excel("Datasets/mayapple_1990_updated.xlsx", na=".")
mayappleData <- mayappleDatasetFull %>%
dplyr::select(COLONY,Lf_90,Sex_90,Sever,Time,brnoB_91,LfB1_91,SexB1_91,LfB2_91,SexB2_91,LfBtot_91,Lftot_91) %>%
filter(!is.na(Lf_90))
mayappleData<- mayappleData %>%
dplyr::mutate(backShootBranchBinary=ifelse(brnoB_91>0,1,0)) %>%
filter(!is.na(backShootBranchBinary))10.6.1 Best model
10.6.2 Estimated marginal means
The main effect of severing
#emmeans(sur_91b_bm, pairwise~Sever, type="response" )
Sever_4 <- emmip(sur_91b_bm, ~ Sever ,type="response",CIs = TRUE )+xlab("")+ylab("Predicted probability")+mdthemes::md_theme_classic()+ggtitle("*Back shoot production in 1991*")The main effect of time
10.7 Back conditional leaf area 1991
## Warning: NAs introduced by coercion10.7.1 Best model
10.7.2 Estimated marginal means
the main effect of sever
#emmeans(b_mod91la.bm, pairwise~Sever, type="response")
Sever_5cond <- emmip(b_mod91la.bm, ~ Sever ,CIs = TRUE )+mdthemes::md_theme_classic()+ggtitle("*Back leaf area in 1991*")## Cannot use mode = "kenward-roger" because *pbkrtest* package is not installed#emmeans(b_mod91la.bm, pairwise~Time, type="response")
Time_5cond <- emmip(b_mod91la.bm, ~ Time ,CIs = TRUE )+ylab("")+mdthemes::md_theme_classic()## Cannot use mode = "kenward-roger" because *pbkrtest* package is not installed10.8 Back leaf area 1991
mayappleData$LfBtot_91<-as.numeric(mayappleData$LfBtot_91)
mayappleData <- mayappleData %>%
filter(!is.na(LfBtot_91))10.8.1 Best model
## fixed-effect model matrix is rank deficient so dropping 3 columns / coefficients10.8.2 Estimated marginal means
the main effect of sever
#emmeans(b_mod91la.bm, pairwise~Sever, type="response")
Sever_5 <- emmip(b_mod91la.bm, ~ Sever ,CIs = TRUE )+mdthemes::md_theme_classic()+ggtitle("*Back leaf area in 1991*")## Cannot use mode = "kenward-roger" because *pbkrtest* package is not installed## NOTE: Results may be misleading due to involvement in interactionsthe main effect of time
#emmeans(b_mod91la.bm, pairwise~Time, type="response")
Time_5 <- emmip(b_mod91la.bm, ~ Time ,CIs = TRUE )+ylab("")+mdthemes::md_theme_classic()## Cannot use mode = "kenward-roger" because *pbkrtest* package is not installed## NOTE: Results may be misleading due to involvement in interactionsthe sever x time interaction
#emmeans(b_mod91la.bm, pairwise~Time|Sever, type="response")
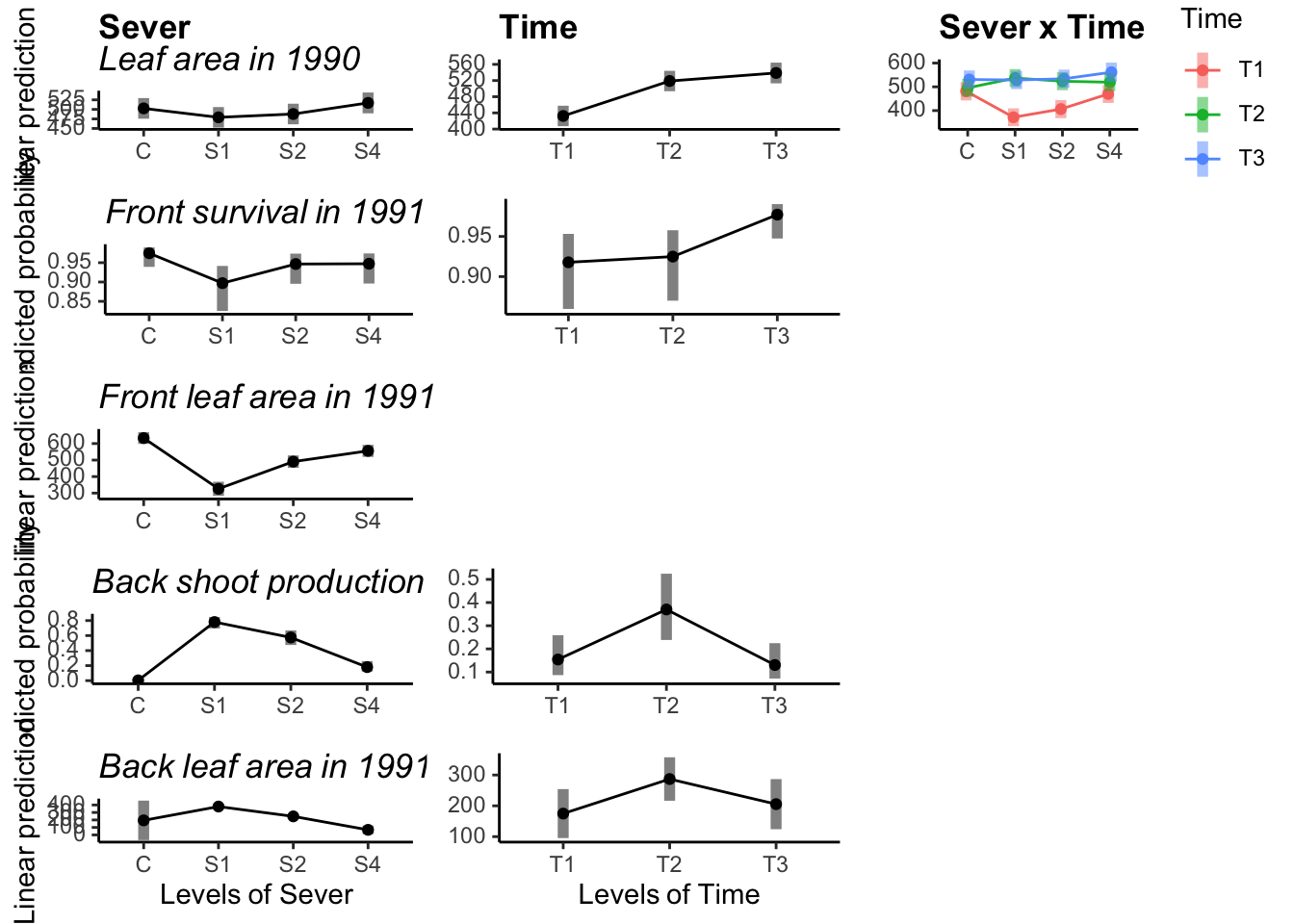
Int_5 <- emmip(b_mod91la.bm, Time ~ Sever ,CIs = TRUE )+ylab("")+mdthemes::md_theme_classic()## Cannot use mode = "kenward-roger" because *pbkrtest* package is not installed10.9 Multipanel figure
library("cowplot")
ggdraw() +
draw_plot(Sever_1, x=0, y=0.8, width=0.33, height=0.2)+
draw_plot(Sever_2, x=0, y=0.6,width=0.33, height=0.2)+
draw_plot(Sever_3, x=0, y=0.4,width=0.33, height=0.2)+
draw_plot(Sever_4, x=0, y=0.2,width=0.33, height=0.2)+
draw_plot(Sever_5, x=0, y=0.0,width=0.33, height=0.2)+
draw_plot(Time_1, x=0.33, y=0.8, width=0.33, height=0.2)+
draw_plot(Time_2, x=0.33, y=0.6,width=0.33, height=0.2)+
draw_plot(Time_3, x=0.33, y=0.4,width=0.33, height=0.2)+
draw_plot(Time_4, x=0.33, y=0.2,width=0.33, height=0.2)+
draw_plot(Time_5, x=0.33, y=0.0,width=0.33, height=0.2)+
draw_plot(Int_1, x=0.67, y=0.8, width=0.33, height=0.2)+
draw_plot(Int_5, x=0.67, y=0.0, width=0.33, height=0.2)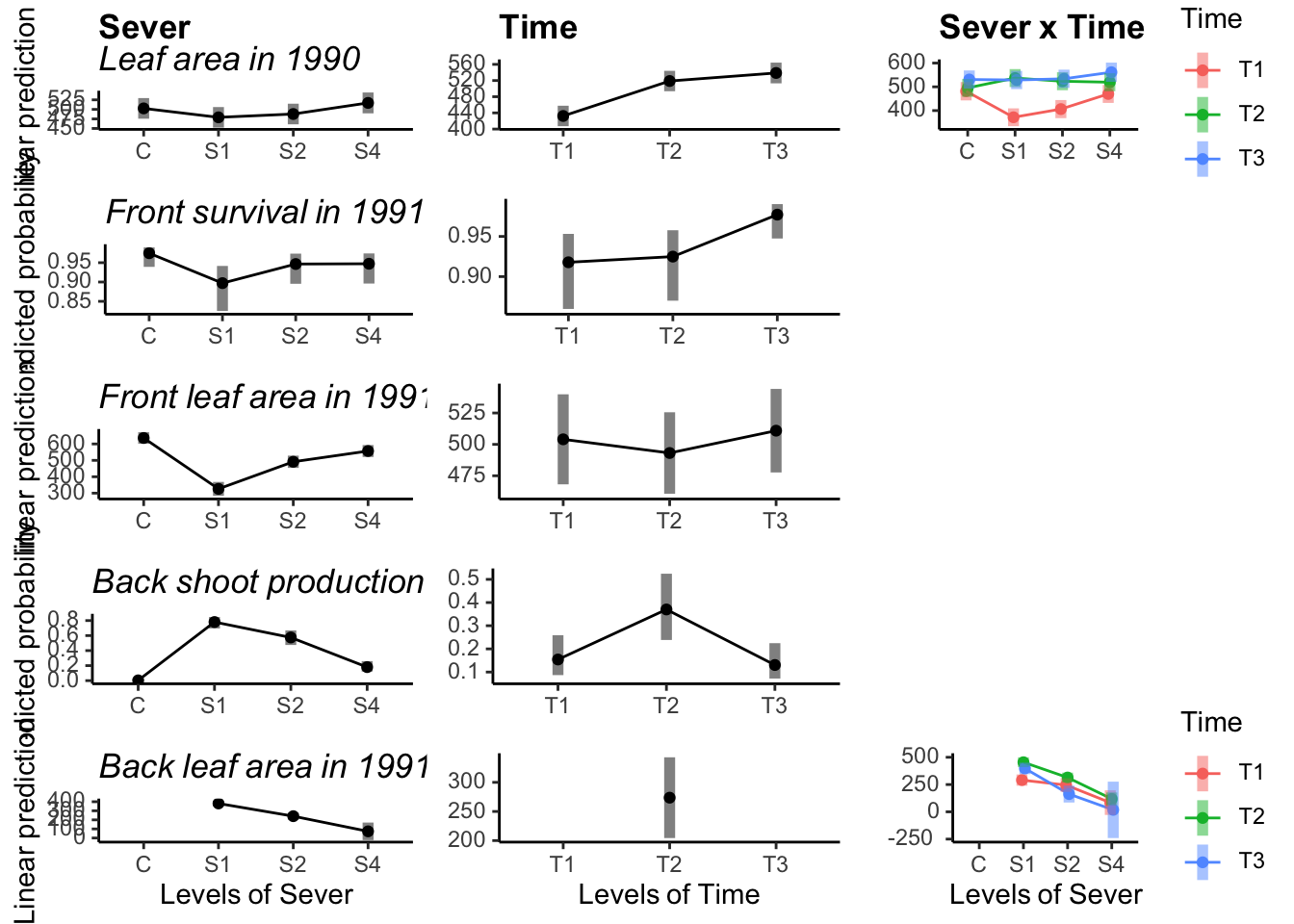
#For conditional leaf area:
ggdraw() +
draw_plot(Sever_1, x=0, y=0.8, width=0.33, height=0.2)+
draw_plot(Sever_2, x=0, y=0.6,width=0.33, height=0.2)+
draw_plot(Sever_3cond, x=0, y=0.4,width=0.33, height=0.2)+
draw_plot(Sever_4, x=0, y=0.2,width=0.33, height=0.2)+
draw_plot(Sever_5cond, x=0, y=0.0,width=0.33, height=0.2)+
draw_plot(Time_1, x=0.33, y=0.8, width=0.33, height=0.2)+
draw_plot(Time_2, x=0.33, y=0.6,width=0.33, height=0.2)+
draw_plot(Time_4, x=0.33, y=0.2,width=0.33, height=0.2)+
draw_plot(Time_5cond, x=0.33, y=0.0,width=0.33, height=0.2)+
draw_plot(Int_1, x=0.67, y=0.8, width=0.33, height=0.2)