Chapter 2 Working with Data in R
R would be almost useless without data. Therefore, it is important to become familiar with conventions that allow users to import, examine, and edit data. In this chapter, we will cover (a) how to install packages, where many open-source datasets are hosted; (b) how to import your own data; (c) how to identify and examine important characteristics of datasets, regardless of their point of origination; and (d) wrangle and modify datasets without leaving the IDE.
2.1 Installing and Calling Packages
BaseR comes with a lot of power. However, packages make R even more
powerful. In essence, packages are the fundamental unit of shareable
code in R. Packages help standardize tools and conventions and save time
as they are often created by data professionals around the world to
solve specific problems. For example, this guidebook relies heavily on
the tidyverse package, which contains a set of R packages that “share
an underlying design philosophy, grammar, and data structure” for the
purpose of data wrangling, analysis, and visualization (Wickham et al.,
2019). These include (a) ggplot2 for data visualization, (b) dplyr
for data manipulation, (c) tidyr for cleaning data, (d) purrr for
consistent treatment of functions and vectors, (e) tibble for fast and
accurate data uploading, (e) stringr for string or text manipulation,
and (f) forcats, which supports categorical variables such as
race/ethnicity or gender. For more information on tidyverse, type the
function help(tidyverse) into a code chunk or script file.
There is a package for almost anything you can think of. You can even make your own packages if you find yourself using R for a unique work flow.

In this chapter, we will be using data from Nate Silver’s statistical
analysis website—fivethirtyeight (Kim et al., 2018). The
fivethirtyeight package includes 128 callable datasets. For a full
list of datasets, follow this link:
<https://cran.r-project.org/web/packages/fivethirtyeight/vignettes/fivethirtyeight.html>.
To see all datasets that are available to you as callable objects, run
the function data()
To install packages, use the code below. Remember to wrap the package in quotes when you are installing it. Then, you will need to call the packages using the library() function. You do not need to use quotes when calling packages you have already installed.
install.packages("tidyverse")
install.packages("fivethirtyeight")library(tidyverse)
library(fivethirtyeight)2.2 Importing and Exporting Data
We will be using a built-in dataset from the fivethirtyeight package
that contains information on US Births from 1994 to 2003. Call the
dataset with the object named 'US_births_1994_2003 and assign it to
the object data with the <- operator. This code can be read as
“create a new object ‘data’ identical to the dataset
‘US_births_1994_2003’.” The purpose of renaming this data object is to
simplify subsequent coding practices. That said, you can choose to
rename your dataset anything you’d like, or keep the name as it was
originally stated.
2.2.1 R-based Data
data <- US_births_1994_2003Practice calling another dataset. Try calling the data()function or
visiting the fivethirtyeight website and select a dataset that you are
interested in examining. Once you have chosen a dataset, you can assign
that data object to another name, or simply type in the name of the
dataset into a code chunk. Alternatively, you can use the data()
funciton with the name of the dataset inside the parenthesis to specify
which dataset you are calling in.
If you would like to import your own data, use the tidyverse
conventions below. Remember, you must have the tidyverse library
installed and called for this function to work.
2.2.2 CSV Data
data <- read_csv('filename.csv')Use caution with xlsx file types that contain multiple sheets. CSVs are
utilized more frequently than xlsx files as tidyverse handles them
quickly and simply. To load an xlsx file, you will need to explicitly
install the “readxl” package. You can then call the specific sheet you
want to load using the sheet argument. You will need to assign that
sheet to an object to save that particular sheet for further examination
and manipulation.
2.2.3 XLSX Data
install.packages('readxl')
library(readxl)
data <- read_excel(spreadsheet, sheet = "sheet1")Haven is a package that is associated with, but not inherently a part
of, the tidyverse package for SPSS, SAS, and STATA filetypes. You will
need to follow similar conventions as with the readxl library. You can
either define the path using an object, or explictly state the path in
the ultimate function’s parenthesis.
2.2.4 SPSS “.sav” Data
install.packages('haven')
library(haven)
path <- file.path("example.sav")
data <- read_sav(path)2.2.7 Exporting Data
You can export data similarly to how you imported data. See the code chunk below for examples of export functions for each data filetype. It is important to note that each of these are exported from the data object, as it is assumed you will have edited that data object at some point while using R. Each of these functions will save a dataset in your working directory based on the characteristics of the object “data” at the time the function was run.
write_csv(data)
write_excel(data)
write_sav(data)
write_sas(data)
write_dta(data)2.3 Working With Datasets
Whether you are using your own dataset or conducting secondary data analysis, it is important to understand the data that are available to you. There are several ways to go about this in R.
2.3.1 Viewing the Dataset
First, consider the head() and tail() functions which display the
first and last six rows of a dataframe, respectively. Place your data
object within the function’s parenthesis, and then run the code to get a
snapshot of the dataset. Here, you might notice the output is a
tibble. Although there are other distinctions, the key difference
between a tibble and a dataframe is that tibbles display data types
along with data (e.g., integer, date, numerial). You can see an example
of this under the each column name below:
head(data)## # A tibble: 6 × 6
## year month date_of_month date day_of_week births
## <int> <int> <int> <date> <ord> <int>
## 1 1994 1 1 1994-01-01 Sat 8096
## 2 1994 1 2 1994-01-02 Sun 7772
## 3 1994 1 3 1994-01-03 Mon 10142
## 4 1994 1 4 1994-01-04 Tues 11248
## 5 1994 1 5 1994-01-05 Wed 11053
## 6 1994 1 6 1994-01-06 Thurs 11406In some cases a dataset contains dozens of variables in columns, which
may be difficult to assess using the data viewer. To investigate the
names of all columns, run the function colnames()
colnames(data)## [1] "year" "month" "date_of_month" "date"
## [5] "day_of_week" "births"For a summary of each column of data, call the summary() function.
summary(data)## year month date_of_month date
## Min. :1994 Min. : 1.000 Min. : 1.00 Min. :1994-01-01
## 1st Qu.:1996 1st Qu.: 4.000 1st Qu.: 8.00 1st Qu.:1996-07-01
## Median :1998 Median : 7.000 Median :16.00 Median :1998-12-31
## Mean :1998 Mean : 6.524 Mean :15.73 Mean :1998-12-31
## 3rd Qu.:2001 3rd Qu.:10.000 3rd Qu.:23.00 3rd Qu.:2001-07-01
## Max. :2003 Max. :12.000 Max. :31.00 Max. :2003-12-31
##
## day_of_week births
## Sun :522 Min. : 6443
## Mon :522 1st Qu.: 8844
## Tues :522 Median :11615
## Wed :522 Mean :10877
## Thurs:521 3rd Qu.:12274
## Fri :521 Max. :14540
## Sat :522You can also see the entire dataset in a separate pane within R using
view()
view(data)2.3.2 Data Types
Most R workflows involve one of five basic data types. These include (a) integers (e.g., 2); (b) numeric values, which include decimals (e.g., 2.5); (c) factors, or categorical variables with more than one level (e.g., gender); (d) logical, or Boolean values (i.e., True/False); and (e) characters or strings (e.g., “Thanks for all the fish”). Some functions will only work when a certain data type is present in the data set. For instance, if you plan to conduct an analysis of variance (ANOVA), you will need a factor to be present in your data. Similarly, a logistic regression typically requires Boolean values, and text analysis tends to require string data.
The following functions are useful to examine different facets related
to data types present in the dataset. The length() function returns
the number of elements or components in a dataset.
length(data)## [1] 6The str() function returns the structure of the dataset. The output of
this function includes example data from each column, the data type, as
well as the class of the data object.
str(data)## tibble [3,652 × 6] (S3: tbl_df/tbl/data.frame)
## $ year : int [1:3652] 1994 1994 1994 1994 1994 1994 1994 1994 1994 1994 ...
## $ month : int [1:3652] 1 1 1 1 1 1 1 1 1 1 ...
## $ date_of_month: int [1:3652] 1 2 3 4 5 6 7 8 9 10 ...
## $ date : Date[1:3652], format: "1994-01-01" "1994-01-02" ...
## $ day_of_week : Ord.factor w/ 7 levels "Sun"<"Mon"<"Tues"<..: 7 1 2 3 4 5 6 7 1 2 ...
## $ births : int [1:3652] 8096 7772 10142 11248 11053 11406 11251 8653 7910 10498 ...If you are only interested in the class of the object, the class()
function refers to the object itself, which in this case is a tibble.
class(data)## [1] "tbl_df" "tbl" "data.frame"The names() function, much like col_names() returns a list of the
column names.
names(data)## [1] "year" "month" "date_of_month" "date"
## [5] "day_of_week" "births"The nrow() function returns the number of rows in the dataset.
nrow(data)## [1] 3652While the dim() function returns the number of rows and columns
simultaneously.
dim(data)## [1] 3652 6Sometimes when your data is loaded, R will recognize a column with an incorrect structure. For example, you can change the data type of the month column in US Births dataset ‘month’ integer, which tends to include decimals, to numeric using the function as.numeric(). The $ sign signifies that you are attempting to access a column based on its name in the dataset.
The code below explicitly converts the month column in the data object
to a numeric data type using the function “as.numeric().” Follow up this
conversion with the str() function to assess whether the change was
made.
data$month <- as.numeric(data$month)
str(data)## tibble [3,652 × 6] (S3: tbl_df/tbl/data.frame)
## $ year : int [1:3652] 1994 1994 1994 1994 1994 1994 1994 1994 1994 1994 ...
## $ month : num [1:3652] 1 1 1 1 1 1 1 1 1 1 ...
## $ date_of_month: int [1:3652] 1 2 3 4 5 6 7 8 9 10 ...
## $ date : Date[1:3652], format: "1994-01-01" "1994-01-02" ...
## $ day_of_week : Ord.factor w/ 7 levels "Sun"<"Mon"<"Tues"<..: 7 1 2 3 4 5 6 7 1 2 ...
## $ births : int [1:3652] 8096 7772 10142 11248 11053 11406 11251 8653 7910 10498 ...When operationalizing factors for quantitative data analysis, you might want to convert a string based factor to numeric values (e.g., creating dummy variables out of categorical data). The code below converts the string data in the day of the week column into a numeric vector. When running factorial/categorical-based analyses (such as ANOVA variants or Chi-Square tests), R will create dummy variables to factors of data type “string.” Each factor will be labeled in the order it uniquely appears in the dataset, row-wise.
In this example, we assigned the function to a new data object. As a result, we can manipulate and analyze the new data object while retaining the original. The creation of new objects will be useful as we begin to subset data for analysis.
num_day_of_Week <- as.numeric(data$day_of_week)2.3.3 Examining Vectors and Variables
Object-oriented programming and statistics both tend to rely on the
analysis of vectors and variables. The unique() function displays
unique values in a column. Use this function to answer the question: Are
all 12 months of the year represented in this dataset? This is also a
great way to troubleshoot extant factor levels that are actually
misspellings, or extra white space in the cell. For instance, if a cell
in the month column mistakenly included 13, it would be visible in the
output below.
# Get the unique months present in the data
unique(data$month)## [1] 1 2 3 4 5 6 7 8 9 10 11 12Furthermore, you can use max() and min() to identify the largest and
smallest values in a column. Other functions help with descriptive
statistics, such as mean() and sd() Descriptive statistics will be
covered in more depth in Chapter Four.
max(data$births)## [1] 14540min(data$births)## [1] 6443mean(data$births)## [1] 10876.82sd(data$births)## [1] 1858.5672.3.4 Filtering Data
To subset the data for births from 1995 alone, use the filter()
function to create a new object with the data you specify. The data are
subset by the argument “year == 1995” which indicates that only
observations that contain 1995 in the year column should be retained. In
the code below, the filtered dataset is assigned to a new data object
“births_1995.” We then use the head() function to examine the first
six instances in the dataset.
births_1995 <- filter(data, year == 1995)
head(births_1995)## # A tibble: 6 × 6
## year month date_of_month date day_of_week births
## <int> <dbl> <int> <date> <ord> <int>
## 1 1995 1 1 1995-01-01 Sun 7828
## 2 1995 1 2 1995-01-02 Mon 7883
## 3 1995 1 3 1995-01-03 Tues 9999
## 4 1995 1 4 1995-01-04 Wed 11315
## 5 1995 1 5 1995-01-05 Thurs 11243
## 6 1995 1 6 1995-01-06 Fri 11506Base R allows you to find the index of one column where a maximum occurs
in another. For example, the following code uses the max() and
which.max() functions to identify the date when the maximum number of
births took place.
# Find the date of the maximum births
maxindex <- which.max(data$births)
maxdate <- data$date[maxindex]
maxdate## [1] "1999-09-09"However, you can use the filter() function from the tidyverse to
achieve the same result in one line of code. You can read this aloud as
“filter the object data where the births variable is equal to the
maximum number present in the births column.”
filter(data, births == max(births))## # A tibble: 1 × 6
## year month date_of_month date day_of_week births
## <int> <dbl> <int> <date> <ord> <int>
## 1 1999 9 9 1999-09-09 Thurs 14540Imagine that you want to identify the max number of births for a Sunday
in particular in any year. This code uses the filter() function to
create a new data object called sundays, which only includes data for
observations where the day of the week was the string ‘Sun.’ In the next
line of code, the filter() function is utilized on the newly created
data object “sundays” to identify the observation with the maximum
number of births.
# this is what filter is doing on groups
sundays <- filter(data, day_of_week == 'Sun')
filter(sundays, births==max(births))## # A tibble: 1 × 6
## year month date_of_month date day_of_week births
## <int> <dbl> <int> <date> <ord> <int>
## 1 1994 8 14 1994-08-14 Sun 8926If you are only interested in specific variables or vectors within a
data frame you can use the select() function and list the data object,
then the column names that you are interested in seeing. Additionally,
notice how the head() function is wrapped around the select()
function in this example to limit the output to the first six
observations.
head(select(data, date, births))## # A tibble: 6 × 2
## date births
## <date> <int>
## 1 1994-01-01 8096
## 2 1994-01-02 7772
## 3 1994-01-03 10142
## 4 1994-01-04 11248
## 5 1994-01-05 11053
## 6 1994-01-06 11406To remove a column from the data object, place a minus sign before it in
the column argument space within the select() function.
head(select(data, -date))## # A tibble: 6 × 5
## year month date_of_month day_of_week births
## <int> <dbl> <int> <ord> <int>
## 1 1994 1 1 Sat 8096
## 2 1994 1 2 Sun 7772
## 3 1994 1 3 Mon 10142
## 4 1994 1 4 Tues 11248
## 5 1994 1 5 Wed 11053
## 6 1994 1 6 Thurs 114062.3.5 Grouping Data
Alternatively, you can use the group_by() function while filtering the
dataset by max births to display the maximum number of births on each
day of the week for any year. The group_by() function groups the data
object by the observations in the called column, which in this case is
“day_of_week.” Then, the grouped data are filtered to with the same
max() function. Grouping the data retains the factor structure of the
“day_of_week” vector in subsequent functions. The grouped and filtered
dataset are assigned to a new object “maxbirths” for future reference.
data_grouped <- group_by(data, day_of_week)
maxbirths <- filter(data_grouped, births == max(births))
maxbirths## # A tibble: 7 × 6
## # Groups: day_of_week [7]
## year month date_of_month date day_of_week births
## <int> <dbl> <int> <date> <ord> <int>
## 1 1994 8 14 1994-08-14 Sun 8926
## 2 1994 9 17 1994-09-17 Sat 9779
## 3 1999 9 9 1999-09-09 Thurs 14540
## 4 2001 12 28 2001-12-28 Fri 13918
## 5 2003 9 3 2003-09-03 Wed 14119
## 6 2003 12 22 2003-12-22 Mon 12967
## 7 2003 12 30 2003-12-30 Tues 14438You can also group by more than one column. This code expands on the previous example to include the maximum number of births on each day of the week within each year present in the dataset.
# Add year as another grouping variable
data_grouped <- group_by(data, day_of_week, year)
data_max_day_year <- filter(data_grouped, births==max(births))
head(data_max_day_year)## # A tibble: 6 × 6
## # Groups: day_of_week, year [6]
## year month date_of_month date day_of_week births
## <int> <dbl> <int> <date> <ord> <int>
## 1 1994 7 6 1994-07-06 Wed 13086
## 2 1994 7 7 1994-07-07 Thurs 13049
## 3 1994 8 14 1994-08-14 Sun 8926
## 4 1994 9 16 1994-09-16 Fri 12884
## 5 1994 9 17 1994-09-17 Sat 9779
## 6 1994 11 21 1994-11-21 Mon 11807Arguments within functions are oftentimes objects themselves. As such,
you can replace the “data_grouped” object in the previous chunk with its
assigned group_by() function to simplify your code while achieving the
same result.
data_max_day_year <- filter(group_by(data, day_of_week, year), births==max(births))
head(data_max_day_year)## # A tibble: 6 × 6
## # Groups: day_of_week, year [6]
## year month date_of_month date day_of_week births
## <int> <dbl> <int> <date> <ord> <int>
## 1 1994 7 6 1994-07-06 Wed 13086
## 2 1994 7 7 1994-07-07 Thurs 13049
## 3 1994 8 14 1994-08-14 Sun 8926
## 4 1994 9 16 1994-09-16 Fri 12884
## 5 1994 9 17 1994-09-17 Sat 9779
## 6 1994 11 21 1994-11-21 Mon 118072.3.6 Altering Data
There are many useful functions for altering your data frame. In this
section you will start to see the %>%operator. You can read this
operator as the word ‘then’ or ‘where’ when you are reading code to
yourself (e.g., take the original dataset, then arrange it by births in
descending order)
You can use this operator to apply functions to your dataset. For
instance, the code below reorders the observations in the data by births
in descending order. To arrange in ascending order simply remove the
additional desc() function within the arrange() function as the
default arrangement is ascending order.
# Order by number of births
data <- data %>% arrange(desc(births))
head(data)## # A tibble: 6 × 6
## year month date_of_month date day_of_week births
## <int> <dbl> <int> <date> <ord> <int>
## 1 1999 9 9 1999-09-09 Thurs 14540
## 2 2003 12 30 2003-12-30 Tues 14438
## 3 2003 9 16 2003-09-16 Tues 14145
## 4 2003 9 3 2003-09-03 Wed 14119
## 5 2003 9 23 2003-09-23 Tues 14036
## 6 2002 9 12 2002-09-12 Thurs 13982You can also rename specific columns of your data without opening and
editing the file using the rename() function. You can read the code
below aloud saying “create a new object data from the old object data
where you rename date_of_month as day, and day_of_week as weekday.”
# Rename day_of_month and day_of_week columns
data <- data %>% rename(day = date_of_month, weekday = day_of_week)
head(data)## # A tibble: 6 × 6
## year month day date weekday births
## <int> <dbl> <int> <date> <ord> <int>
## 1 1999 9 9 1999-09-09 Thurs 14540
## 2 2003 12 30 2003-12-30 Tues 14438
## 3 2003 9 16 2003-09-16 Tues 14145
## 4 2003 9 3 2003-09-03 Wed 14119
## 5 2003 9 23 2003-09-23 Tues 14036
## 6 2002 9 12 2002-09-12 Thurs 13982The examples below utilize the %>% operator to conditionally select
observations within the data object. The first example reads, “create a
new object july_data” based on the old object data where you filter for
observations where the month column value is 7.” The second example
extends this concept by including a second condition. The ampersand in
the filter() function allows for the identification of observations
only in the month of January, 2000.
# Select days only in the month of July
july_data <- data %>% filter(month == 7)
# Select days only in the month of january in the year 2000
jan_2000_data <- data %>% filter(month == 1 & year == 2000)Recall our use of sequences in Chapter One. In this example, we filter
out the data for the first half of the year (January through June) using
a colon in the subsetting argument rather than individual numbers. Then,
use the max() function to check whether the the filtering function
worked.
janthrujune <- data %>% filter(month == 1:6)
max(janthrujune$month)## [1] 6You can also add an additional column to the data frame based on
conditional values of other columns using the mutate() function. The
example below creates a vector Boolean values which indicates whether
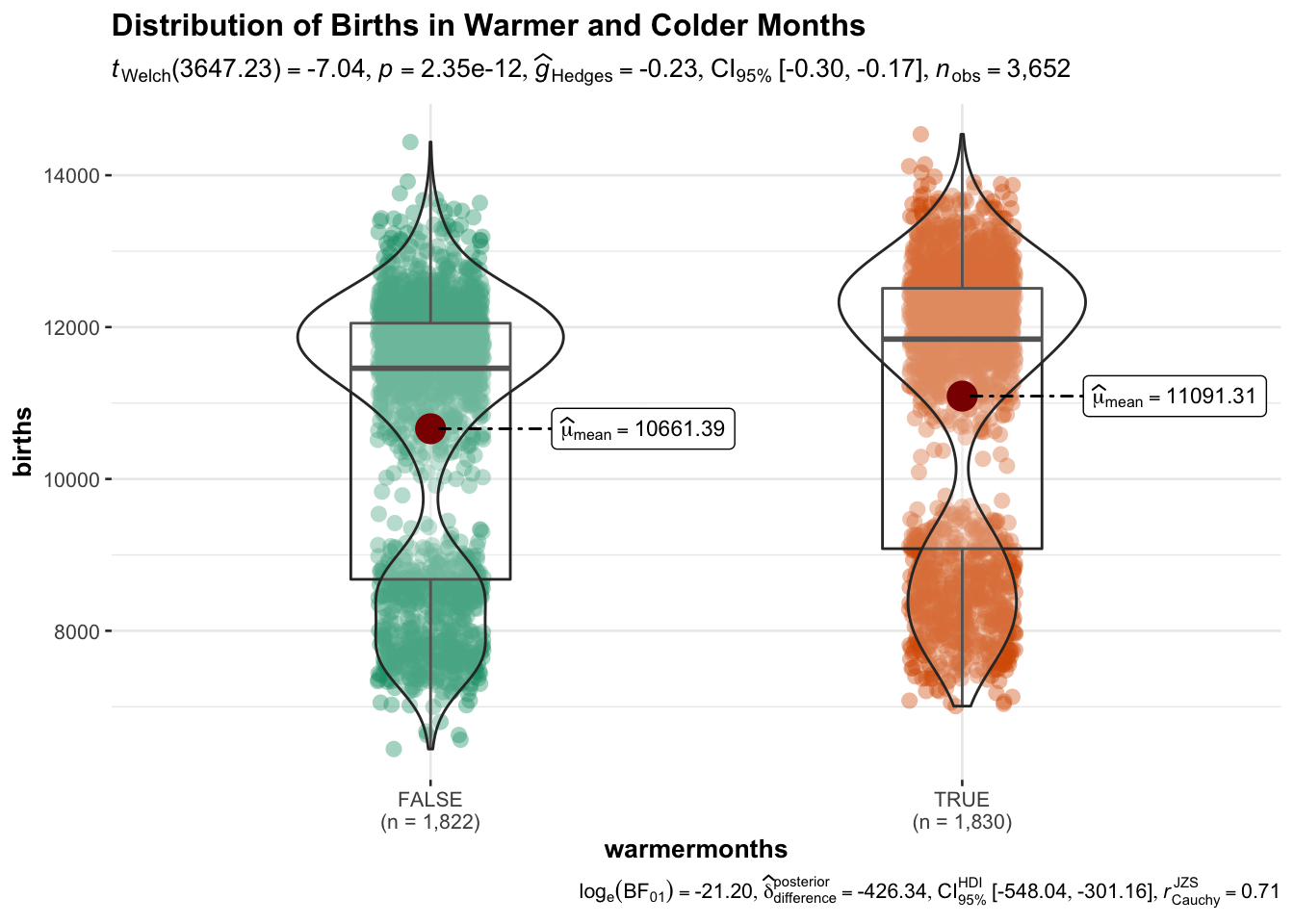
the observation occurred during a warmer month. The subsequent plot
serves as a preview of the next chapter on Data Visualization, but also
demonstrates a practical use of the mutate function in statistical
modeling. The grouped violin plot answers the question: Were there more
births in warmer months than in colder months?
data <- data %>% mutate(warmermonths = between(month, 4, 9))
str(data)## tibble [3,652 × 7] (S3: tbl_df/tbl/data.frame)
## $ year : int [1:3652] 1999 2003 2003 2003 2003 2002 2001 2003 2002 2002 ...
## $ month : num [1:3652] 9 12 9 9 9 9 12 9 9 9 ...
## $ day : int [1:3652] 9 30 16 3 23 12 28 10 24 17 ...
## $ date : Date[1:3652], format: "1999-09-09" "2003-12-30" ...
## $ weekday : Ord.factor w/ 7 levels "Sun"<"Mon"<"Tues"<..: 5 3 3 4 3 5 6 4 3 3 ...
## $ births : int [1:3652] 14540 14438 14145 14119 14036 13982 13918 13908 13884 13883 ...
## $ warmermonths: logi [1:3652] TRUE FALSE TRUE TRUE TRUE TRUE ...library(ggstatsplot)
ggbetweenstats(
data = data,
x = warmermonths,
y = births,
title = "Distribution of Births in Warmer and Colder Months"
)
2.4 Case Study
Use the following exercises to learn more about the US_births_1994_2003 dataset and practice the skills you acquired in this chapter.
Using the
filter(),min(), and/orgroup_by()functions, identify the Wednesday and Friday in the dataset with the least total number of births.Find the number of births for January 1st across the dataset.
Subset the dataset to only include observations that occur in winter (December, January, February, and March in North America)
Subset the previous winter births dataset further by only allowing instances after 2001 to remain in the dataset.
2.5 Review
In this chapter, we covered how to install and use libraries, how to import data from several formats, and how packages support several basic data science tasks. To make sure you understand this material, there is a practice assessment to go along with this chapter at https://jayholster.shinyapps.io/IntrotoRAssessment/.
2.6 References
Kim A.Y., Ismay C., & Chunn, J. (2018). “The fivethirtyeight R Package: ‘Tame Data’ Principles for Introductory Statistics and Data Science Courses.” Technology Innovations in Statistics Education, 11. https://escholarship.org/uc/item/0rx1231m.
Patil, I. (2021). Visualizations with statistical details: The ‘ggstatsplot’ approach. Journal of Open Source Software, 6(61), 3167. https://doi.org/10.21105/joss.03167
Wickham H., Bryan, J. (2022). readxl: Read Excel Files. https://readxl.tidyverse.org, https://github.com/tidyverse/readxl.
Wickahm, H., & Miller, E., (2022) haven. https://cran.r-project.org/web/packages/haven/index.html
Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L.D., François, R., Grolemund, G., Hayes, A., Henry, L., Hester, J., Kuhn, M., Pedersen, T.L., Miller, E., Bache, S.M., Müller, K., Ooms, J., Robinson, D., Seidel, D.P., Spinu, V., Takahashi, K., Vaughan, D., Wilke, C., Woo, K., & Yutani, H. (2019). Welcome to the tidyverse. Journal of Open Source Software, 4(43), 1686. https://doi.org/10.21105/joss.01686.
2.6.1 R Short Course Series
Video lectures of each guidebook chapter can be found at https://osf.io/6jb9t/. For this chapter, find the follow the folder path Intro to R -> AY 2021-2022 Spring and access the video files, r markdown documents, and other materials for each short course.
2.6.2 Acknowledgements
This guidebook was created with support from the Center for Research Data and Digital Scholarship and the Laboratory for Interdisciplinary Statistical Analaysis at the University of Colorado Boulder, as well as the U.S. Agency for International Development under cooperative agreement #7200AA18CA00022. Individuals who contributed to materials related to this project include Jacob Holster, Eric Vance, Michael Ramsey, Nicholas Varberg, and Nickoal Eichmann-Kalwara.