2 The tidyverse
2.1 Connect to SQLite database
library('tidyverse')
#> -- Attaching packages --------------------------------------- tidyverse 1.3.1 --
#> v ggplot2 3.3.3 v purrr 0.3.4
#> v tibble 3.1.2 v dplyr 1.0.6
#> v tidyr 1.1.3 v stringr 1.4.0
#> v readr 1.4.0 v forcats 0.5.1
#> -- Conflicts ------------------------------------------ tidyverse_conflicts() --
#> x dplyr::filter() masks stats::filter()
#> x dplyr::lag() masks stats::lag()
NHANES_DB <- src_sqlite(
"C:/Users/jkyle/Documents/GitHub/Jeff_Data_Mgt_R_PRV/DATA/sql_db/NHANES_DB.db",
create = FALSE)
#> Warning: `src_sqlite()` was deprecated in dplyr 1.0.0.
#> Please use `tbl()` directly with a database connectionWe see we get a warning so we try to oblige:
ERROR <- tbl("C:/Users/jkyle/Documents/GitHub/Jeff_Data_Mgt_R_PRV/DATA/sql_db/NHANES_DB.db")
#> Error in UseMethod("tbl"): no applicable method for 'tbl' applied to an object of class "character"We post error to GitHub: https://github.com/tidyverse/dplyr/issues/5817 but we can still continue to perform work:
2.2 List of tables
src_tbls(NHANES_DB)
#> [1] "ACQ" "ALB_CR" "ALQ" "ALQY" "AUQ"
#> [6] "BIOPRO" "BMX" "BPQ" "BPX" "CBC"
#> [11] "CDQ" "CMV" "COT" "CRCO" "DBQ"
#> [16] "DEMO" "DEQ" "DIQ" "DLQ" "DPQ"
#> [21] "DR1IFF" "DR1TOT" "DR2IFF" "DR2TOT" "DRXFCD"
#> [26] "DS1IDS" "DS1TOT" "DS2TOT" "DSBI" "DSII"
#> [31] "DSQIDS" "DSQTOT" "DUQ" "DXX" "DXXFEM"
#> [36] "DXXSPN" "Ds2IDS" "ECQ" "FASTQX" "FERTIN"
#> [41] "FETIB" "FOLATE" "FOLFMS" "GHB" "GLU"
#> [46] "HDL" "HEPA" "HEPBD" "HEPC" "HEPE"
#> [51] "HEQ" "HIQ" "HIV" "HOQ" "HSCRP"
#> [56] "HSQ" "HSV" "HUQ" "IHGEM" "IMQ"
#> [61] "INS" "KIQ" "LUX" "MCQ" "METADATA"
#> [66] "OCQ" "OHQ" "OHXDEN" "OHXREF" "OSQ"
#> [71] "PAQ" "PAQY" "PBCD" "PFAS" "PFQ"
#> [76] "PUQMEC" "RHQ" "RXQASA" "RXQ_DRUG" "RXQ_RX"
#> [81] "SLQ" "SMQ" "SMQFAM" "SMQRTU" "SMQSHS"
#> [86] "SSPFAS" "SXQ" "TCHOL" "TFR" "TRIGLY"
#> [91] "UCFLOW" "UCM" "UCPREG" "UHG" "UIO"
#> [96] "UNI" "VIC" "VOCWB" "VTQ" "sqlite_stat1"
#> [101] "sqlite_stat4"
NHANES_Tables <- src_tbls(NHANES_DB) [! src_tbls(NHANES_DB) %in% c("sqlite_stat1" , 'sqlite_stat4', 'METADATA') ]
NHANES_Tables
#> [1] "ACQ" "ALB_CR" "ALQ" "ALQY" "AUQ" "BIOPRO"
#> [7] "BMX" "BPQ" "BPX" "CBC" "CDQ" "CMV"
#> [13] "COT" "CRCO" "DBQ" "DEMO" "DEQ" "DIQ"
#> [19] "DLQ" "DPQ" "DR1IFF" "DR1TOT" "DR2IFF" "DR2TOT"
#> [25] "DRXFCD" "DS1IDS" "DS1TOT" "DS2TOT" "DSBI" "DSII"
#> [31] "DSQIDS" "DSQTOT" "DUQ" "DXX" "DXXFEM" "DXXSPN"
#> [37] "Ds2IDS" "ECQ" "FASTQX" "FERTIN" "FETIB" "FOLATE"
#> [43] "FOLFMS" "GHB" "GLU" "HDL" "HEPA" "HEPBD"
#> [49] "HEPC" "HEPE" "HEQ" "HIQ" "HIV" "HOQ"
#> [55] "HSCRP" "HSQ" "HSV" "HUQ" "IHGEM" "IMQ"
#> [61] "INS" "KIQ" "LUX" "MCQ" "OCQ" "OHQ"
#> [67] "OHXDEN" "OHXREF" "OSQ" "PAQ" "PAQY" "PBCD"
#> [73] "PFAS" "PFQ" "PUQMEC" "RHQ" "RXQASA" "RXQ_DRUG"
#> [79] "RXQ_RX" "SLQ" "SMQ" "SMQFAM" "SMQRTU" "SMQSHS"
#> [85] "SSPFAS" "SXQ" "TCHOL" "TFR" "TRIGLY" "UCFLOW"
#> [91] "UCM" "UCPREG" "UHG" "UIO" "UNI" "VIC"
#> [97] "VOCWB" "VTQ"2.3 Demographics Table
DEMO = tbl(NHANES_DB, "DEMO")Let’s test the glimpse function with the pipe %>% operator:
# this is the same as
# glimpse(DEMO)
DEMO %>%
glimpse()
#> Rows: ??
#> Columns: 175
#> Database: sqlite 3.35.5 [C:\Users\jkyle\Documents\GitHub\Jeff_Data_Mgt_R_PRV\DATA\sql_db\NHANES_DB.db]
#> $ SEQN <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18~
#> $ SDDSRVYR <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1~
#> $ RIDSTATR <dbl> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2~
#> $ RIDEXMON <dbl> 2, 2, 1, 2, 2, 2, 2, 1, 2, 2, 2, 1, 2, 1, 1, 1, 2, 1, 1, 2, 2~
#> $ RIAGENDR <dbl> 2, 1, 2, 1, 1, 2, 2, 1, 2, 1, 1, 1, 1, 1, 2, 2, 1, 2, 1, 2, 1~
#> $ RIDAGEYR <dbl> 2, 77, 10, 1, 49, 19, 59, 13, 11, 43, 15, 37, 70, 81, 38, 85,~
#> $ RIDAGEMN <dbl> 29, 926, 125, 22, 597, 230, 712, 159, 133, 518, 183, 453, 850~
#> $ RIDAGEEX <dbl> 31, 926, 126, 23, 597, 230, 712, 160, 133, 518, 184, 454, 850~
#> $ RIDRETH1 <dbl> 4, 3, 3, 4, 3, 5, 4, 3, 4, 4, 3, 3, 1, 3, 3, 4, 4, 3, 1, 1, 1~
#> $ RIDRETH2 <dbl> 2, 1, 1, 2, 1, 4, 2, 1, 2, 2, 1, 1, 3, 1, 1, 2, 2, 1, 3, 3, 3~
#> $ DMQMILIT <dbl> NA, 1, NA, NA, 1, 2, 2, NA, NA, 1, NA, 1, 2, 1, 2, 2, NA, NA,~
#> $ DMDBORN <dbl> 1, 1, 3, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1~
#> $ DMDCITZN <dbl> 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1~
#> $ DMDYRSUS <dbl> NA, NA, 2, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA~
#> $ DMDEDUC3 <dbl> NA, NA, 3, NA, NA, 15, NA, 5, 5, NA, 8, NA, NA, NA, NA, NA, N~
#> $ DMDEDUC2 <dbl> NA, 5, NA, NA, 5, NA, 2, NA, NA, 3, NA, 4, 1, 2, 5, 1, NA, NA~
#> $ DMDEDUC <dbl> NA, 3, 1, NA, 3, 3, 1, 1, 1, 2, 1, 3, 1, 1, 3, 1, NA, NA, NA,~
#> $ DMDSCHOL <dbl> NA, NA, 1, NA, NA, 1, NA, 1, 1, NA, 1, NA, NA, NA, NA, NA, NA~
#> $ DMDMARTL <dbl> NA, NA, NA, NA, 1, 5, 1, NA, NA, 4, 5, 5, 1, 3, 3, 2, NA, NA,~
#> $ DMDHHSIZ <dbl> 3, 1, 4, 7, 3, 2, 1, 7, 4, 1, 5, 4, 2, 1, 2, 1, 5, 4, 4, 2, 6~
#> $ INDHHINC <dbl> 3, 8, 6, 3, 11, 4, NA, 4, NA, NA, 6, 11, 3, 5, 8, 1, 4, 8, 7,~
#> $ INDFMINC <dbl> 3, 8, 6, 3, 11, 3, NA, 3, 99, 99, 6, 11, 3, 5, 8, 1, 4, 8, 7,~
#> $ INDFMPIR <dbl> 0.86, 5.00, 1.47, 0.57, 5.00, 1.21, NA, 0.53, NA, NA, 1.25, 4~
#> $ RIDEXPRG <dbl> NA, NA, NA, NA, NA, 2, 2, NA, NA, NA, NA, NA, NA, NA, 2, NA, ~
#> $ RIDPREG <dbl> NA, NA, NA, NA, NA, 2, 2, NA, NA, NA, NA, NA, NA, NA, 2, NA, ~
#> $ DMDHRGND <dbl> 2, 1, 1, 2, 2, 2, 2, 2, 2, 1, 1, 2, 2, 1, 2, 2, 2, 1, 1, 1, 1~
#> $ DMDHRAGE <dbl> 27, 77, 37, 34, 42, 19, 59, 30, 37, 43, 50, 32, 75, 81, 38, 8~
#> $ DMDHRBRN <dbl> 1, 1, 3, 1, 3, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1~
#> $ DMDHREDU <dbl> 3, 5, 4, 3, 4, 4, 2, 1, 2, 3, 3, 2, 1, 2, 5, 1, 2, 5, 3, 2, 1~
#> $ DMDHRMAR <dbl> 5, NA, 1, 4, 1, 5, 1, 3, 4, 4, 1, 1, NA, 3, 3, 2, 1, NA, NA, ~
#> $ DMDHSEDU <dbl> NA, NA, 3, NA, NA, NA, NA, NA, NA, NA, 3, 5, NA, NA, NA, NA, ~
#> $ WTINT2YR <dbl> 9727.079, 26678.636, 43621.681, 10346.119, 91050.847, 36508.2~
#> $ WTINT4YR <dbl> 4291.490, 14203.336, 20123.764, 4582.132, 44161.868, 16850.97~
#> $ WTMEC2YR <dbl> 10982.899, 28325.385, 46192.257, 10251.260, 99445.066, 39656.~
#> $ WTMEC4YR <dbl> 4456.207, 15336.200, 21258.467, 4562.389, 45985.968, 18337.31~
#> $ SDMVPSU <dbl> 1, 3, 2, 1, 2, 2, 2, 1, 2, 1, 2, 2, 2, 1, 2, 1, 1, 1, 2, 2, 2~
#> $ SDMVSTRA <dbl> 5, 1, 7, 2, 8, 2, 4, 6, 9, 7, 1, 6, 13, 12, 11, 11, 5, 8, 13,~
#> $ SDJ1REPN <dbl> 19, 36, 30, 47, 41, 7, 13, 9, 2, 5, 15, 39, 34, 28, 38, 17, 2~
#> $ DMAETHN <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMARACE <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ WTMREP01 <dbl> 11045.789, 28595.040, 46829.651, 10301.751, 100344.689, 40567~
#> $ WTMREP02 <dbl> 11537.655, 28487.810, 46309.918, 10693.774, 100213.183, 39838~
#> $ WTMREP03 <dbl> 11052.382, 28277.212, 46191.688, 10316.455, 99530.738, 39656.~
#> $ WTMREP04 <dbl> 10981.995, 28428.863, 46603.586, 10250.416, 100036.558, 39900~
#> $ WTMREP05 <dbl> 11304.028, 28689.510, 46638.380, 10765.240, 100789.825, 40055~
#> $ WTMREP06 <dbl> 11788.404, 28554.443, 47073.298, 10631.594, 101061.235, 41273~
#> $ WTMREP07 <dbl> 10982.144, 28953.832, 47186.987, 10250.555, 101883.027, 0.000~
#> $ WTMREP08 <dbl> 11882.033, 28865.193, 46720.914, 10846.616, 101117.975, 40140~
#> $ WTMREP09 <dbl> 11082.382, 28816.149, 47371.667, 10389.726, 100869.040, 40415~
#> $ WTMREP10 <dbl> 10910.758, 29009.335, 46703.109, 10419.682, 101768.139, 40206~
#> $ WTMREP11 <dbl> 11152.686, 29511.004, 48675.069, 10410.644, 102611.942, 40768~
#> $ WTMREP12 <dbl> 11368.379, 28799.451, 47567.231, 10470.139, 101829.652, 39675~
#> $ WTMREP13 <dbl> 11639.066, 28520.262, 47042.789, 10780.564, 100098.756, 40563~
#> $ WTMREP14 <dbl> 10982.086, 28824.622, 47095.644, 10250.501, 102463.971, 41015~
#> $ WTMREP15 <dbl> 10968.688, 29838.479, 49158.400, 10237.996, 105667.423, 41974~
#> $ WTMREP16 <dbl> 11014.937, 29014.725, 48733.459, 10266.450, 103689.537, 41730~
#> $ WTMREP17 <dbl> 11786.119, 28886.146, 46662.025, 10776.922, 101531.848, 40415~
#> $ WTMREP18 <dbl> 11158.423, 28570.071, 47012.402, 10385.185, 100611.626, 40666~
#> $ WTMREP19 <dbl> 0.000, 29175.483, 47088.485, 10373.025, 101703.619, 39790.591~
#> $ WTMREP20 <dbl> 10982.854, 28406.076, 46842.111, 10251.218, 99669.792, 40145.~
#> $ WTMREP21 <dbl> 11079.764, 28540.729, 46711.955, 10444.979, 101050.558, 40218~
#> $ WTMREP22 <dbl> 10981.332, 28382.113, 46191.733, 10249.797, 99652.447, 39656.~
#> $ WTMREP23 <dbl> 10997.244, 28669.314, 47000.059, 10264.650, 100733.782, 40387~
#> $ WTMREP24 <dbl> 10981.440, 29011.197, 46411.984, 10249.898, 100808.842, 39920~
#> $ WTMREP25 <dbl> 10979.467, 28374.504, 46191.178, 10248.057, 99407.847, 39655.~
#> $ WTMREP26 <dbl> 11608.248, 28662.460, 46430.750, 10803.698, 100746.356, 39850~
#> $ WTMREP27 <dbl> 10987.996, 29861.915, 48456.319, 10241.340, 103704.536, 41785~
#> $ WTMREP28 <dbl> 11347.582, 28771.558, 47537.523, 10440.221, 102490.776, 41173~
#> $ WTMREP29 <dbl> 11354.100, 28798.756, 46390.532, 10574.667, 100457.203, 38299~
#> $ WTMREP30 <dbl> 11168.792, 28876.999, 0.000, 10402.873, 101960.985, 40481.733~
#> $ WTMREP31 <dbl> 10988.949, 28493.224, 46265.549, 10201.964, 100093.124, 39722~
#> $ WTMREP32 <dbl> 11108.697, 29903.412, 48571.404, 10343.808, 104829.440, 42857~
#> $ WTMREP33 <dbl> 11057.813, 29153.225, 46904.271, 10524.956, 100117.849, 40429~
#> $ WTMREP34 <dbl> 11120.884, 28503.965, 46411.338, 10345.463, 99981.654, 39816.~
#> $ WTMREP35 <dbl> 11885.457, 28443.983, 47430.196, 11236.715, 102828.393, 40514~
#> $ WTMREP36 <dbl> 11007.347, 0.000, 46740.889, 10274.079, 101294.751, 40098.944~
#> $ WTMREP37 <dbl> 10819.875, 28467.041, 46191.711, 10288.781, 99809.107, 39656.~
#> $ WTMREP38 <dbl> 11179.917, 29037.401, 46798.279, 10454.650, 101633.388, 40121~
#> $ WTMREP39 <dbl> 11031.809, 29042.380, 46940.618, 10263.960, 101718.690, 40274~
#> $ WTMREP40 <dbl> 11002.862, 28435.895, 46353.765, 10276.953, 100105.211, 39792~
#> $ WTMREP41 <dbl> 11475.124, 29110.361, 47676.541, 10670.986, 0.000, 41084.442,~
#> $ WTMREP42 <dbl> 10937.054, 29373.665, 47629.579, 10279.077, 103833.940, 40088~
#> $ WTMREP43 <dbl> 11431.549, 28924.957, 46579.640, 10601.255, 102254.002, 40223~
#> $ WTMREP44 <dbl> 11197.627, 29016.386, 46707.762, 10425.580, 99399.906, 40081.~
#> $ WTMREP45 <dbl> 10976.247, 29874.144, 49637.450, 10245.051, 104410.645, 42429~
#> $ WTMREP46 <dbl> 11372.168, 29559.796, 47746.235, 10542.918, 103032.836, 41344~
#> $ WTMREP47 <dbl> 11323.564, 29874.486, 46727.779, 0.000, 101665.126, 40993.482~
#> $ WTMREP48 <dbl> 11145.176, 28364.510, 46686.213, 10431.038, 99740.420, 40051.~
#> $ WTMREP49 <dbl> 10990.797, 28525.592, 47504.119, 10530.152, 100050.072, 40798~
#> $ WTMREP50 <dbl> 11728.074, 28965.388, 47206.144, 10753.406, 101733.346, 40328~
#> $ WTMREP51 <dbl> 10927.616, 29143.510, 48250.804, 10479.951, 103321.769, 41468~
#> $ WTMREP52 <dbl> 11655.083, 28705.129, 46363.582, 10910.424, 100585.330, 39965~
#> $ WTIREP01 <dbl> 9787.451, 26923.621, 44489.187, 10401.970, 92772.795, 37416.2~
#> $ WTIREP02 <dbl> 10190.660, 26824.718, 43736.673, 10798.969, 91672.817, 36679.~
#> $ WTIREP03 <dbl> 9789.596, 26631.356, 43621.681, 10412.616, 91122.756, 36508.2~
#> $ WTIREP04 <dbl> 9727.079, 26777.075, 43964.278, 10346.119, 91506.263, 36722.0~
#> $ WTIREP05 <dbl> 10129.044, 27018.552, 43956.597, 10868.584, 92345.587, 36793.~
#> $ WTIREP06 <dbl> 10284.424, 26904.615, 44396.201, 10734.330, 92565.814, 37486.~
#> $ WTIREP07 <dbl> 9727.079, 27241.302, 44432.410, 10346.119, 93036.324, 0.000, ~
#> $ WTIREP08 <dbl> 10436.054, 27198.787, 44133.063, 10932.389, 92649.682, 36936.~
#> $ WTIREP09 <dbl> 9816.986, 27099.418, 44684.839, 10487.390, 92223.816, 37217.2~
#> $ WTIREP10 <dbl> 9664.797, 27293.981, 44304.079, 10517.698, 93249.667, 37003.0~
#> $ WTIREP11 <dbl> 9845.871, 27701.659, 45711.849, 10473.304, 93778.678, 37601.6~
#> $ WTIREP12 <dbl> 10011.322, 27220.559, 44886.928, 10568.957, 93052.787, 36558.~
#> $ WTIREP13 <dbl> 10234.820, 26875.742, 44507.450, 10865.804, 91695.619, 37407.~
#> $ WTIREP14 <dbl> 9727.079, 27203.215, 45325.284, 10346.119, 93379.450, 37984.4~
#> $ WTIREP15 <dbl> 9727.079, 28289.914, 46199.091, 10346.119, 96241.873, 38528.7~
#> $ WTIREP16 <dbl> 9757.028, 27504.821, 45798.642, 10363.133, 94316.889, 38316.3~
#> $ WTIREP17 <dbl> 10257.406, 27171.694, 44086.711, 10829.241, 92900.828, 37063.~
#> $ WTIREP18 <dbl> 9884.906, 26894.202, 44397.650, 10483.498, 91968.726, 37349.9~
#> $ WTIREP19 <dbl> 0.000, 27424.680, 44698.518, 10470.417, 92883.436, 36632.100,~
#> $ WTIREP20 <dbl> 9727.079, 26730.099, 44174.268, 10346.119, 91175.971, 36923.6~
#> $ WTIREP21 <dbl> 9789.576, 26885.349, 44171.404, 10526.899, 92439.001, 37074.1~
#> $ WTIREP22 <dbl> 9727.079, 26732.989, 43621.681, 10346.119, 91243.872, 36508.2~
#> $ WTIREP23 <dbl> 9727.079, 26939.627, 44326.007, 10346.119, 92173.287, 37170.2~
#> $ WTIREP24 <dbl> 9727.079, 27208.479, 43816.319, 10346.119, 92329.665, 36643.1~
#> $ WTIREP25 <dbl> 9727.079, 26716.514, 43621.681, 10346.119, 91020.040, 36508.2~
#> $ WTIREP26 <dbl> 10244.119, 26979.278, 43880.912, 10890.470, 92251.677, 36725.~
#> $ WTIREP27 <dbl> 9757.028, 27961.236, 45853.268, 10363.133, 94752.727, 38561.0~
#> $ WTIREP28 <dbl> 9953.666, 27519.241, 44814.552, 10540.663, 93798.390, 37833.4~
#> $ WTIREP29 <dbl> 10026.451, 27120.349, 43885.338, 10677.140, 91963.518, 37058.~
#> $ WTIREP30 <dbl> 9897.641, 27208.404, 0.000, 10518.053, 93141.879, 37229.126, ~
#> $ WTIREP31 <dbl> 9731.354, 26827.816, 43691.069, 10294.999, 91594.731, 36566.3~
#> $ WTIREP32 <dbl> 9836.711, 28153.657, 45952.825, 10437.526, 95832.366, 38651.8~
#> $ WTIREP33 <dbl> 10104.422, 27401.252, 44150.487, 10607.707, 93541.934, 37052.~
#> $ WTIREP34 <dbl> 9835.452, 26823.593, 43800.456, 10438.987, 91488.015, 36642.6~
#> $ WTIREP35 <dbl> 10438.851, 27228.740, 44709.565, 11355.685, 93511.220, 37215.~
#> $ WTIREP36 <dbl> 9749.358, 0.000, 44389.377, 10369.817, 93485.478, 36946.104, ~
#> $ WTIREP37 <dbl> 9796.890, 26814.711, 43621.681, 10385.568, 91393.204, 36508.2~
#> $ WTIREP38 <dbl> 9903.459, 27224.137, 44068.616, 10552.965, 92984.640, 36678.0~
#> $ WTIREP39 <dbl> 9761.917, 27343.860, 44332.639, 10365.898, 93161.374, 37011.8~
#> $ WTIREP40 <dbl> 9745.269, 26786.595, 43767.009, 10372.592, 91575.035, 36629.8~
#> $ WTIREP41 <dbl> 10183.043, 27404.256, 44886.361, 10778.674, 0.000, 37756.206,~
#> $ WTIREP42 <dbl> 9694.540, 27595.650, 45015.721, 10382.678, 94484.113, 37894.3~
#> $ WTIREP43 <dbl> 10094.017, 27186.729, 43993.193, 10702.307, 93164.782, 36948.~
#> $ WTIREP44 <dbl> 9912.462, 27324.345, 44075.386, 10531.444, 92119.609, 36855.2~
#> $ WTIREP45 <dbl> 9727.079, 28099.664, 46642.564, 10346.119, 95388.490, 38998.4~
#> $ WTIREP46 <dbl> 10041.524, 27757.067, 44967.682, 10636.063, 94131.384, 38022.~
#> $ WTIREP47 <dbl> 9953.956, 28049.286, 44572.482, 0.000, 95297.810, 37541.486, ~
#> $ WTIREP48 <dbl> 9857.382, 26716.602, 44087.946, 10533.109, 91325.082, 36871.2~
#> $ WTIREP49 <dbl> 9865.152, 26877.705, 44831.371, 10654.750, 91640.586, 37520.6~
#> $ WTIREP50 <dbl> 10327.993, 27268.025, 44480.987, 10851.024, 92817.927, 37151.~
#> $ WTIREP51 <dbl> 9809.165, 27406.384, 45389.113, 10564.981, 94282.855, 38016.5~
#> $ WTIREP52 <dbl> 10323.316, 26984.813, 43781.906, 11012.530, 91993.251, 36710.~
#> $ yr_range <chr> "1999-2000", "1999-2000", "1999-2000", "1999-2000", "1999-200~
#> $ SIALANG <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ SIAPROXY <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ SIAINTRP <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ FIALANG <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ FIAPROXY <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ FIAINTRP <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ MIALANG <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ MIAPROXY <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ MIAINTRP <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ AIALANG <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDFMSIZ <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDBORN2 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ INDHHIN2 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ INDFMIN2 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDHRBR2 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ RIDRETH3 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ RIDEXAGY <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ RIDEXAGM <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMQMILIZ <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMQADFC <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDBORN4 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ AIALANGA <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDHHSZA <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDHHSZB <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDHHSZE <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDHRBR4 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDHRAGZ <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDHREDZ <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDHRMAZ <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DMDHSEDZ <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~We can use the pipe along with other dplyr functions
DEMO %>%
summarise(cnt = n_distinct(SEQN))
#> # Source: lazy query [?? x 1]
#> # Database: sqlite 3.35.5
#> # [C:\Users\jkyle\Documents\GitHub\Jeff_Data_Mgt_R_PRV\DATA\sql_db\NHANES_DB.db]
#> cnt
#> <int>
#> 1 101316\(~\)
\(~\)
2.4 dplyr vocabulary
| function | purpose | examples |
|---|---|---|
select |
selects or drops columns |
|
mutate |
adds columns | DEMO %>% mutate(Gender = if_else(RIAGENDR == 2 , "Female", "Male")) |
arrange |
orders data |
|
filter |
filters by condition | DEMO %>% filter(RIDAGEYR > 17) |
group_by |
group data in meaningful ways | |
summarise |
compute summary statistics |
|
The pipe %>% operator takes the information on the left and passes it to the information on the right: x %>% f() is equivalent to f(x). You can think of the pipe as saying “next apply”.
DEMO %>%
mutate(Gender = if_else(RIAGENDR == 2 , "Female", "Male")) %>%
filter(RIDAGEYR > 17) %>%
group_by(Gender) %>%
summarise(
mean_age = mean(RIDAGEYR, na.rm = TRUE),
sd_age = sd(RIDAGEYR, na.rm =TRUE)
)
#> # Source: lazy query [?? x 3]
#> # Database: sqlite 3.35.5
#> # [C:\Users\jkyle\Documents\GitHub\Jeff_Data_Mgt_R_PRV\DATA\sql_db\NHANES_DB.db]
#> Gender mean_age sd_age
#> <chr> <dbl> <dbl>
#> 1 Female 47.5 19.5
#> 2 Male 47.9 19.52.5 dplyr %>% SQL
If you are more familiar with SQL then dplyr is writing SQL for you, for instance, if we pipe a dplyr string into the show_query function we can get equivalent SQL for the dplyr pipeline :
DEMO %>%
select(SEQN, RIAGENDR) %>%
mutate(Gender = if_else(RIAGENDR == 2 , "Female", "Male")) %>%
filter(RIDAGEYR > 17) %>%
group_by(Gender) %>%
summarise(
mean_age = mean(RIDAGEYR, na.rm = TRUE),
sd_age = sd(RIDAGEYR, na.rm =TRUE)
) %>%
show_query()
#> <SQL>
#> SELECT `Gender`, AVG(`RIDAGEYR`) AS `mean_age`, STDEV(`RIDAGEYR`) AS `sd_age`
#> FROM (SELECT `SEQN`, `RIAGENDR`, CASE WHEN (`RIAGENDR` = 2.0) THEN ('Female') WHEN NOT(`RIAGENDR` = 2.0) THEN ('Male') END AS `Gender`
#> FROM `DEMO`)
#> WHERE (`RIDAGEYR` > 17.0)
#> GROUP BY `Gender`Another thing to note is that the dplyr pipe and SQL statement are somewhat “inverted”:
dplyr |
SQL |
|---|---|
mutate(Gender = if_else(RIAGENDR == 2 , "Female", "Male")) |
|
filter(RIDAGEYR > 17) |
WHERE ('RIDAGEYR' > 17.0) |
group_by(Gender) |
GROUP BY 'Gender' |
|
AVG('RIDAGEYR') AS 'mean_age', STDEV('RIDAGEYR') AS 'sd_age' |
- dplyr
mutate(Gender = if_else(RIAGENDR == 2 , "Female", "Male"))corresponds withCASE WHEN ('RIAGENDR' = 2.0) THEN ('Female') WHEN NOT('RIAGENDR' = 2.0) THEN ('Male') END AS 'Gender'in SQL which is the inner most sub-query. - dplyr
filter(RIDAGEYR > 17)corresponds withWHERE ('RIDAGEYR' > 17.0)in SQL - dplyr
group_by(Gender)corresponds withGROUP BY 'Gender'in SQL - dplyr
summarise(mean_age = mean(RIDAGEYR, na.rm = TRUE),sd_age = sd(RIDAGEYR, na.rm =TRUE))corresponds withAVG('RIDAGEYR') AS 'mean_age', STDEV('RIDAGEYR') AS 'sd_age'which is within the first line of theSELECTstatement of SQL
\(~\)
\(~\)
2.6 JOINS
Joins are performed on two datasets x and y when there is related information in two tables kept in separate locations. Typically, joins are used to add additional columns and suragate keys are needed to perform the join.
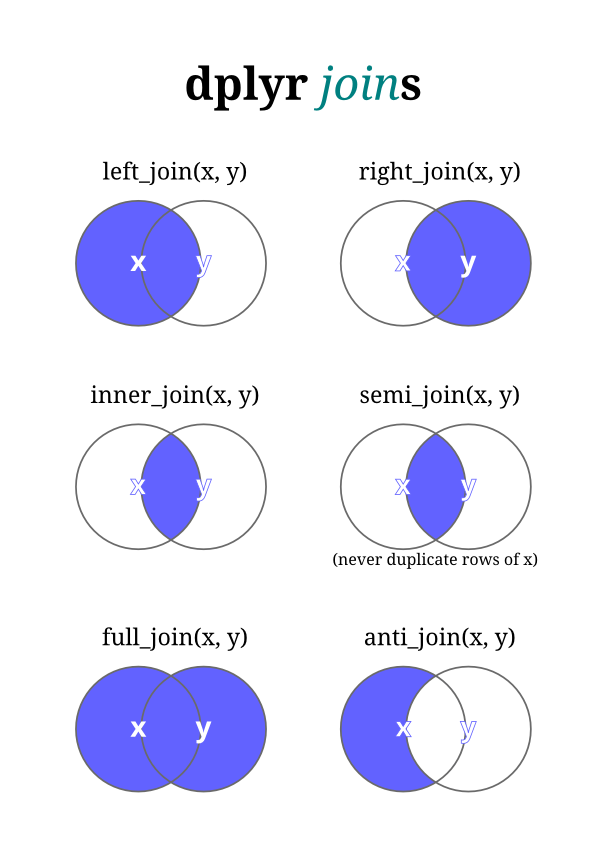
2.6.1 DPLYR JOINS
The types of dplyr join types from https://twitter.com/yutannihilation/status/551572539697143808
\(~\)
\(~\)
2.6.1.1 Quick Examples
Suppose
A <- tibble(num = 1:6,
char = letters[1:6])
A
#> # A tibble: 6 x 2
#> num char
#> <int> <chr>
#> 1 1 a
#> 2 2 b
#> 3 3 c
#> 4 4 d
#> 5 5 e
#> 6 6 f
evens <- seq(from=2, to=6, by =2)
B <- tibble(num = evens,
Upper_char = LETTERS[evens])
B
#> # A tibble: 3 x 2
#> num Upper_char
#> <dbl> <chr>
#> 1 2 B
#> 2 4 D
#> 3 6 FWe will briefly review the different join types and what they do using these two small example datasets, we will assume that num is the suragate key.
2.6.1.1.1 Left Join
Let’s look at
A %>%
left_join(B) %>%
knitr::kable(booktabs = TRUE, caption = 'A lef_join B')
#> Joining, by = "num"| num | char | Upper_char |
|---|---|---|
| 1 | a | NA |
| 2 | b | B |
| 3 | c | NA |
| 4 | d | D |
| 5 | e | NA |
| 6 | f | F |
Note, above that every row in A (our x) is kept while adding the additional column from the B dataset, the columns are “joined” by the surrogate key (num)
Now, compare that to
B %>%
left_join(A)
#> Joining, by = "num"
#> # A tibble: 3 x 3
#> num Upper_char char
#> <dbl> <chr> <chr>
#> 1 2 B b
#> 2 4 D d
#> 3 6 F fAbove, every record in B is kept however, the records in A that do not correspond to those in B by the surrogate key are not because B is our x in this instance.
2.6.1.1.2 Right Join
As another way to get to the same data in @ref(tab:A-left_join-B) would be
B %>%
right_join(A)
#> Joining, by = "num"
#> # A tibble: 6 x 3
#> num Upper_char char
#> <dbl> <chr> <chr>
#> 1 2 B b
#> 2 4 D d
#> 3 6 F f
#> 4 1 <NA> a
#> 5 3 <NA> c
#> 6 5 <NA> eNote the results are out of order and the columns in B come first, however, could fix that with a simple arrange and select statements:
B %>%
right_join(A) %>%
select(num, char, Upper_char) %>%
arrange(num) %>%
knitr::kable(booktabs = TRUE, caption = 'B right_join A reformatted')
#> Joining, by = "num"| num | char | Upper_char |
|---|---|---|
| 1 | a | NA |
| 2 | b | B |
| 3 | c | NA |
| 4 | d | D |
| 5 | e | NA |
| 6 | f | F |
2.6.1.1.3 Inner Join
For inner joins, only records that have a surrogate keys that is in both x and y are maintained, the corresponding columns of y are still appended:
A %>%
inner_join(B)
#> Joining, by = "num"
#> # A tibble: 3 x 3
#> num char Upper_char
#> <dbl> <chr> <chr>
#> 1 2 b B
#> 2 4 d D
#> 3 6 f F2.6.1.1.4 Semi Join
Similar to the inner_join above, the semi_join will keep only the records and columns contained within x so long as there is a corresponding record with a surrogate key in y:
A %>%
semi_join(B)
#> Joining, by = "num"
#> # A tibble: 3 x 2
#> num char
#> <int> <chr>
#> 1 2 b
#> 2 4 d
#> 3 6 f2.6.1.1.5 Anti Join
Anti Joins can be used to find all of the members of x not in y, (again this is done by the surggate key) note that no additional columns are added here, instead the records removed are the ones that are not in y.
A %>%
anti_join(B)
#> Joining, by = "num"
#> # A tibble: 3 x 2
#> num char
#> <int> <chr>
#> 1 1 a
#> 2 3 c
#> 3 5 e2.6.1.1.6 Full Join
Full Joins will all keep all records and columns from both x and y, if there is a matching surrogate key in x and y then the columns of x and y are adjoined by the surrogate key:
A_less_than_3 <- A %>%
filter(num < 3)
A_less_than_3
#> # A tibble: 2 x 2
#> num char
#> <int> <chr>
#> 1 1 a
#> 2 2 b
A_less_than_3 %>%
full_join(B)
#> Joining, by = "num"
#> # A tibble: 4 x 3
#> num char Upper_char
#> <dbl> <chr> <chr>
#> 1 1 a <NA>
#> 2 2 b B
#> 3 4 <NA> D
#> 4 6 <NA> F2.7 Left join Example
We will go over an example of left_join using the DIQ Table.
2.7.1 DIQ Table Example:
https://wwwn.cdc.gov/Nchs/Nhanes/2017-2018/DIQ_J.htm
DIQ = tbl(NHANES_DB, "DIQ")
DIQ %>%
glimpse()
#> Rows: ??
#> Columns: 81
#> Database: sqlite 3.35.5 [C:\Users\jkyle\Documents\GitHub\Jeff_Data_Mgt_R_PRV\DATA\sql_db\NHANES_DB.db]
#> $ SEQN <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18~
#> $ DIQ010 <dbl> 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 1, 2, 2, 2, 2, 2, 2, 2, 2~
#> $ DIQ040G <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 1, NA, NA, NA~
#> $ DIQ040Q <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 67, NA, NA, N~
#> $ DIQ050 <dbl> NA, NA, 2, NA, 2, NA, 2, 2, 2, 2, 2, 2, 2, 2, NA, 2, NA, NA, ~
#> $ DIQ060G <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ060Q <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ060U <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ070 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 1, NA, NA, NA~
#> $ DIQ080 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, 2, NA, NA, NA~
#> $ DIQ090 <dbl> NA, 2, NA, NA, 2, NA, 2, NA, NA, 1, NA, NA, 2, 2, NA, 2, NA, ~
#> $ DIQ100 <dbl> NA, 2, NA, NA, 2, NA, 1, NA, NA, 2, NA, NA, 1, 2, NA, 2, NA, ~
#> $ DIQ110 <dbl> NA, NA, NA, NA, NA, NA, 1, NA, NA, NA, NA, NA, 2, NA, NA, NA,~
#> $ DIQ120 <dbl> NA, 2, NA, NA, 2, NA, 2, NA, NA, 9, NA, NA, 2, 2, NA, 2, NA, ~
#> $ DIQ130 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ140 <dbl> NA, 2, NA, NA, 2, NA, 2, NA, NA, 2, NA, NA, 2, 2, NA, 2, NA, ~
#> $ DIQ150 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ yr_range <chr> "1999-2000", "1999-2000", "1999-2000", "1999-2000", "1999-200~
#> $ DID040G <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID040Q <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID060G <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID060Q <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID040 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ220 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ160 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ170 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ180 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ190A <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ190B <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ190C <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ200A <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ200B <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ200C <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID060 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID070 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ230 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ240 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID250 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID260 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ260U <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID270 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ280 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ290 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ300S <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ300D <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID310S <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID310D <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID320 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID330 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID340 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID350 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ350U <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ360 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DID341 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ172 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175A <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175B <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175C <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175D <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175E <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175F <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175G <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175H <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175I <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175J <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175K <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175L <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175M <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175N <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175O <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175P <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175Q <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175R <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175S <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175T <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175U <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175V <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175W <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ275 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ291 <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~
#> $ DIQ175X <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N~DEMO.DIQ <- DEMO %>%
left_join(DIQ)
#> Joining, by = c("SEQN", "yr_range")One thing to notice in the eaxmples above is that we did not specify by in left_join - R searched both tables for column names and made an assumption. This can be both beneficial and dangerous if users are unfamiliar with the data.
If we wanted to be more explicit or exhibit a different level of control we can join by a specific column:
DEMO.DIQ_by_yr_range <- DEMO %>%
left_join(DIQ, by = c("yr_range"="yr_range"))
DEMO.DIQ_by_yr_range %>%
select(contains('SEQN')) %>%
colnames()
#> [1] "SEQN.x" "SEQN.y"You can see in this case R will produce two columns in the output labeled with .x and .y buy default. At times, it might be advantageous to control that as well, it can be accomplished with the suffix parameter:
DEMO.DIQ_by_yr_range <- DEMO %>%
left_join(DIQ,
by = c("yr_range"="yr_range"),
suffix = c("_DEMO","_DIQ"))
DEMO.DIQ_by_yr_range %>%
select(contains('SEQN')) %>%
colnames()
#> [1] "SEQN_DEMO" "SEQN_DIQ"DEMO.DIQ_by_SEQN <- DEMO %>%
left_join(DIQ,
by = c("SEQN"="SEQN"),
suffix = c("_DEMO","_DIQ"))
DEMO.DIQ_by_SEQN %>%
select(contains('yr_range')) %>%
colnames()
#> [1] "yr_range_DEMO" "yr_range_DIQ"DEMO.DIQ_by_SEQN_yr_range <- DEMO %>%
left_join(DIQ, by=c("SEQN"="SEQN", 'yr_range'='yr_range'))2.7.2 Question
Which of the above would you use for what situation? Why?
\(~\)
\(~\)
2.8 NHANES Table Helpers
2.8.1 NHANES_table_helper
To get started learning how to write your own helper functions you can review this example here. We provide the function NHANES_table_helper with a Table_Name and it will return for us a text url link url_link:
NHANES_table_helper <- function(Table_Name){
url_link <- paste0('https://wwwn.cdc.gov/Nchs/Nhanes/2017-2018/',Table_Name,'_J.htm')
return(url_link)
}2.8.1.1 Test out function:
NHANES_table_helper('DR1TOT')NHANES_table_helper(NHANES_Tables[2])2.8.2 Open_NHANES_table_help
Open_NHANES_table_help <- function(Table_Name){
if(Table_Name %in% NHANES_Tables){
return(browseURL(NHANES_table_helper(Table_Name)))
} else {
ln1 <- paste0("ERROR : ", Table_Name, " is not a valid table name! \n")
ln2 <- 'Please Choose one of : \n'
ln3 <- NHANES_Tables
cat(ln1)
cat(ln2)
cat(ln3)
}
}Open_NHANES_table_help('FROG')
#> ERROR : FROG is not a valid table name!
#> Please Choose one of :
#> ACQ ALB_CR ALQ ALQY AUQ BIOPRO BMX BPQ BPX CBC CDQ CMV COT CRCO DBQ DEMO DEQ DIQ DLQ DPQ DR1IFF DR1TOT DR2IFF DR2TOT DRXFCD DS1IDS DS1TOT DS2TOT DSBI DSII DSQIDS DSQTOT DUQ DXX DXXFEM DXXSPN Ds2IDS ECQ FASTQX FERTIN FETIB FOLATE FOLFMS GHB GLU HDL HEPA HEPBD HEPC HEPE HEQ HIQ HIV HOQ HSCRP HSQ HSV HUQ IHGEM IMQ INS KIQ LUX MCQ OCQ OHQ OHXDEN OHXREF OSQ PAQ PAQY PBCD PFAS PFQ PUQMEC RHQ RXQASA RXQ_DRUG RXQ_RX SLQ SMQ SMQFAM SMQRTU SMQSHS SSPFAS SXQ TCHOL TFR TRIGLY UCFLOW UCM UCPREG UHG UIO UNI VIC VOCWB VTQOpen_NHANES_table_help('DEMO')2.9 (APPENDIX) Code Appendix For Part I
\(~\)
\(~\)
library('tidyverse')
NHANES_DB <- src_sqlite(
"C:/Users/jkyle/Documents/GitHub/Jeff_Data_Mgt_R_PRV/DATA/sql_db/NHANES_DB.db",
create = FALSE)
ERROR <- tbl("C:/Users/jkyle/Documents/GitHub/Jeff_Data_Mgt_R_PRV/DATA/sql_db/NHANES_DB.db")
src_tbls(NHANES_DB)
NHANES_Tables <- src_tbls(NHANES_DB) [! src_tbls(NHANES_DB) %in% c("sqlite_stat1" , 'sqlite_stat4', 'METADATA') ]
NHANES_Tables
DEMO = tbl(NHANES_DB, "DEMO")
# this is the same as
# glimpse(DEMO)
DEMO %>%
glimpse()
DEMO %>%
summarise(cnt = n_distinct(SEQN))
DEMO %>%
mutate(Gender = if_else(RIAGENDR == 2 , "Female", "Male")) %>%
filter(RIDAGEYR > 17) %>%
group_by(Gender) %>%
summarise(
mean_age = mean(RIDAGEYR, na.rm = TRUE),
sd_age = sd(RIDAGEYR, na.rm =TRUE)
)
DEMO %>%
select(SEQN, RIAGENDR) %>%
mutate(Gender = if_else(RIAGENDR == 2 , "Female", "Male")) %>%
filter(RIDAGEYR > 17) %>%
group_by(Gender) %>%
summarise(
mean_age = mean(RIDAGEYR, na.rm = TRUE),
sd_age = sd(RIDAGEYR, na.rm =TRUE)
) %>%
show_query()
A <- tibble(num = 1:6,
char = letters[1:6])
A
evens <- seq(from=2, to=6, by =2)
B <- tibble(num = evens,
Upper_char = LETTERS[evens])
B
A %>%
left_join(B) %>%
knitr::kable(booktabs = TRUE, caption = 'A lef_join B')
B %>%
left_join(A)
B %>%
right_join(A)
B %>%
right_join(A) %>%
select(num, char, Upper_char) %>%
arrange(num) %>%
knitr::kable(booktabs = TRUE, caption = 'B right_join A reformatted')
A %>%
inner_join(B)
A %>%
semi_join(B)
A %>%
anti_join(B)
A_less_than_3 <- A %>%
filter(num < 3)
A_less_than_3
A_less_than_3 %>%
full_join(B)
DIQ = tbl(NHANES_DB, "DIQ")
DIQ %>%
glimpse()
DEMO.DIQ <- DEMO %>%
left_join(DIQ)
DEMO.DIQ_by_yr_range <- DEMO %>%
left_join(DIQ, by = c("yr_range"="yr_range"))
DEMO.DIQ_by_yr_range %>%
select(contains('SEQN')) %>%
colnames()
DEMO.DIQ_by_yr_range <- DEMO %>%
left_join(DIQ,
by = c("yr_range"="yr_range"),
suffix = c("_DEMO","_DIQ"))
DEMO.DIQ_by_yr_range %>%
select(contains('SEQN')) %>%
colnames()
DEMO.DIQ_by_SEQN <- DEMO %>%
left_join(DIQ,
by = c("SEQN"="SEQN"),
suffix = c("_DEMO","_DIQ"))
DEMO.DIQ_by_SEQN %>%
select(contains('yr_range')) %>%
colnames()
DEMO.DIQ_by_SEQN_yr_range <- DEMO %>%
left_join(DIQ, by=c("SEQN"="SEQN", 'yr_range'='yr_range'))
NHANES_table_helper <- function(Table_Name){
url_link <- paste0('https://wwwn.cdc.gov/Nchs/Nhanes/2017-2018/',Table_Name,'_J.htm')
return(url_link)
}
NHANES_table_helper('DR1TOT')
NHANES_table_helper(NHANES_Tables[2])
Open_NHANES_table_help <- function(Table_Name){
if(Table_Name %in% NHANES_Tables){
return(browseURL(NHANES_table_helper(Table_Name)))
} else {
ln1 <- paste0("ERROR : ", Table_Name, " is not a valid table name! \n")
ln2 <- 'Please Choose one of : \n'
ln3 <- NHANES_Tables
cat(ln1)
cat(ln2)
cat(ln3)
}
}
Open_NHANES_table_help('FROG')
Open_NHANES_table_help('DEMO')