7 1992 back system analyses
7.1 Leaf area
This analysis uses all systems that have a non-NA value for total leaf area of the front and back in 1991 and in 1992, excluding those systems for which 1990 leaf area was NA.
7.1.1 Data & visualization
Read in data
# set na.strings="." to recode '.' to NA
mayappleDatasetFull<-readxl::read_excel("Datasets/mayapple_1990_updated_version2.xlsx", na=".")Select the total leaf area in front in 1992 variable, and the independent variables. Also filtering out rows that don’t have a leaf area record in 1990. We need to do this in order to be able to compare models from model set A (which don’t include leaf area, and so wouldn’t naturally be affected by the missing data) with models from model set B (which do include leaf area and would be affected by the missing data). In other words, removing these rows means that model set A and model set B will run using the exact same dataset, which is necessary in order for us to compare the models.
# select variables for analysis and filter out rows that don't have a leaf area recorded in 1990
mayappleData <- mayappleDatasetFull %>%
dplyr::select(COLONY,Lf_90,Sex_90,Sever,Time,LftotB_92, Lftot_91, LfBtot_91, LfFtot_91) %>%
mutate(LftotB_92=parse_double(LftotB_92), Lftot_91=parse_double(Lftot_91), LfBtot_91=parse_double(LfBtot_91), LfFtot_91=parse_double(LfFtot_91) ) %>%
filter(!is.na(Lf_90)) %>%
filter(!is.na(Lftot_91)) %>%
filter(!is.na(LftotB_92))
ggplot(data=mayappleData, aes(x=Sever, y=LftotB_92, fill = Time)) +
stat_summary(fun = mean, geom = "bar",position="dodge") +
stat_summary(fun.data = mean_se, geom = "errorbar", position="dodge")+
facet_wrap(~Sex_90) +
ylab("Mean total leaf area \n in back shoots in 1992") +
scale_fill_brewer(palette="Set1")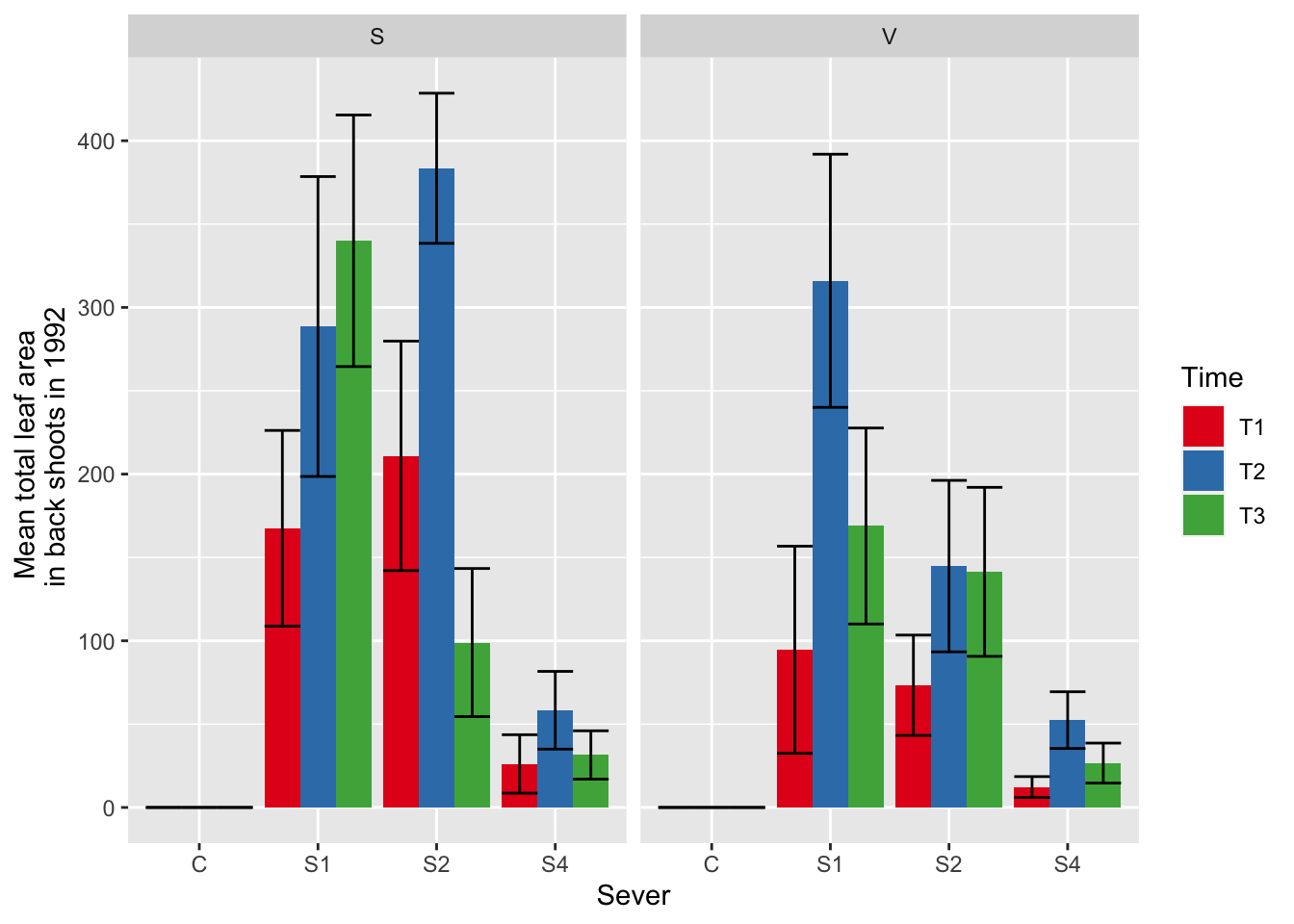
7.1.2 Model selection with leaf area in 1990 and 1991
d_mod92la.0l<-lmer(LftotB_92~Sex_90*Sever*Time +LfBtot_91+Lf_90+(1|COLONY), data = mayappleData, na.action ="na.fail",REML=FALSE)
#this is the global model
#MuMIn::getAllTerms(d_mod92la.0l) #this lets us see how to specify terms in the model
dr<-dredge(d_mod92la.0l, fixed=~Lf_90+LfBtot_91)
tab <- head(dr, n=10) %>% gt() %>%
fmt_number(columns = vars(`(Intercept)`, Lf_90, LfBtot_91, logLik, AICc, delta, weight), decimals = 2) %>%
tab_spanner(label = "Fit information", columns = vars(df, logLik, AICc, delta, weight)) %>%
tab_spanner(label = "Non-fixed parameters", columns = vars(`(Intercept)`, Lf_90, LfBtot_91, Sever, Sex_90, Time, `Sever:Sex_90`, `Sever:Time`, `Sex_90:Time`, `Sever:Sex_90:Time`)) %>%
tab_spanner(label = "Fixed parameters", columns = vars(`(Intercept)`,Lf_90, LfBtot_91)) %>%
tab_style(style = cell_fill(color = "lightgrey"), locations = cells_body(columns = everything(),rows = delta <= 2)) %>%
tab_style(style = cell_fill(color = "darkgrey"), locations = cells_body( columns = everything(), rows = 1))
tab %>% gtsave("back_92_a.png", vwidth=1200) 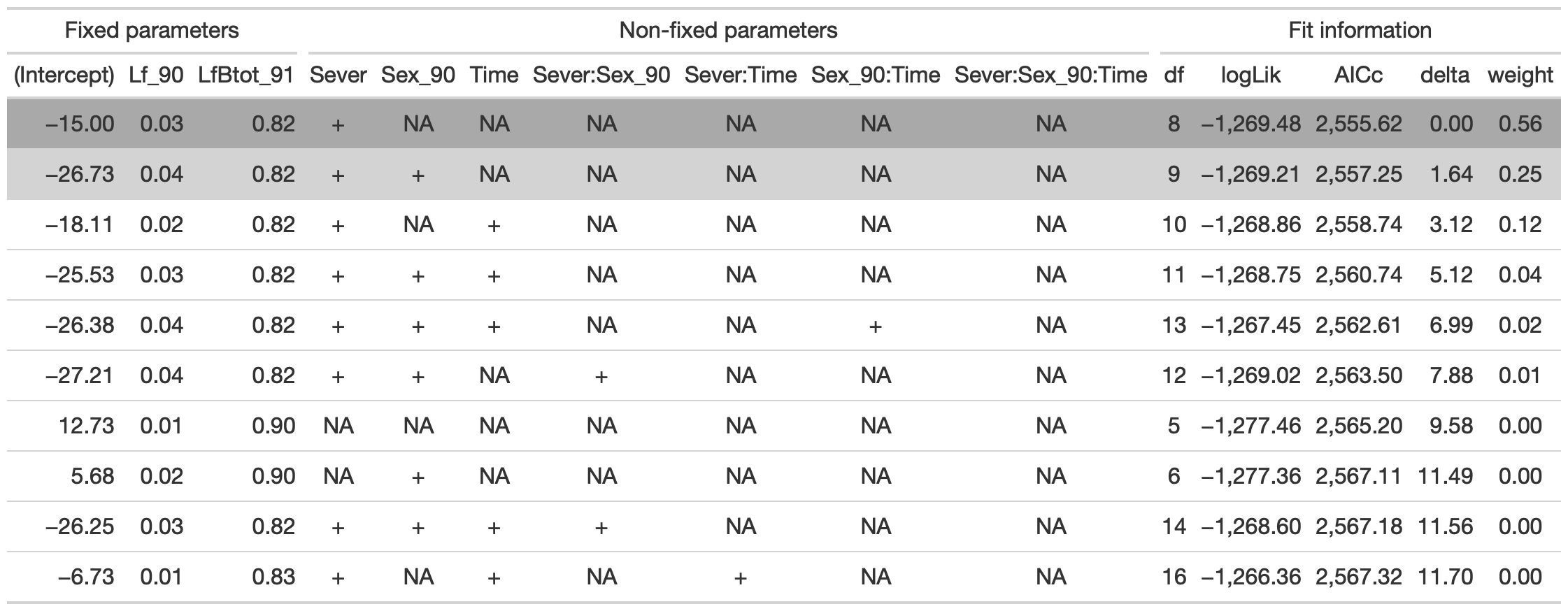
From this round of model selection, when Lf_90 and LfBtot_91 are required to be in the models that are considered, there are two models within two AICc units: (1) Lf_90, LfBtot91, and Sever (AICc 2555.6) (2) Lf_90, LFBtot91, Sever, and Sex_90 (AICc 2557.3)
7.1.3 Model selection with leaf area in 1991
dr<-dredge(d_mod92la.0l, fixed=~LfBtot_91)
tab <- head(dr, n=10) %>% gt() %>%
fmt_number(columns = vars(`(Intercept)`, Lf_90, LfBtot_91, logLik, AICc, delta, weight), decimals = 2) %>%
tab_spanner(label = "Fit information", columns = vars(df, logLik, AICc, delta, weight)) %>%
tab_spanner(label = "Non-fixed parameters", columns = vars(`(Intercept)`, Lf_90, LfBtot_91, Sever, Sex_90, Time, `Sever:Sex_90`, `Sever:Time`, `Sex_90:Time`, `Sever:Sex_90:Time`)) %>%
tab_spanner(label = "Fixed parameters", columns = vars(`(Intercept)`, LfBtot_91)) %>%
tab_style(style = cell_fill(color = "lightgrey"), locations = cells_body(columns = everything(),rows = delta <= 2)) %>%
tab_style(style = cell_fill(color = "darkgrey"), locations = cells_body( columns = everything(), rows = 1))
tab %>% gtsave("back_92_b.png", vwidth=1200) 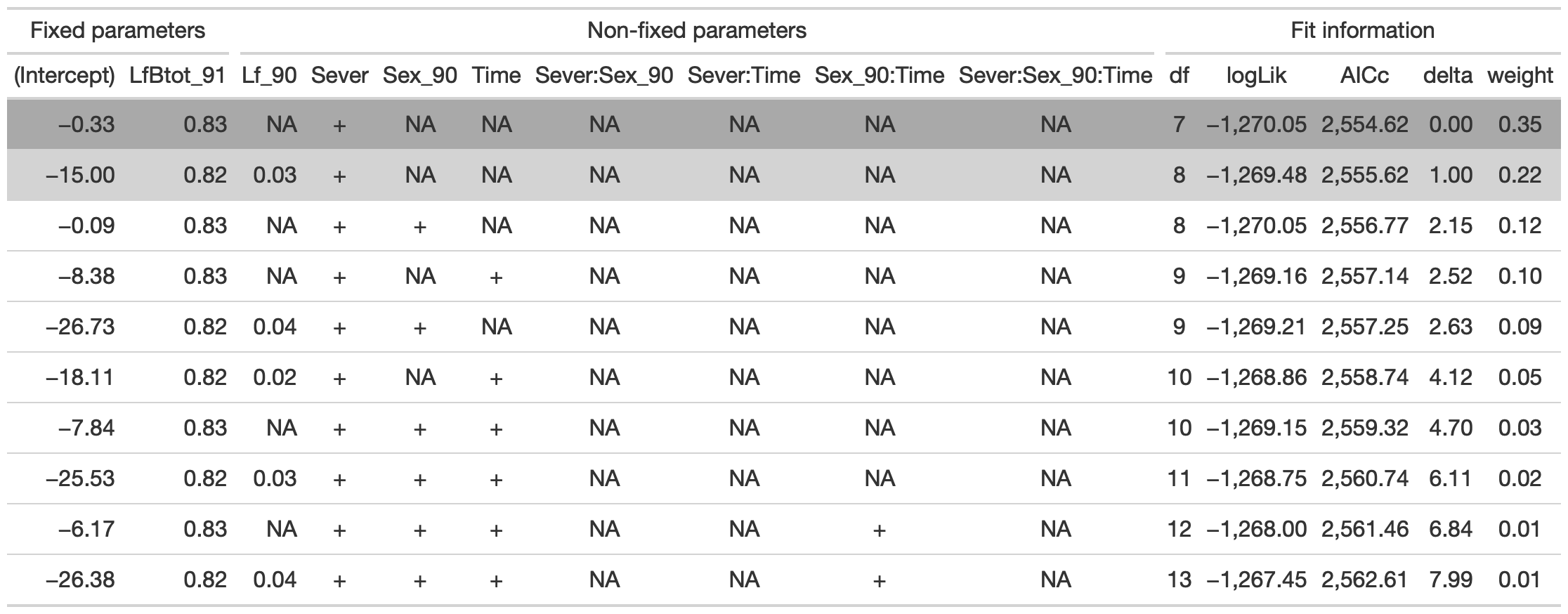
From this round of model selection, when LfFtot_91 only is required to be in the models that are considered, there are two models within two AICc units: (1) LfBtot_91 and Sever (AICc 2554.6) (2) Lf_90, LfBtot_91, and Sever (AICc 2555.6)
7.1.4 Model selection with leaf area in 1990
dr<-dredge(d_mod92la.0l, fixed=~Lf_90)
tab <- head(dr, n=10) %>% gt() %>%
fmt_number(columns = vars(`(Intercept)`, Lf_90, LfBtot_91, logLik, AICc, delta, weight), decimals = 2) %>%
tab_spanner(label = "Fit information", columns = vars(df, logLik, AICc, delta, weight)) %>%
tab_spanner(label = "Non-fixed parameters", columns = vars(`(Intercept)`, Lf_90, LfBtot_91, Sever, Sex_90, Time, `Sever:Sex_90`, `Sever:Time`, `Sex_90:Time`, `Sever:Sex_90:Time`)) %>%
tab_spanner(label = "Fixed parameters", columns = vars(`(Intercept)`, Lf_90)) %>%
tab_style(style = cell_fill(color = "lightgrey"), locations = cells_body(columns = everything(),rows = delta <= 2)) %>%
tab_style(style = cell_fill(color = "darkgrey"), locations = cells_body( columns = everything(), rows = 1))
tab %>% gtsave("back_92_c.png", vwidth=1200)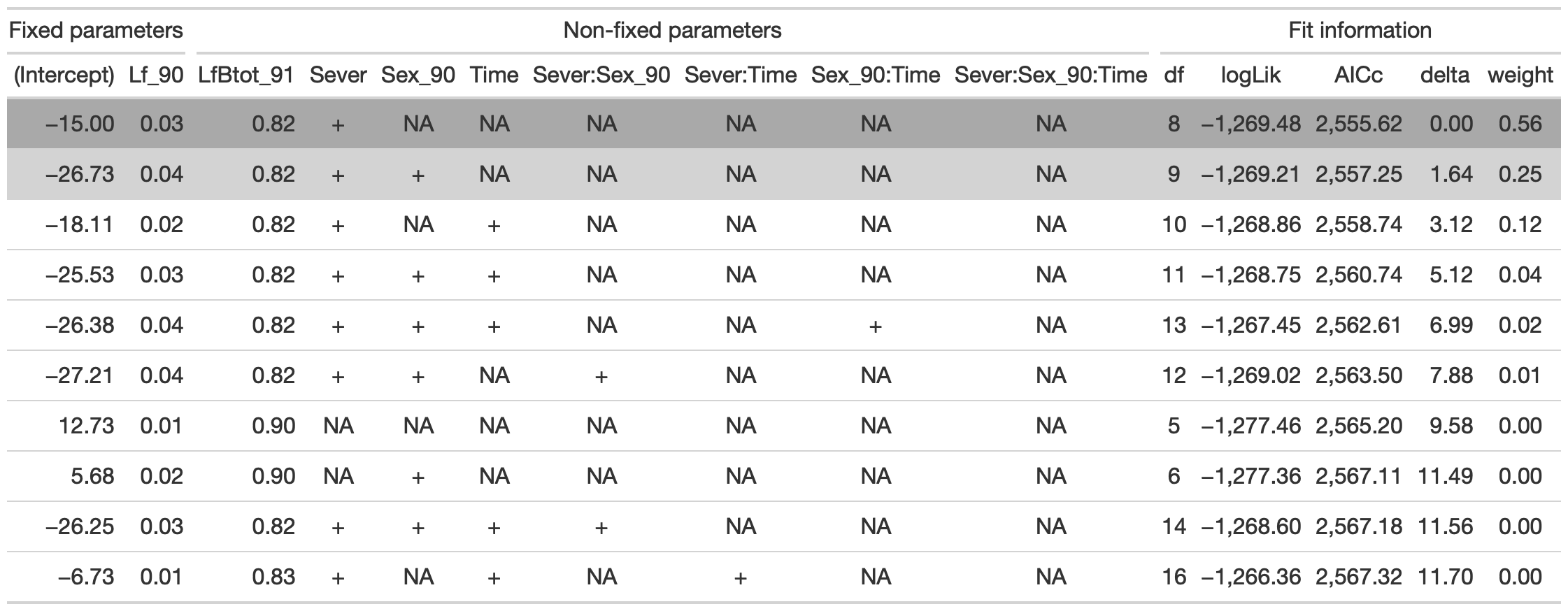
From this round of model selection, when Lf_90 is required to be in the models that are considered, there are two models within two AICc units (the same two as when Lf_90 and LfBtot_91 were both required): (1) Lf_90, LfBtot91, and Sever (AICc 2555.6) (2) Lf_90, LFBtot91, Sever, and Sex_90 (AICc 2557.3)
7.1.5 Model selection without leaf area
## Fixed term is "(Intercept)"tab <- head(dr, n=10) %>% gt() %>%
fmt_number(columns = vars(`(Intercept)`, Lf_90, LfBtot_91, logLik, AICc, delta, weight), decimals = 2) %>%
tab_spanner(label = "Fit information", columns = vars(df, logLik, AICc, delta, weight)) %>%
tab_spanner(label = "Non-fixed parameters", columns = vars(`(Intercept)`, Lf_90, LfBtot_91, Sever, Sex_90, Time, `Sever:Sex_90`, `Sever:Time`, `Sex_90:Time`, `Sever:Sex_90:Time`)) %>%
tab_spanner(label = "Fixed parameter", columns = vars(`(Intercept)`)) %>%
tab_style(style = cell_fill(color = "lightgrey"), locations = cells_body(columns = everything(),rows = delta <= 2)) %>%
tab_style(style = cell_fill(color = "darkgrey"), locations = cells_body( columns = everything(), rows = 1))
tab %>% gtsave("back_92_d.png", vwidth=1200)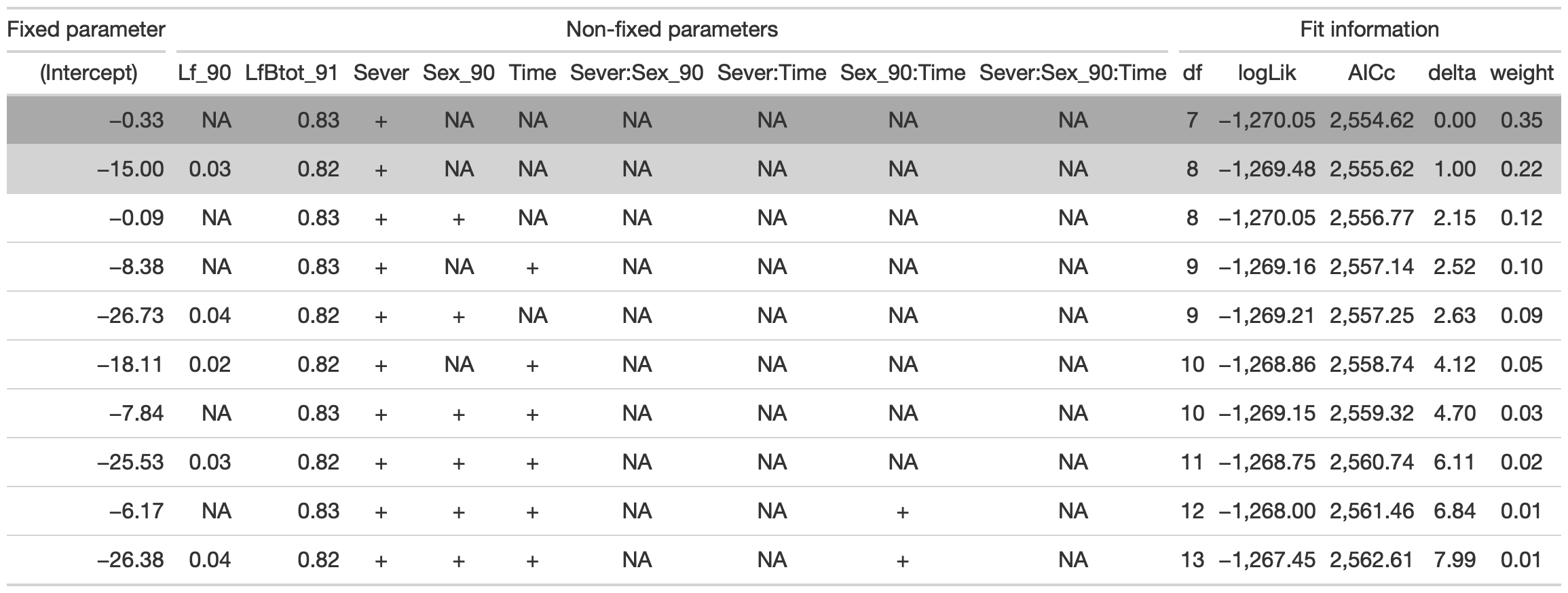
From this round of model selection, when the leaf area effects are not required to be in the models that are considered, there are two models within two AICc units (the same as when only LfBtot_91 was required): (1) LfBtot_91 and Sever (AICc 2554.6) (2) Lf_90, LfBtot_91, and Sever (AICc 2555.6)
7.1.6 Conclusion
The best model has main effects of LfBtot_91 and Sever.
7.1.7 Estimated marginal means
the main effect of sever
g1 <- ggplot(data=mayappleData, aes(x=Sever, y=LftotB_92)) +
geom_bar(stat="summary", fun.y = "mean", position = "dodge") +
ylab("Mean of total leaf area \n for branches in back of plant in 1992")
emmeans(d_mod92la.bm, pairwise~Sever, type="response")## $emmeans
## Sever emmean SE df lower.CL upper.CL
## C 74.4 9.24 91.9 56.1 92.8
## S1 118.1 11.22 136.5 95.9 140.3
## S2 124.3 10.21 120.4 104.1 144.5
## S4 88.9 9.28 94.7 70.5 107.3
##
## Degrees-of-freedom method: satterthwaite
## Confidence level used: 0.95
##
## $contrasts
## contrast estimate SE df t.ratio p.value
## C - S1 -43.66 14.8 222 -2.957 0.0180
## C - S2 -49.87 13.5 218 -3.689 0.0016
## C - S4 -14.48 11.7 216 -1.243 0.6004
## S1 - S2 -6.21 13.6 222 -0.455 0.9685
## S1 - S4 29.17 14.6 223 2.005 0.1892
## S2 - S4 35.39 13.4 217 2.642 0.0435
##
## Degrees-of-freedom method: satterthwaite
## P value adjustment: tukey method for comparing a family of 4 estimates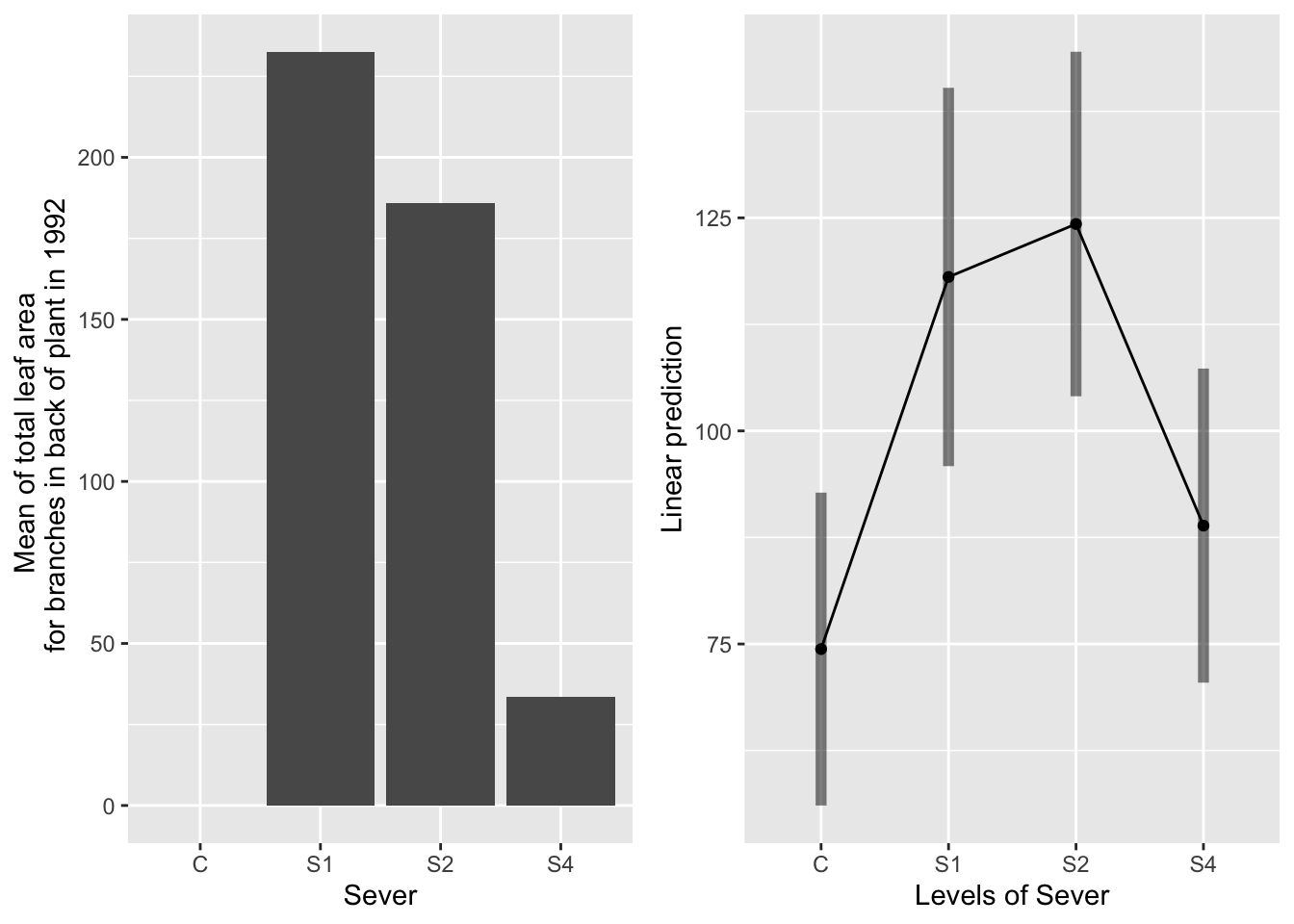
S2 systems have the most leaf area in back in 1992, significantly more than control and S4 systems. S1 systems have marginally more leaf area than control systems.
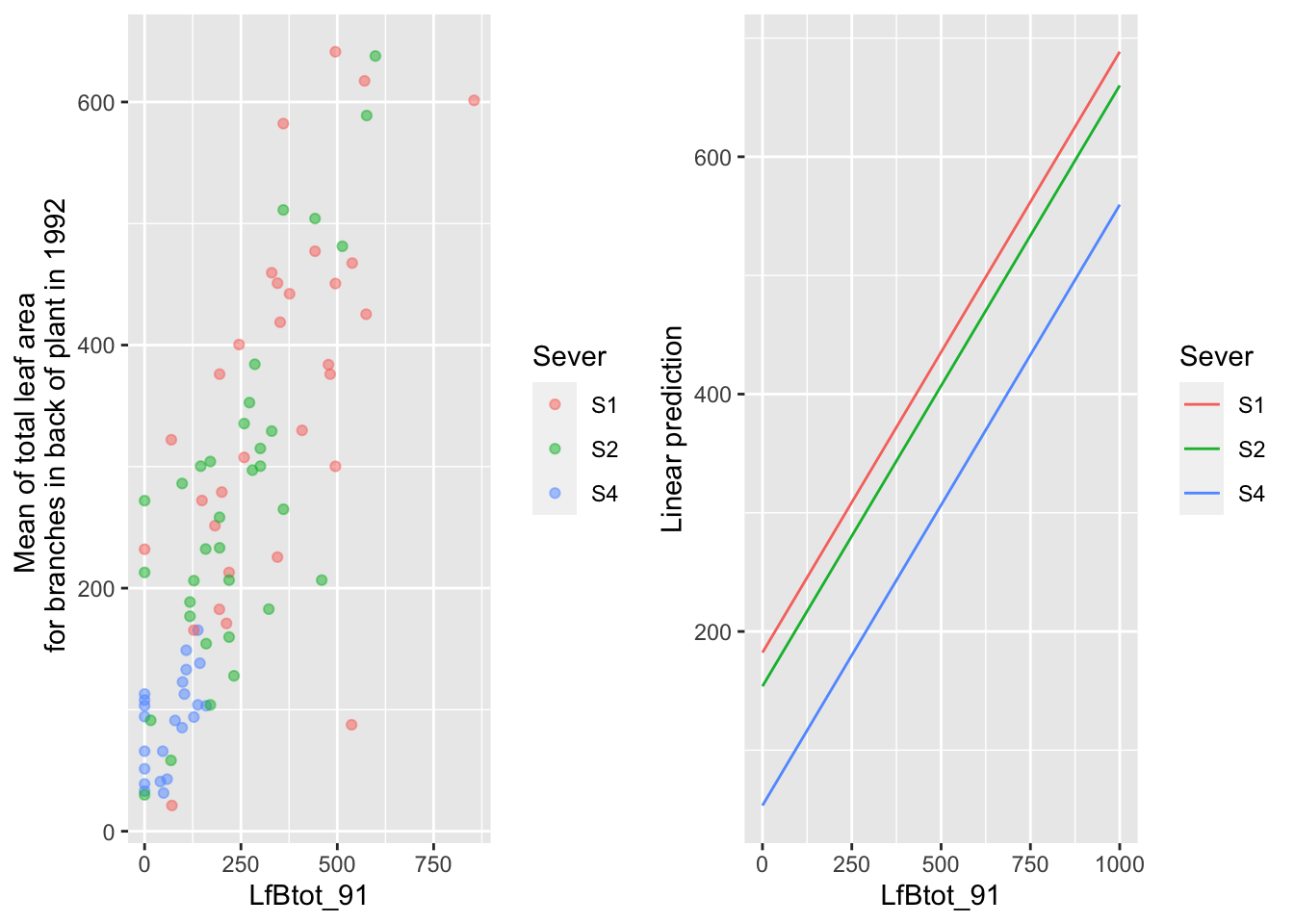
the main effect of back leaf area in 1991
mylist <- list(LfBtot_91=seq(0,1000,by=100))
g1 <- ggplot(data=mayappleData, aes(x=LfBtot_91, y=LftotB_92, color=Sever)) +
geom_point(alpha=0.5) +
ylab("Mean of total leaf area \n for branches in back of plant in 1992")
g2 <- emmip(d_mod92la.bm, Sever~ LfBtot_91 , at=mylist)
gridExtra::grid.arrange(g1,g2,nrow=1)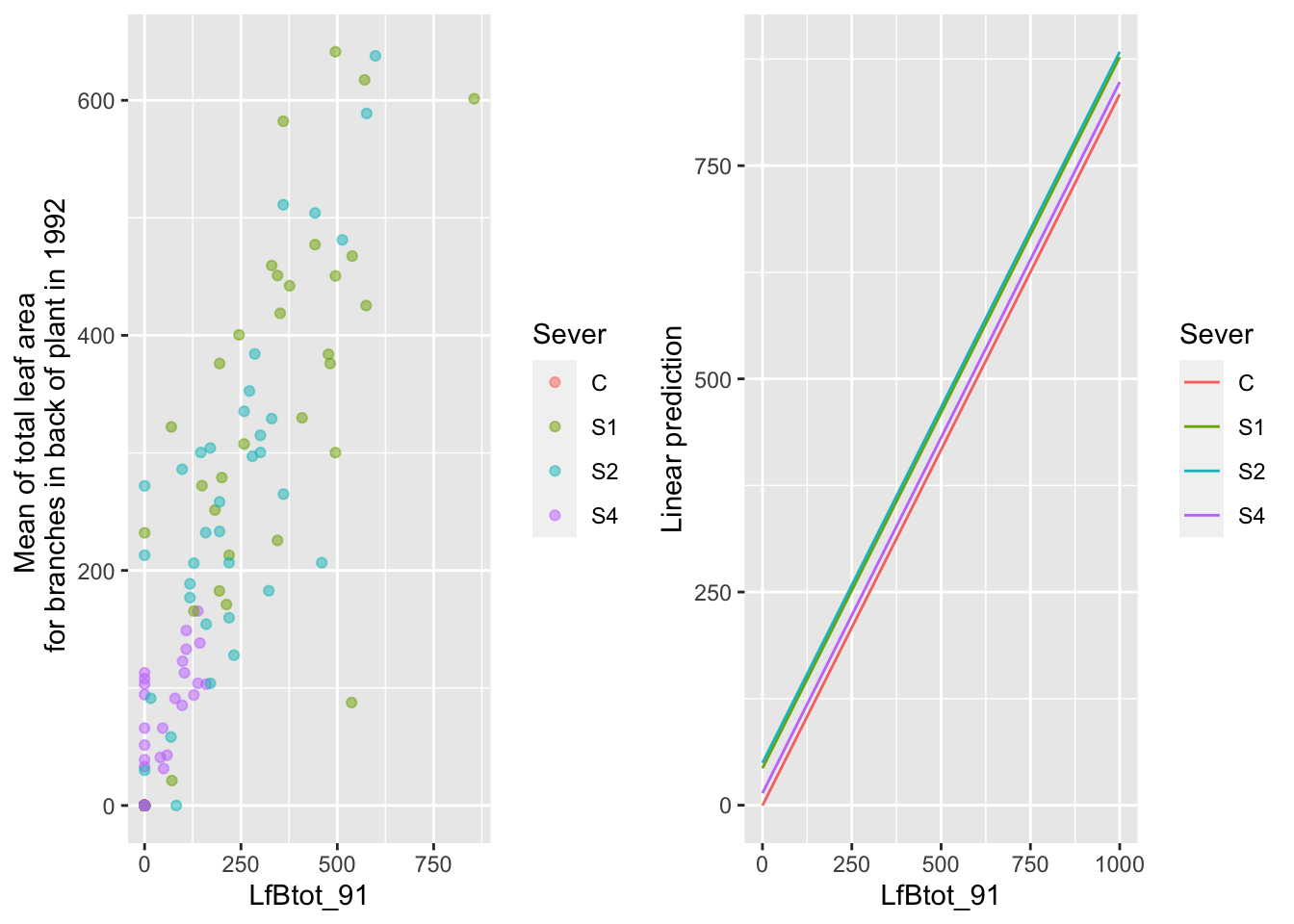
Across all of the severing treatments, there are strong positive relationships with leaf area in 1991.
7.2 Conditional leaf area
This analysis uses all systems that have a non-NA value for total leaf area of the front and back in 1991 and in 1992, excluding those systems for which 1990 leaf area was NA. Furthermore, this analysis excludes any systems with a zero leaf area in 1992.
7.2.1 Data & visualization
Read in data
# set na.strings="." to recode '.' to NA
mayappleDatasetFull<-readxl::read_excel("Datasets/mayapple_1990_updated_version2.xlsx", na=".")Select the total leaf area in front in 1992 variable, and the independent variables. Also filtering out rows that don’t have a leaf area record in 1990. We need to do this in order to be able to compare models from model set A (which don’t include leaf area, and so wouldn’t naturally be affected by the missing data) with models from model set B (which do include leaf area and would be affected by the missing data). In other words, removing these rows means that model set A and model set B will run using the exact same dataset, which is necessary in order for us to compare the models.
# select variables for analysis and filter out rows that don't have a leaf area recorded in 1990
mayappleData <- mayappleDatasetFull %>%
dplyr::select(COLONY,Lf_90,Sex_90,Sever,Time,LftotB_92, Lftot_91, LfBtot_91, LfFtot_91) %>%
mutate(LftotB_92=parse_double(LftotB_92), Lftot_91=parse_double(Lftot_91), LfBtot_91=parse_double(LfBtot_91), LfFtot_91=parse_double(LfFtot_91) ) %>%
filter(!is.na(Lf_90)) %>%
filter(!is.na(Lftot_91)) %>%
filter(!is.na(LftotB_92)) %>%
filter(LftotB_92>0)
ggplot(data=mayappleData, aes(x=Sever, y=LftotB_92, fill = Time)) +
stat_summary(fun = mean, geom = "bar",position="dodge") +
stat_summary(fun.data = mean_se, geom = "errorbar", position="dodge")+
facet_wrap(~Sex_90) +
ylab("Mean total leaf area \n in back shoots in 1992") +
scale_fill_brewer(palette="Set1")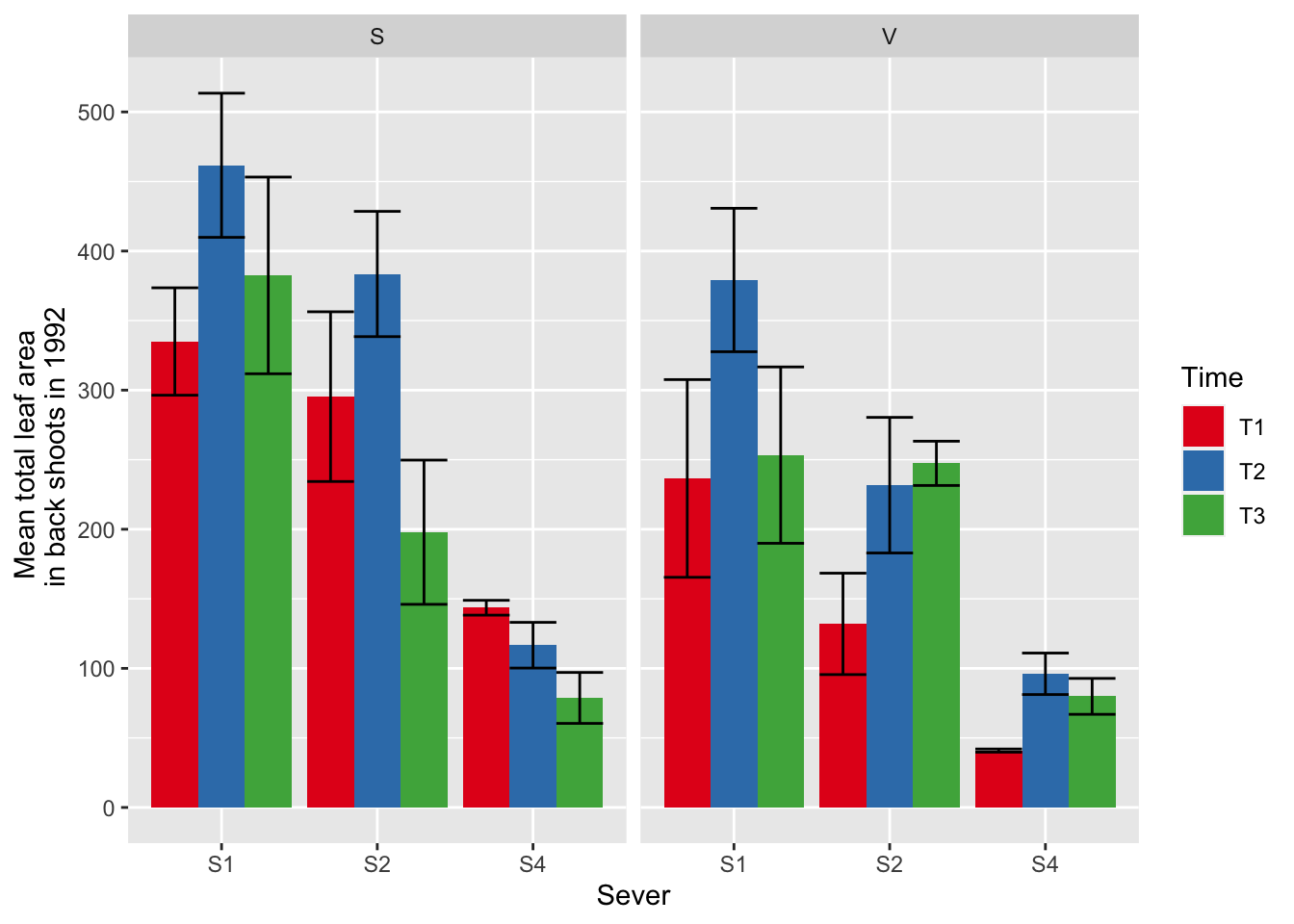
7.2.2 Model selection with leaf area in 1990 and 1991
d_mod92la.0l<-lmer(LftotB_92~Sex_90*Sever*Time +LfBtot_91+Lf_90+(1|COLONY), data = mayappleData, na.action ="na.fail",REML=FALSE)
#this is the global model
#MuMIn::getAllTerms(d_mod92la.0l) #this lets us see how to specify terms in the model
dr<-dredge(d_mod92la.0l)
tab <- head(dr, n=10) %>% gt() %>%
fmt_number(columns = vars(`(Intercept)`, Lf_90, LfBtot_91, logLik, AICc, delta, weight), decimals = 2) %>%
tab_spanner(label = "Fit information", columns = vars(df, logLik, AICc, delta, weight)) %>%
tab_spanner(label = "Non-fixed parameters", columns = vars(`(Intercept)`, Lf_90, LfBtot_91, Sever, Sex_90, Time, `Sever:Sex_90`, `Sever:Time`, `Sex_90:Time`, `Sever:Sex_90:Time`)) %>%
tab_spanner(label = "Fixed parameters", columns = vars(`(Intercept)`,Lf_90, LfBtot_91)) %>%
tab_style(style = cell_fill(color = "lightgrey"), locations = cells_body(columns = everything(),rows = delta <= 2)) %>%
tab_style(style = cell_fill(color = "darkgrey"), locations = cells_body( columns = everything(), rows = 1))
tab %>% gtsave("back_92_conditional.png", vwidth=1200) 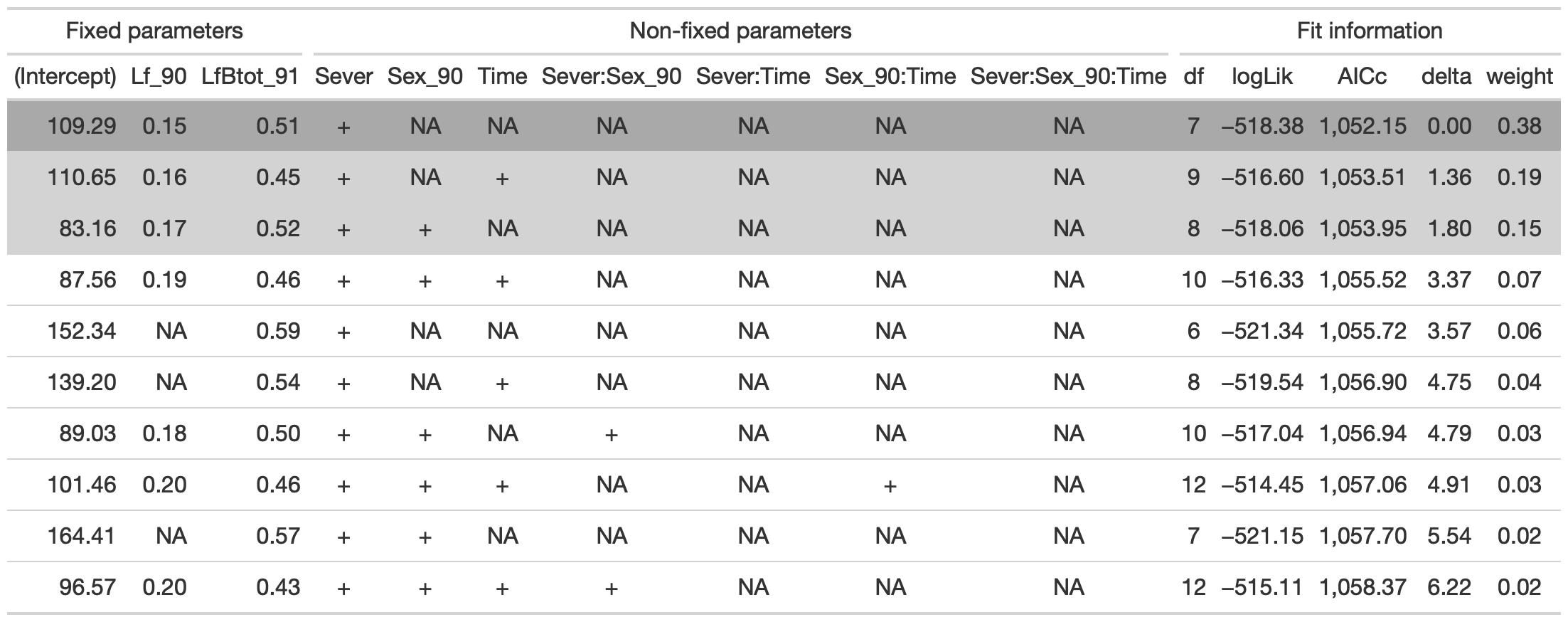
The best model contains fixed effects of Lf_90, leaf area in 1991, and Sever.
7.2.3 Conclusion
the main effect of sever
g1 <- ggplot(data=mayappleData, aes(x=Sever, y=LftotB_92)) +
geom_bar(stat="summary", fun.y = "mean", position = "dodge") +
ylab("Mean of total leaf area \n for branches in back of plant in 1992")
emmeans(d_mod92la.bm, pairwise~Sever, type="response")## $emmeans
## Sever emmean SE df lower.CL upper.CL
## S1 298 20.2 43.8 258 339
## S2 270 17.4 33.0 234 305
## S4 169 24.2 56.6 121 218
##
## Degrees-of-freedom method: satterthwaite
## Confidence level used: 0.95
##
## $contrasts
## contrast estimate SE df t.ratio p.value
## S1 - S2 28.4 22.5 82.8 1.260 0.4218
## S1 - S4 128.9 31.0 84.2 4.162 0.0002
## S2 - S4 100.5 26.1 84.5 3.853 0.0007
##
## Degrees-of-freedom method: satterthwaite
## P value adjustment: tukey method for comparing a family of 3 estimates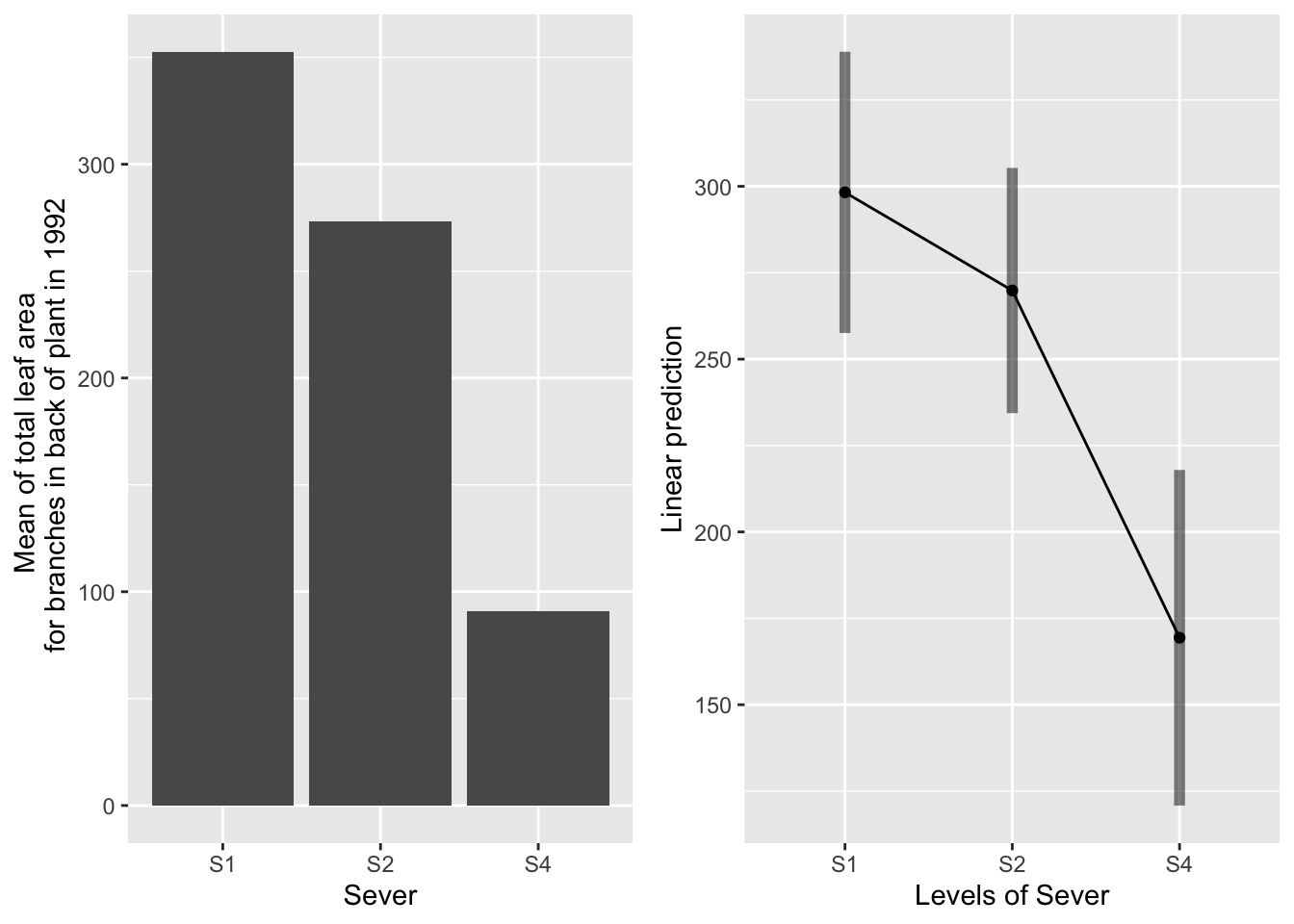
S2 systems have the most leaf area in back in 1992, significantly more than control and S4 systems. S1 systems have marginally more leaf area than control systems.
the main effect of back leaf area in 1990
mylist <- list(Lf_90=seq(0,700,by=100))
g1 <- ggplot(data=mayappleData, aes(x=Lf_90, y=LftotB_92, color=Sever)) +
geom_point(alpha=0.5) +
ylab("Mean of total leaf area \n for branches in back of plant in 1992")
g2 <- emmip(d_mod92la.bm, Sever~ Lf_90 , at=mylist)
gridExtra::grid.arrange(g1,g2,nrow=1)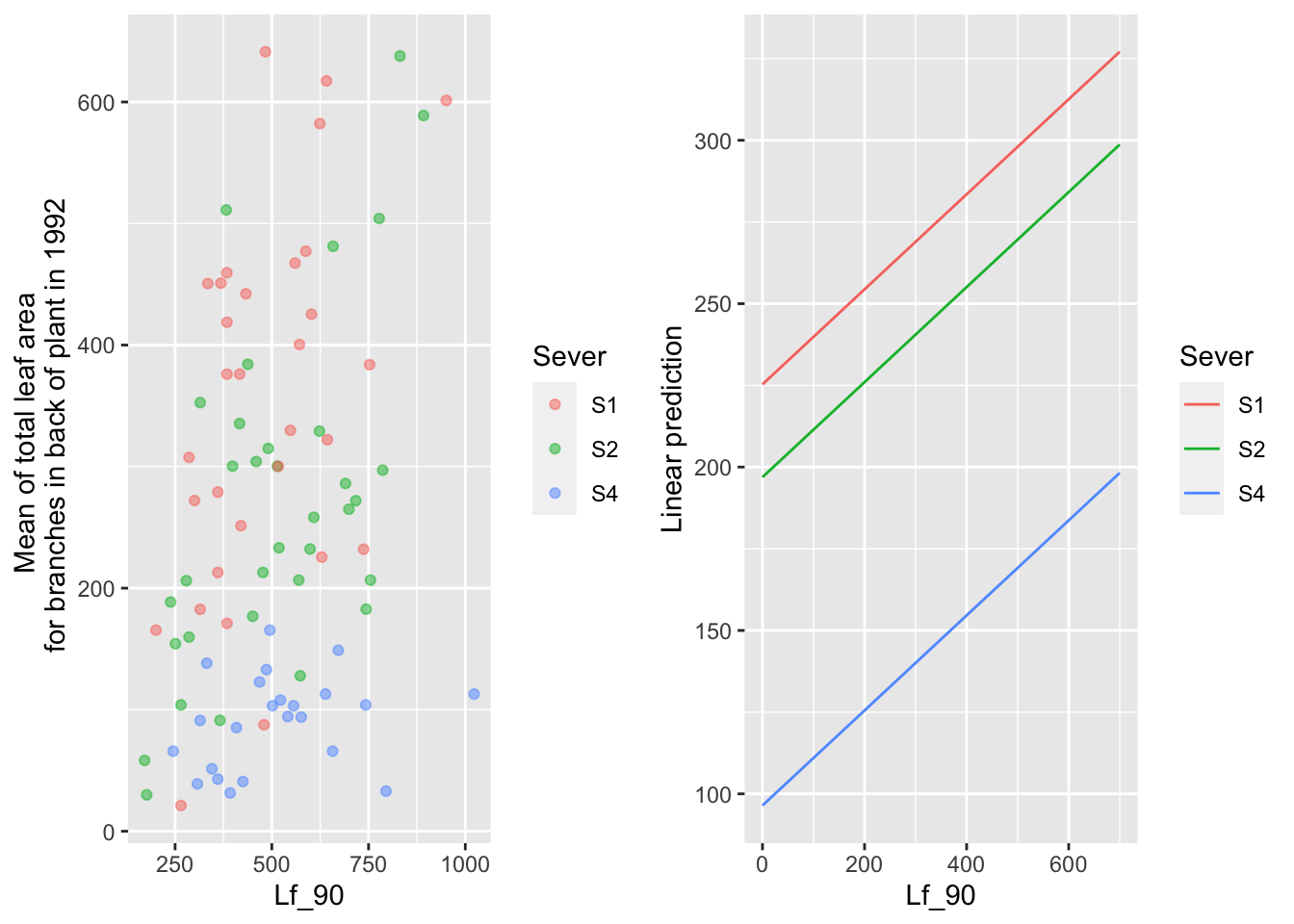
the main effect of back leaf area in 1991
mylist <- list(LfBtot_91=seq(0,1000,by=100))
g1 <- ggplot(data=mayappleData, aes(x=LfBtot_91, y=LftotB_92, color=Sever)) +
geom_point(alpha=0.5) +
ylab("Mean of total leaf area \n for branches in back of plant in 1992")
g2 <- emmip(d_mod92la.bm, Sever~ LfBtot_91 , at=mylist)
gridExtra::grid.arrange(g1,g2,nrow=1)