1 Day 1 (January 21)
1.1 Welcome and preliminaries
Teaching Assistant
-
- How I will use Canvas
- Grades, journal and project submissions only
- How I will use Canvas
-
- Required and Recomended material
- Statistical programming languages
- Reproducibility requirement (data analysis and computing can be successfully repeated)
- Academic Honesty: working in groups, sharing code, etc.
- Grades
- Topics
Who is in this class?
- Group work and collaboration
url <- "https://www.dropbox.com/scl/fi/yy44rp2bx263d9byltk3m/students_STAT_768.csv?rlkey=zbt60fpkl9ta9d3uqysu3vbtf&dl=1"
df <- read.csv(url)
par(mar=c(13,2,2,2))
plot(rev(sort(table(df$degreeProgram))),las=2,xlab="",ylab="Number of students",ylim=c(0,8))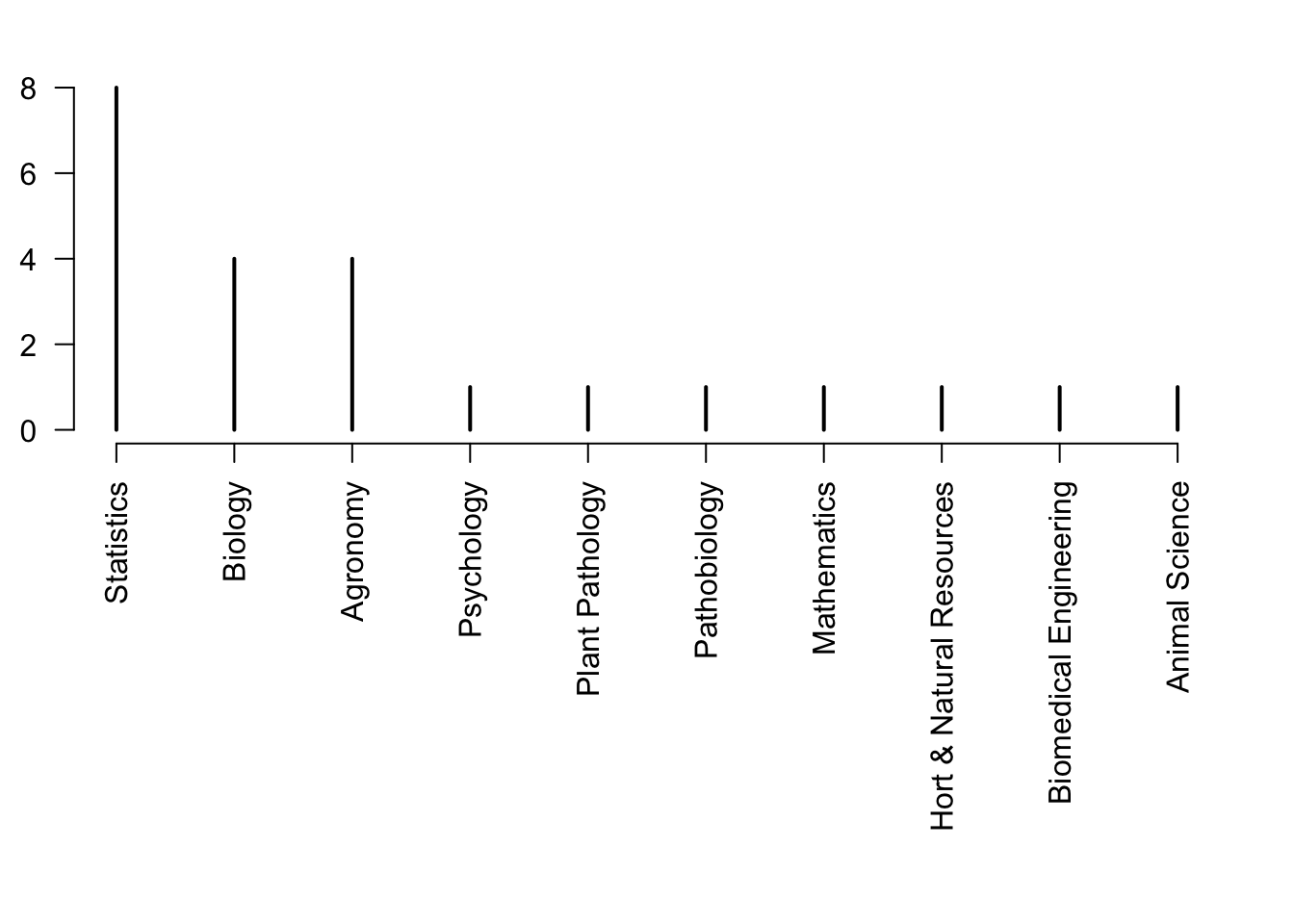
par(mar=c(13,2,2,2))
plot(rev(sort(table(df$classLevel))),las=2,xlab="",ylab="Number of students",ylim=c(0,15))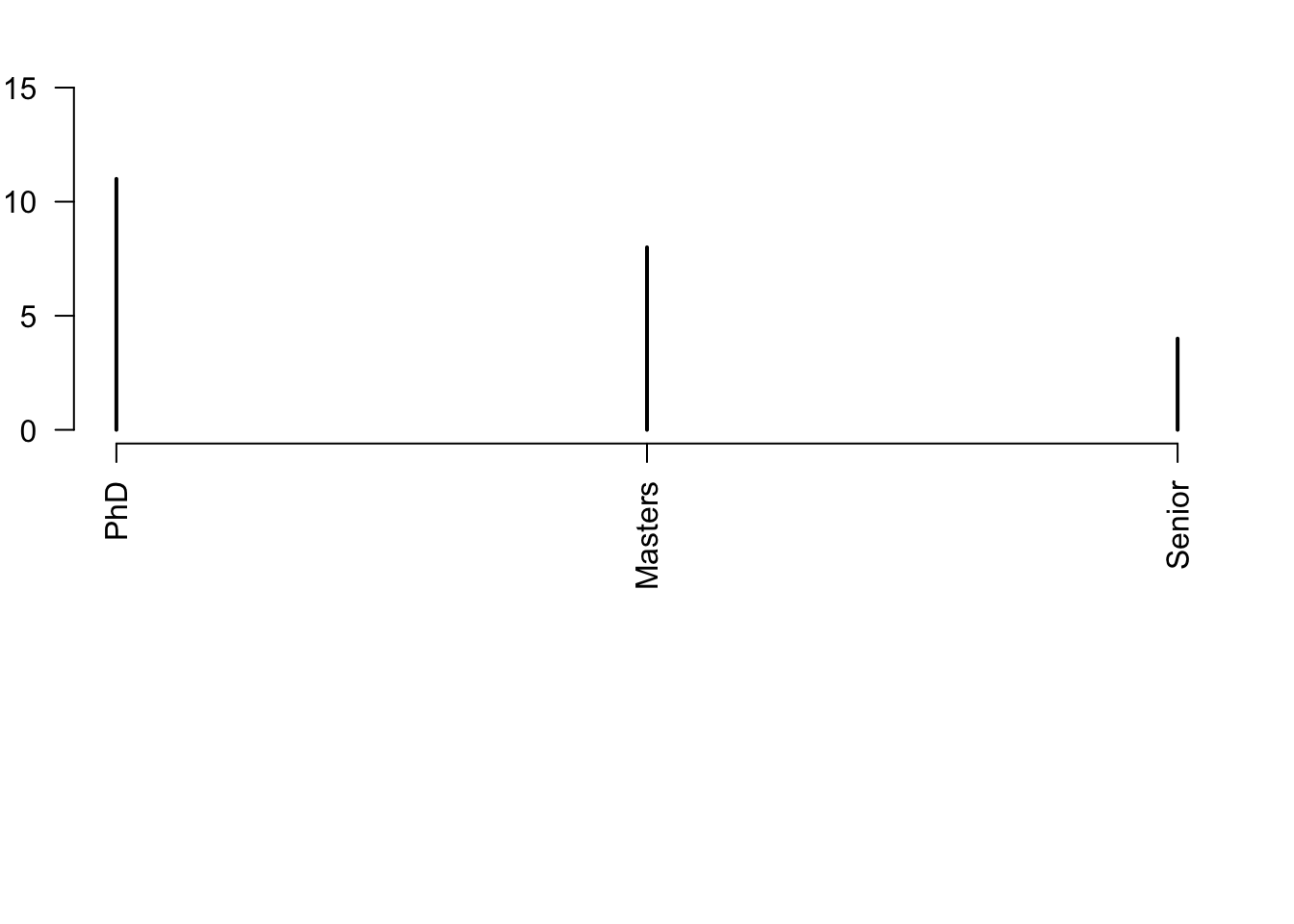
1.2 Intro to Bayesian statistical modelling
- What is data?
- Something in the real world that you can, in some way, observe and measure with or without error
- What is a statistic?
- A function of the data
- What is a model?
- Mathematical models
- Statistical models
- What is a parameter?
- Part of a statistical model that is usually unknown and must be assumed or estimated.
- What is the goal of Bayesian statistics?
- Obtain the distribution of potentially unrecordable random variables given recorded random variables
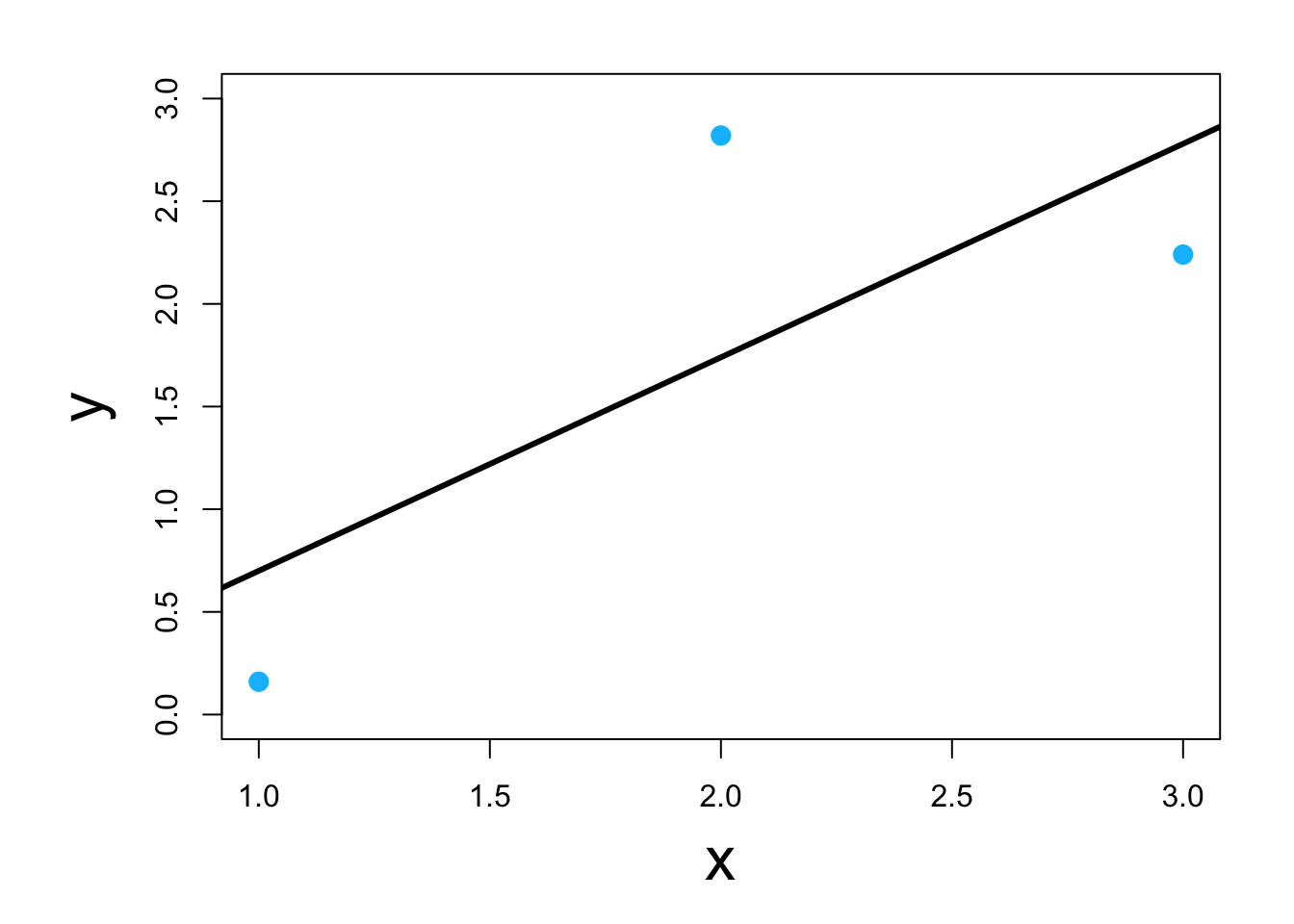
1.3 Example with linear models
What is a model?
What is a linear model?
Most widely used model in science, engineering, and statistics.
Scalar form: \(y_i=\beta_{0}+\beta_{1}x_{i}+\varepsilon_i\)
Which part of the model is the mathematical model.
Which part of the model makes the linear model a “statistical” model.
Visual
1.4 Estimation and inference
- Three options to estimate \(\beta_0\) and \(\beta_1\)
- Minimize a loss function
- Maximize a likelihood function
- Find the posterior distribution
- Each option requires different assumptions
1.5 Loss function approach
- Define a measure of discrepancy between the data and the mathematical model
- Find the values of \(\beta_0\) and \(\beta_1\) that makes \(\beta_{0}+\beta_{1}x_i\) “closest” to \(y_i\)
- Visual
- Real data example
- Details from Palmero et al. (2024)
# Preliminary steps url <- "https://www.dropbox.com/scl/fi/2qph4g9vnacibr73edrsb/Fig3_data.csv?rlkey=n48lbrv2zf2z5k1uja1393sof&dl=1" df.all <- read.csv(url) df.fp <- df.all[which(df.all$Scenario=="Scenario A"),] # Plot data for field pea plot(df.fp$Ndfa,df.fp$PartNbalance, xlab="Ndfa (%)",ylab="Partial N balance (kg/ha)", xlim=c(0,110),ylim=c(-100,200),main="Field pea") abline(a=0,b=0,col="gold",lwd=3)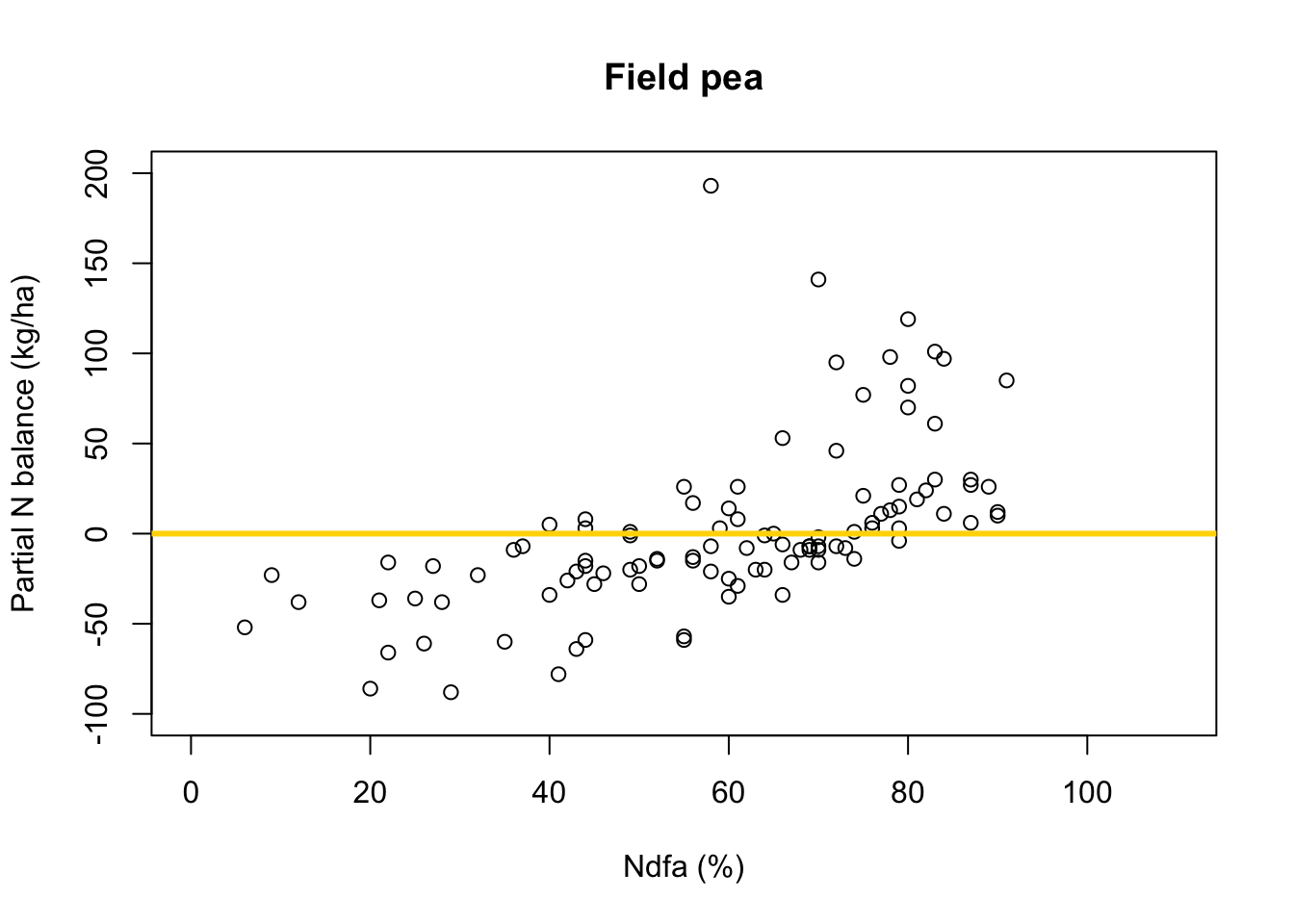
- Fit linear model to data using least squares
- What value of Ndfa is needed to achieve a neutral N balance?
beta0.hat <- as.numeric(coef(m1)[1]) beta1.hat <- as.numeric(coef(m1)[2]) theta.hat <- -beta0.hat/beta1.hat theta.hat## [1] 58.26843- Visual representation of \(\theta\)
# Plot data, line of best fit and theta plot(df.fp$Ndfa,df.fp$PartNbalance, xlab="Ndfa (%)",ylab="Partial N balance (kg/ha)", xlim=c(0,110),ylim=c(-100,200),main="Field pea") abline(a=0,b=0,col="gold",lwd=3) abline(m1,col="red",lwd=3) abline(v=58,lwd=3,lty=2,col="green")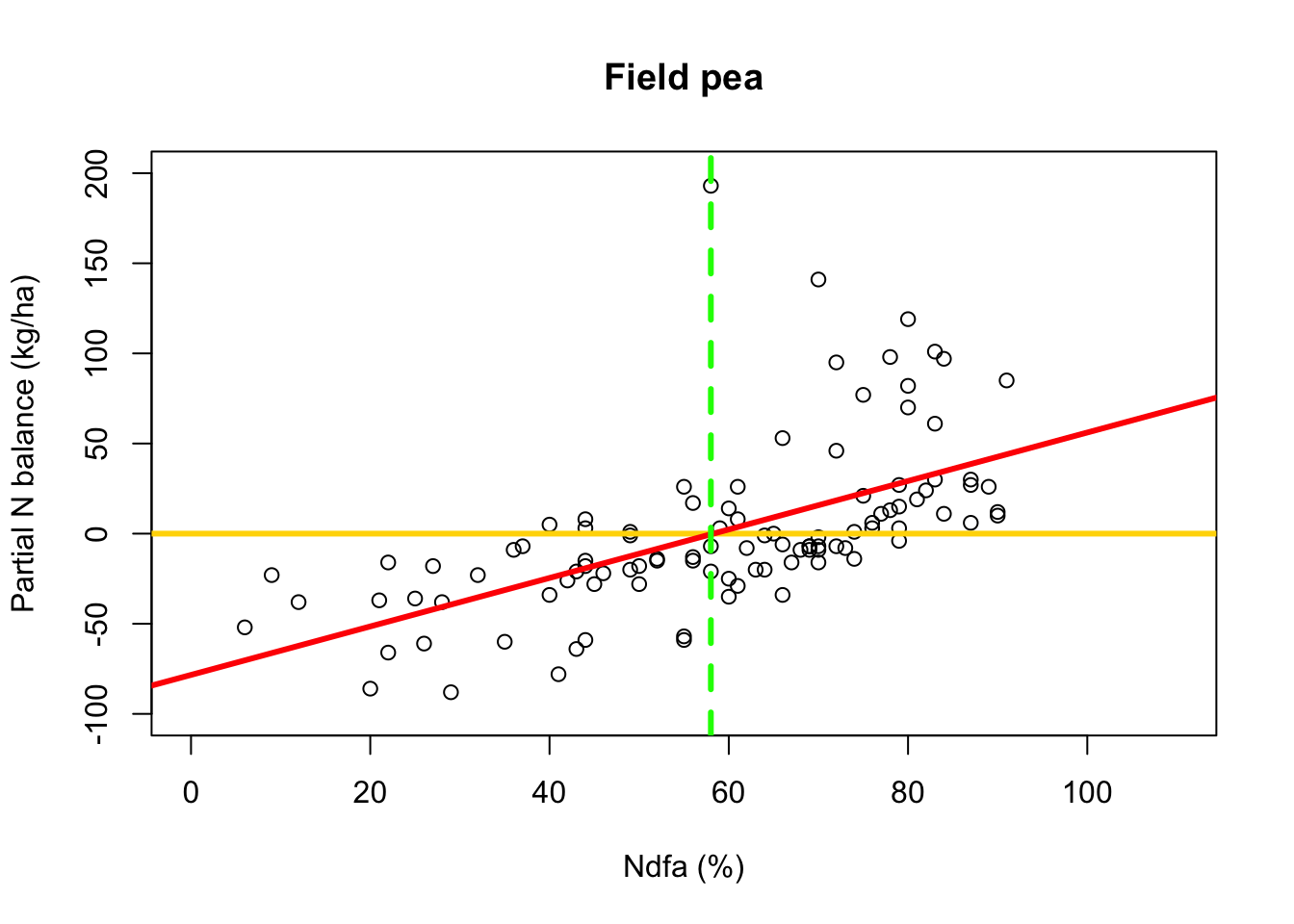
1.6 Likelihood-based approach
- Assume that \(y_i=\beta_{0}+\beta_{1}x_{i}+\varepsilon_i\) and \(\varepsilon_i\sim \text{N}(0,\sigma^2)\)
- Maximum likelihood estimation for the linear model
- Visual
- We added assumptions to our model, so what else do we get?
- Full likelihood-based statistical inference (e.g, p-values, confidence intervals, prediction intervals, etc)
- Real data example
- Fit linear model to data using using maximum likelihood estimation
library(nlme) # Fit simple linear regression model using maximum likelihood estimation m2 <- gls(PartNbalance ~ Ndfa,data=df.fp,method="ML")- What value of Ndfa is needed to achieve a neutral N balance?
# Use maximum likelihood estimate (MLE) to obtain estimate of theta beta0.hat <- as.numeric(coef(m2)[1]) beta1.hat <- as.numeric(coef(m2)[2]) theta.hat <- -beta0.hat/beta1.hat theta.hat## [1] 58.26843# Use delta method to obtain approximate approximate standard errors and # then construct Wald-type confidence intervals library(msm) theta.se <- deltamethod(~-x1/x2, mean=coef(m2), cov=vcov(m2)) theta.ci <- c(theta.hat-1.96*theta.se,theta.hat+1.96*theta.se) theta.ci## [1] 52.88317 63.65370- Visual representation of \(\theta\)
# Plot data, line of best fit and theta plot(df.fp$Ndfa,df.fp$PartNbalance, xlab="Ndfa (%)",ylab="Partial N balance (kg/ha)", xlim=c(0,110),ylim=c(-100,200),main="Field pea") abline(a=0,b=0,col="gold",lwd=3) abline(m1,col="red",lwd=3) abline(v=58.3,lwd=3,lty=2,col="green") abline(v=52.9,lwd=1,lty=2,col="green") abline(v=63.7,lwd=1,lty=2,col="green")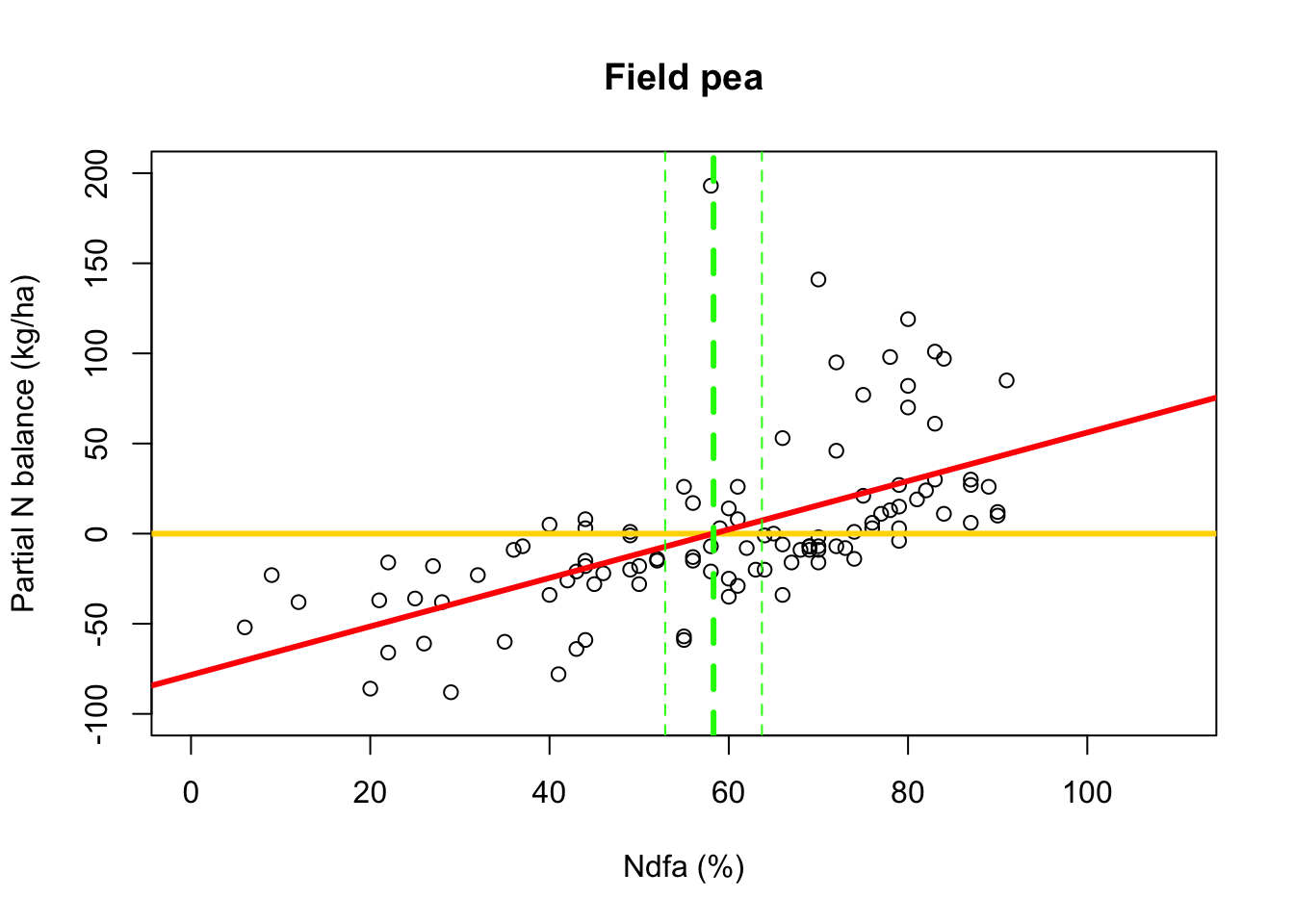
1.7 Bayesian approach
Assume that \(y_i=\beta_{0}+\beta_{1}x_{i}+\varepsilon_i\) with \(\varepsilon_i\sim \text{N}(0,\sigma^2)\), \(\beta_{0}\sim \text{N}(0,10^6)\) and \(\beta_{1}\sim \text{N}(0,10^6)\)
Statistical inference
- Using Bayes rule (Bayes 1763) we can obtain the joint posterior distribution
\[[\beta_0,\beta_1,\sigma_{\varepsilon}^{2}|\mathbf{y}]=\frac{[\mathbf{y}|\beta_0,\beta_1,\sigma_{\varepsilon}^{2}][\beta_0][\beta_1][\sigma_{\varepsilon}^{2}]}{\int\int\int [\mathbf{y}|\beta_0,\beta_1,\sigma_{\varepsilon}^{2}][\beta_0][\beta_1][\sigma_{\varepsilon}^{2}]d\beta_0d\beta_1,d\sigma_{\varepsilon}^{2}}\]
- Statistical inference about a paramters is obtained from the marginal posterior distributions \[[\beta_0|\mathbf{y}]=\int[\beta_0,\beta_1,\sigma_{\varepsilon}^{2}|\mathbf{y}]d\beta_1d\sigma_{\varepsilon}^{2}\] \[[\beta_1|\mathbf{y}]=\int[\beta_0,\beta_1,\sigma_{\varepsilon}^{2}|\mathbf{y}]d\beta_0d\sigma_{\varepsilon}^{2}\] \[[\sigma_{\varepsilon}^{2}|\mathbf{y}]=\int[\beta_0,\beta_1,\sigma_{\varepsilon}^{2}|\mathbf{y}]d\beta_0d\beta_1\]
- Derived quantities can be obtained by transformations of the joint posterior
- Using Bayes rule (Bayes 1763) we can obtain the joint posterior distribution
\[[\beta_0,\beta_1,\sigma_{\varepsilon}^{2}|\mathbf{y}]=\frac{[\mathbf{y}|\beta_0,\beta_1,\sigma_{\varepsilon}^{2}][\beta_0][\beta_1][\sigma_{\varepsilon}^{2}]}{\int\int\int [\mathbf{y}|\beta_0,\beta_1,\sigma_{\varepsilon}^{2}][\beta_0][\beta_1][\sigma_{\varepsilon}^{2}]d\beta_0d\beta_1,d\sigma_{\varepsilon}^{2}}\]
Computations
- Using a Markov chain Monte Carlo algorithm (see Ch.11 in Hooten and Hefley 2019)
norm.reg.mcmc <- function(y,X,beta.mn,beta.var,s2.mn,s2.sd,n.mcmc){ # # Code Box 11.1 # ### ### Subroutines ### library(mvtnorm) invgammastrt <- function(igmn,igvar){ q <- 2+(igmn^2)/igvar r <- 1/(igmn*(q-1)) list(r=r,q=q) } invgammamnvr <- function(r,q){ # This fcn is not necessary mn <- 1/(r*(q-1)) vr <- 1/((r^2)*((q-1)^2)*(q-2)) list(mn=mn,vr=vr) } ### ### Hyperpriors ### n=dim(X)[1] p=dim(X)[2] r=invgammastrt(s2.mn,s2.sd^2)$r q=invgammastrt(s2.mn,s2.sd^2)$q Sig.beta.inv=diag(p)/beta.var beta.save=matrix(0,p,n.mcmc) s2.save=rep(0,n.mcmc) Dbar.save=rep(0,n.mcmc) y.pred.mn=rep(0,n) ### ### Starting Values ### beta=solve(t(X)%*%X)%*%t(X)%*%y ### ### MCMC Loop ### for(k in 1:n.mcmc){ ### ### Sample s2 ### tmp.r=(1/r+.5*t(y-X%*%beta)%*%(y-X%*%beta))^(-1) tmp.q=n/2+q s2=1/rgamma(1,tmp.q,,tmp.r) ### ### Sample beta ### tmp.var=solve(t(X)%*%X/s2 + Sig.beta.inv) tmp.mn=tmp.var%*%(t(X)%*%y/s2 + Sig.beta.inv%*%beta.mn) beta=as.vector(rmvnorm(1,tmp.mn,tmp.var,method="chol")) ### ### Save Samples ### beta.save[,k]=beta s2.save[k]=s2 } ### ### Write Output ### list(beta.save=beta.save,s2.save=s2.save,y=y,X=X,n.mcmc=n.mcmc,n=n,r=r,q=q,p=p) }Model fitting
- MCMC
samples <- norm.reg.mcmc(y = df.fp$PartNbalance,X = model.matrix(~ Ndfa,data=df.fp), beta.mn = c(0,0),beta.var=c(10^6,10^6), s2.mn = 10, s2.sd = 10^6, n.mcmc = 5000) burn.in <- 1000 # Look a histograms of posterior distributions par(mfrow=c(2,1),mar=c(5,6,1,1)) hist(samples$beta.save[1,-c(1:1000)],xlab=expression(beta[0]*"|"*bold(y)),ylab=expression("["*beta[0]*"|"*bold(y)*"]"),freq=FALSE,col="grey",main="",breaks=30) hist(samples$beta.save[2,-c(1:1000)],xlab=expression(beta[1]*"|"*bold(y)),ylab=expression("["*beta[1]*"|"*bold(y)*"]"),freq=FALSE,col="grey",main="",breaks=30)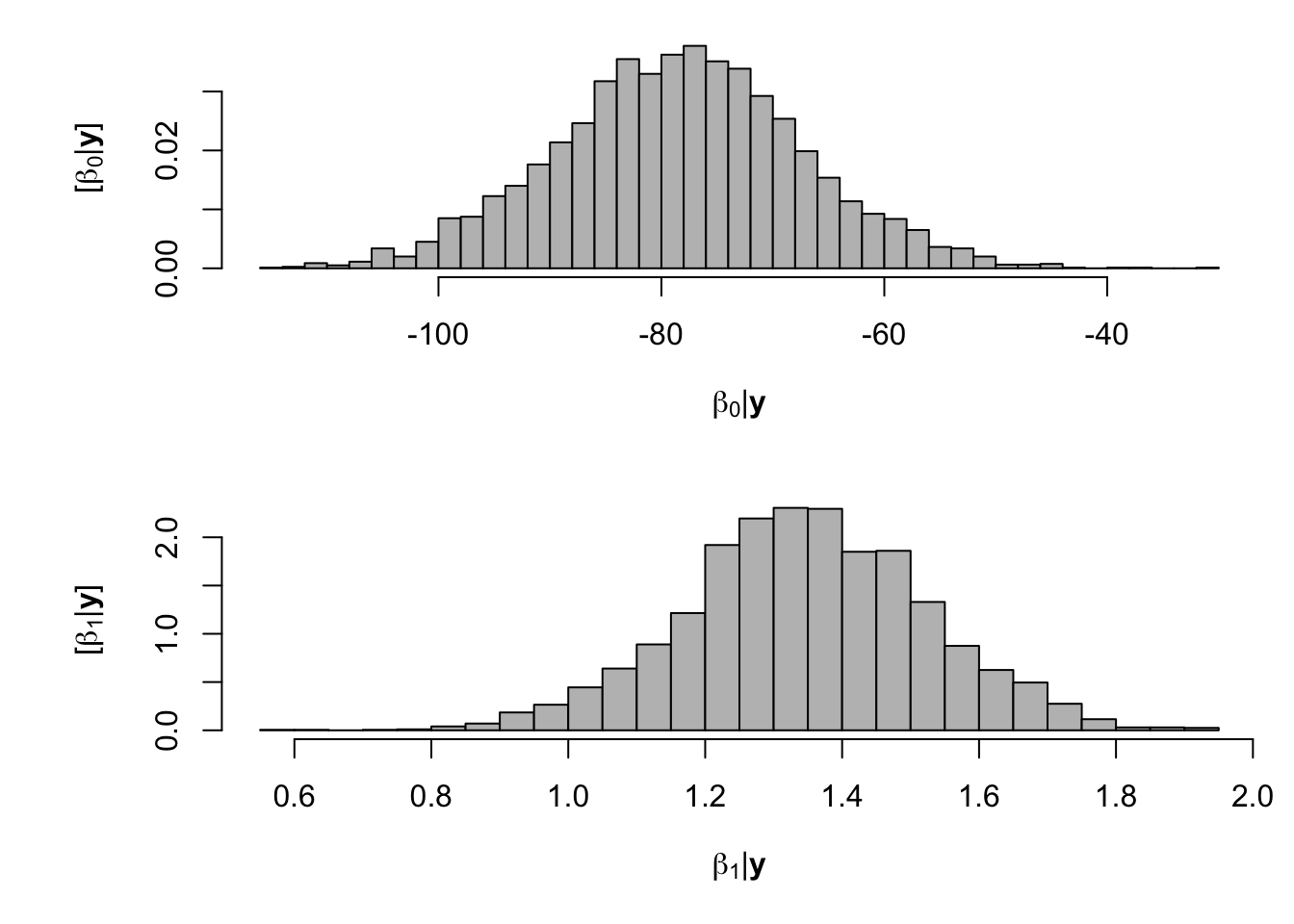
What value of Ndfa is needed to achieve a neutral N balance?
hist(-samples$beta.save[1,-c(1:1000)]/samples$beta.save[2,-c(1:1000)],xlab=expression(theta*"|"*bold(y)),ylab=expression("["*theta*"|"*bold(y)*"]"),freq=FALSE,col="grey",main="",breaks=30)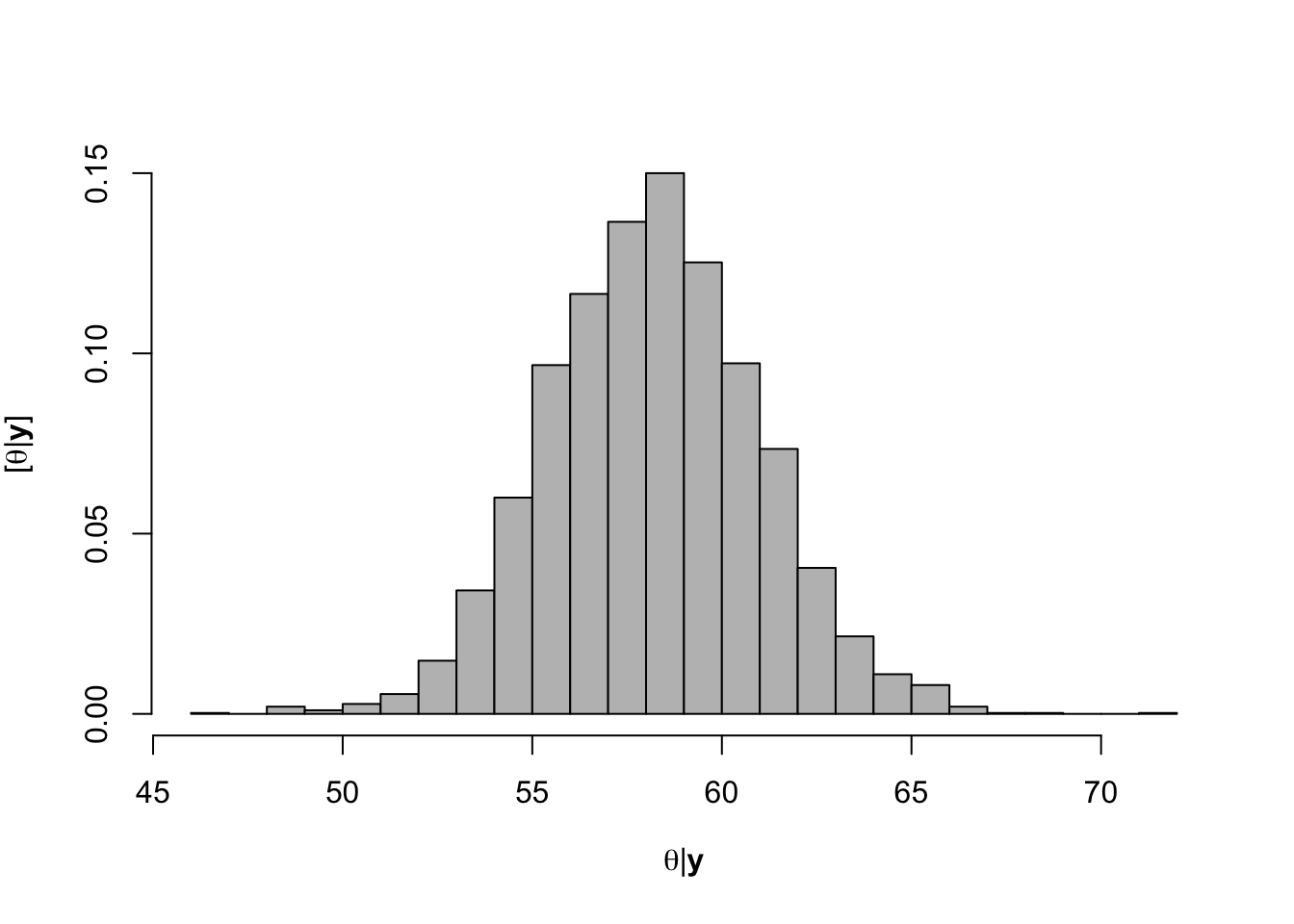
## [1] 58.21184# 95% credible intervals for theta quantile(-samples$beta.save[1,]/samples$beta.save[2,],prob=c(0.025,0.975))## 2.5% 97.5% ## 52.96028 63.83604- Visual representation of posterior distribuiton of theta \(\theta\)
# Plot data and theta plot(df.fp$Ndfa,df.fp$PartNbalance, xlab="Ndfa (%)",ylab="Partial N balance (kg/ha)", xlim=c(0,110),ylim=c(-100,200),main="Field pea") abline(a=0,b=0,col="gold",lwd=3) abline(v=58.3,lwd=3,lty=2,col="green") abline(v=52.7,lwd=1,lty=2,col="green") abline(v=63.7,lwd=1,lty=2,col="green") rug(-samples$beta.save[1,]/samples$beta.save[2,],col=gray(0.5,0.03))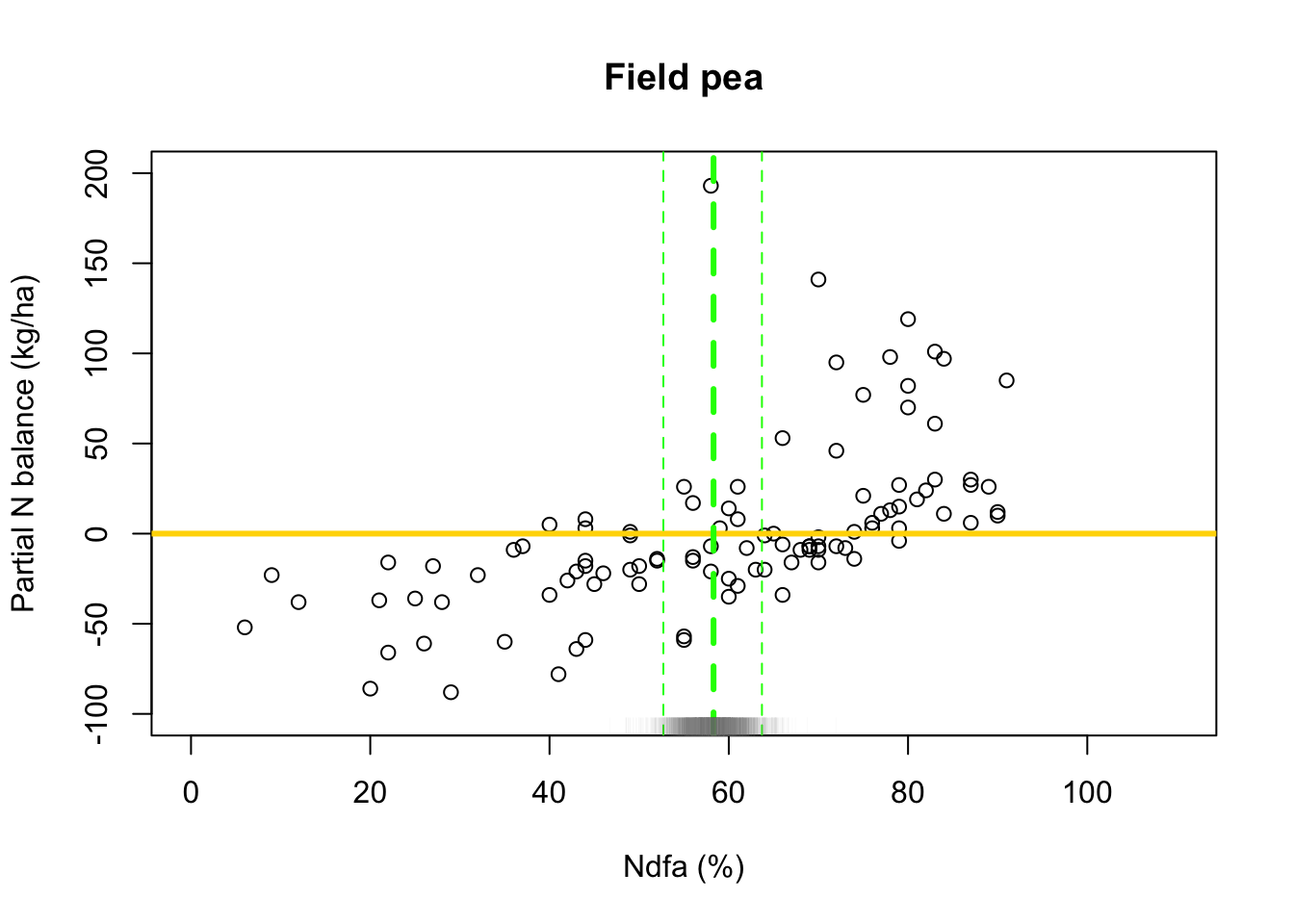
1.8 Low information content data
What value of Ndfa is needed to achieve a neutral N balance?
df.wl <- df.all[which(df.all$Scenario=="Scenario B"),] plot(df.wl$Ndfa,df.wl$PartNbalance, xlab="Ndfa (%)",ylab="Partial N balance (kg/ha)", xlim=c(0,110),ylim=c(-100,200),main="White lupin") abline(a=0,b=0,col="gold",lwd=3)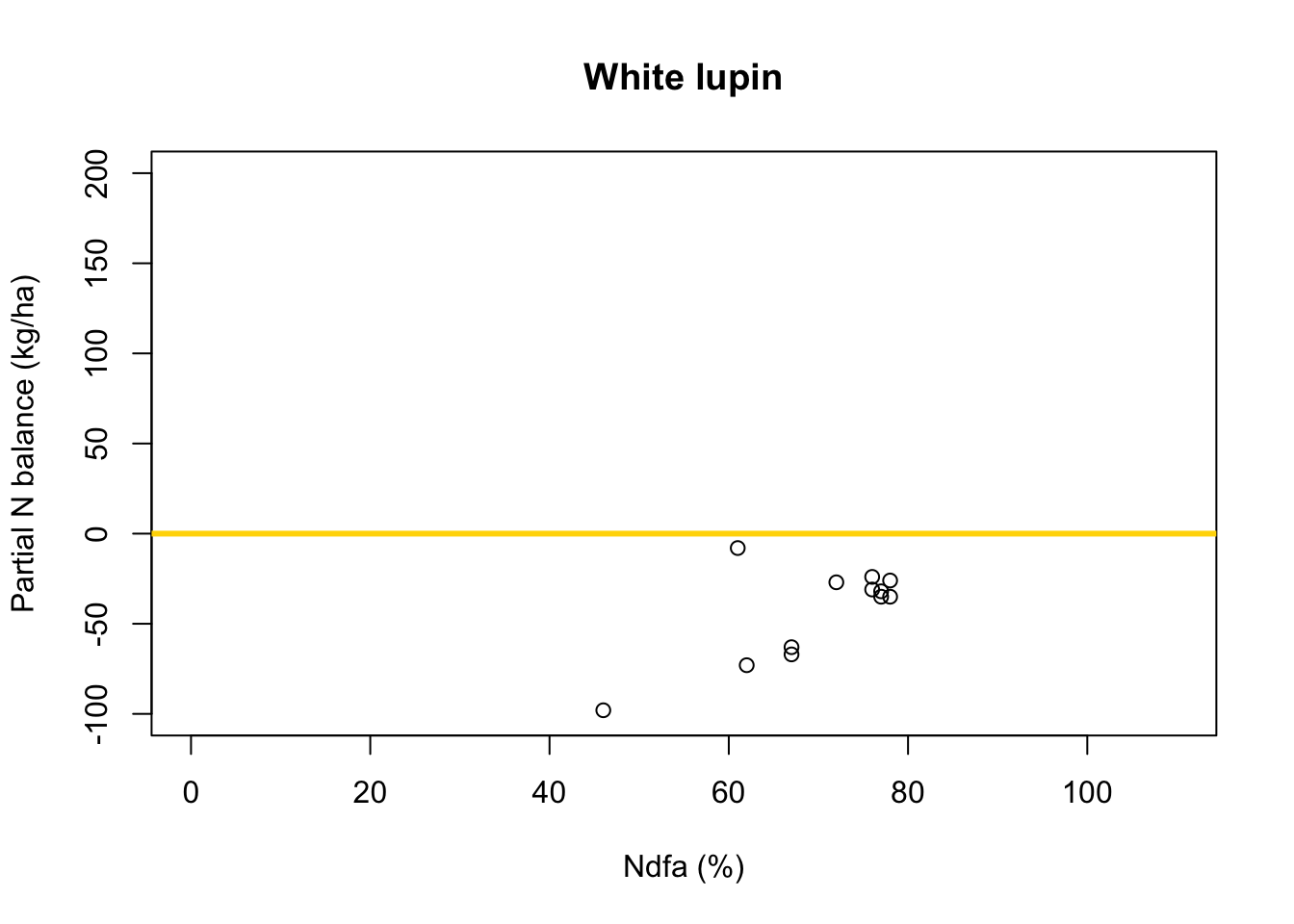
Using least squares
- What value of Ndfa is needed to achieve a neutral N balance?
beta0.hat <- as.numeric(coef(m1)[1]) beta1.hat <- as.numeric(coef(m1)[2]) theta.hat <- -beta0.hat/beta1.hat theta.hat## [1] 93.09456- Visual representation of \(\theta\)
# Plot data, line of best fit and theta plot(df.wl$Ndfa,df.wl$PartNbalance, xlab="Ndfa (%)",ylab="Partial N balance (kg/ha)", xlim=c(0,110),ylim=c(-100,200),main="White lupin") abline(a=0,b=0,col="gold",lwd=3) abline(m1,col="red",lwd=3) abline(v=93.1,lwd=3,lty=2,col="green")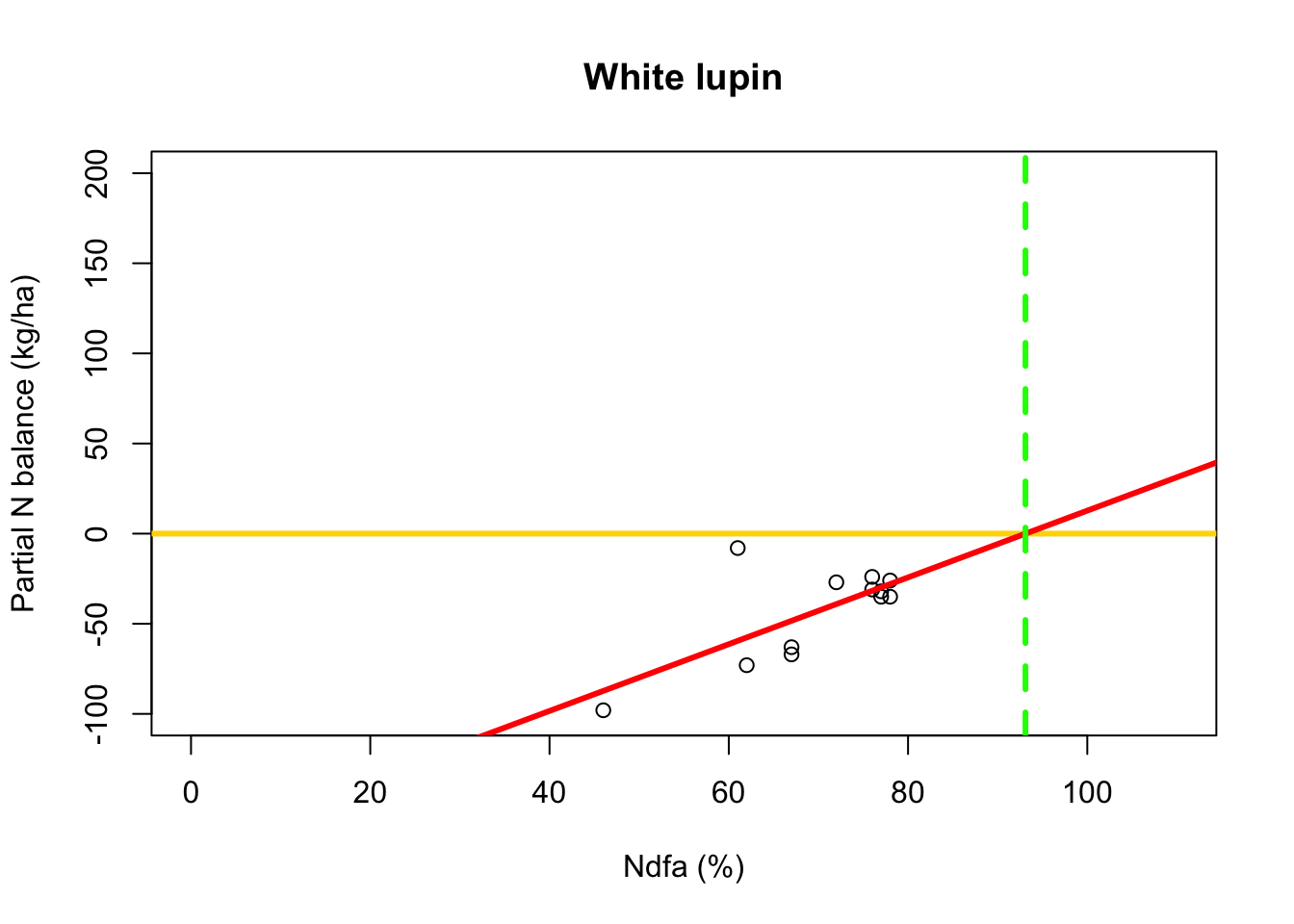
Using likelihood-based inference
- Fit linear model to data using using maximum likelihood estimation
library(nlme) # Fit simple linear regression model using maximum likelihood estimation m2 <- gls(PartNbalance ~ Ndfa,data=df.wl,method="ML")- What value of Ndfa is needed to achieve a neutral N balance?
# Use maximum likelihood estimate (MLE) to obtain estimate of theta beta0.hat <- as.numeric(coef(m2)[1]) beta1.hat <- as.numeric(coef(m2)[2]) theta.hat <- -beta0.hat/beta1.hat theta.hat## [1] 93.09456# Use delta method to obtain approximate approximate standard errors and # then construct Wald-type confidence intervals library(msm) theta.se <- deltamethod(~-x1/x2, mean=coef(m2), cov=vcov(m2)) theta.ci <- c(theta.hat-1.96*theta.se,theta.hat+1.96*theta.se) theta.ci## [1] 76.91135 109.27778- Visual representation of \(\theta\)
# Plot data, line of best fit and theta plot(df.wl$Ndfa,df.wl$PartNbalance, xlab="Ndfa (%)",ylab="Partial N balance (kg/ha)", xlim=c(0,110),ylim=c(-100,200),main="White lupin") abline(a=0,b=0,col="gold",lwd=3) abline(m1,col="red",lwd=3) abline(v=93.1,lwd=3,lty=2,col="green") abline(v=76.9,lwd=1,lty=2,col="green") abline(v=109.2,lwd=1,lty=2,col="green")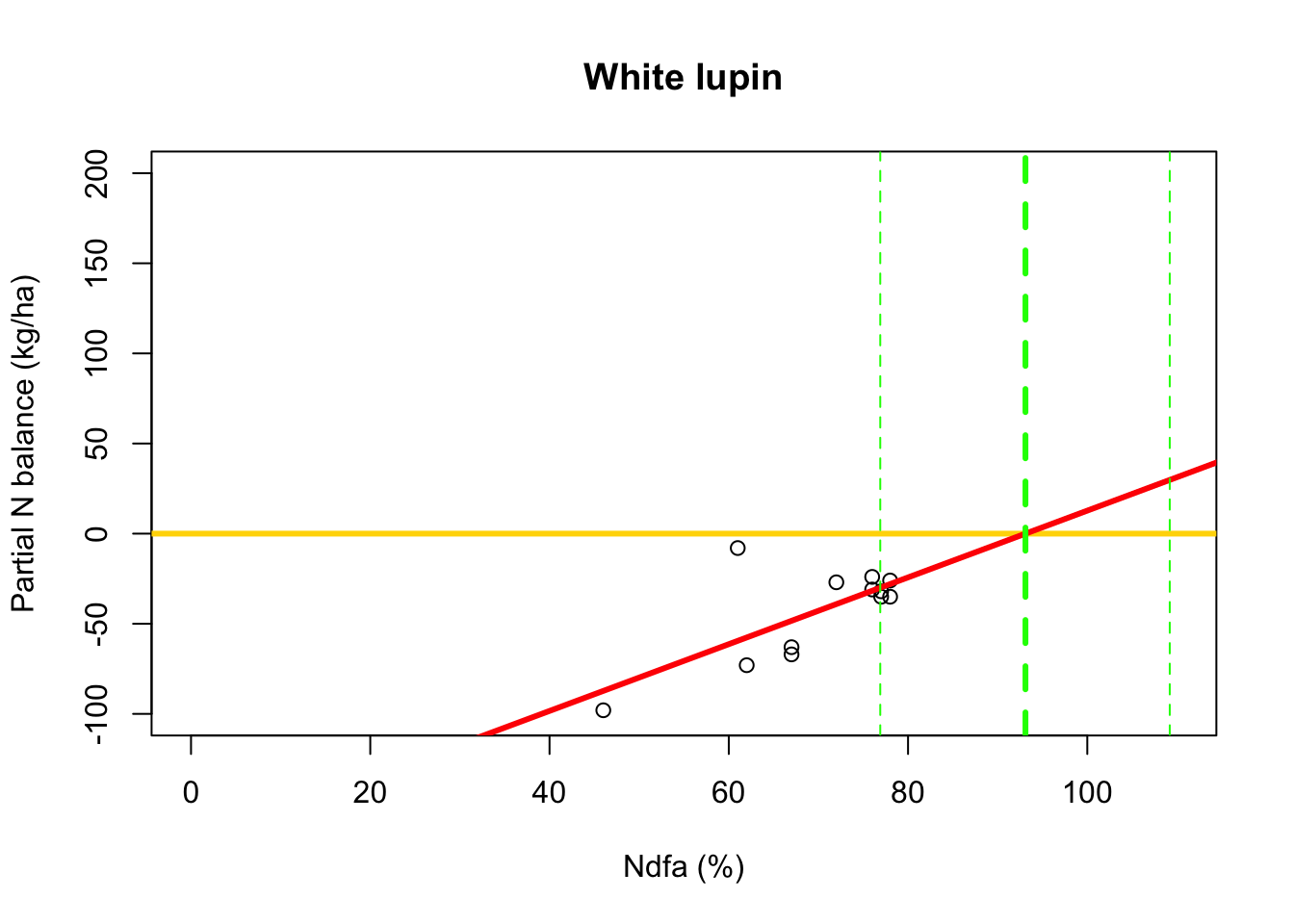
Using Bayesian inference
- Assume that \(y_i=\beta_{0}+\beta_{1}x_{i}+\varepsilon_i\) with \(\varepsilon_i\sim \text{N}(0,\sigma^2)\), \(\beta_{0}\sim \text{N}(0,10^6)\) and \(\beta_{1}\sim \text{N}(2.5,0.1)\)
- Model fitting
samples <- norm.reg.mcmc(y = df.wl$PartNbalance,X = model.matrix(~ Ndfa,data=df.wl), beta.mn = c(0,2.5),beta.var=c(10^6,0.1), s2.mn = 10, s2.sd = 10^6, n.mcmc = 5000) burn.in <- 1000 # Look a histograms of posterior distributions par(mfrow=c(2,1),mar=c(5,6,1,1)) hist(samples$beta.save[1,-c(1:1000)],xlab=expression(beta[0]*"|"*bold(y)),ylab=expression("["*beta[0]*"|"*bold(y)*"]"),freq=FALSE,col="grey",main="",breaks=30) hist(samples$beta.save[2,-c(1:1000)],xlab=expression(beta[1]*"|"*bold(y)),ylab=expression("["*beta[1]*"|"*bold(y)*"]"),freq=FALSE,col="grey",main="",breaks=30)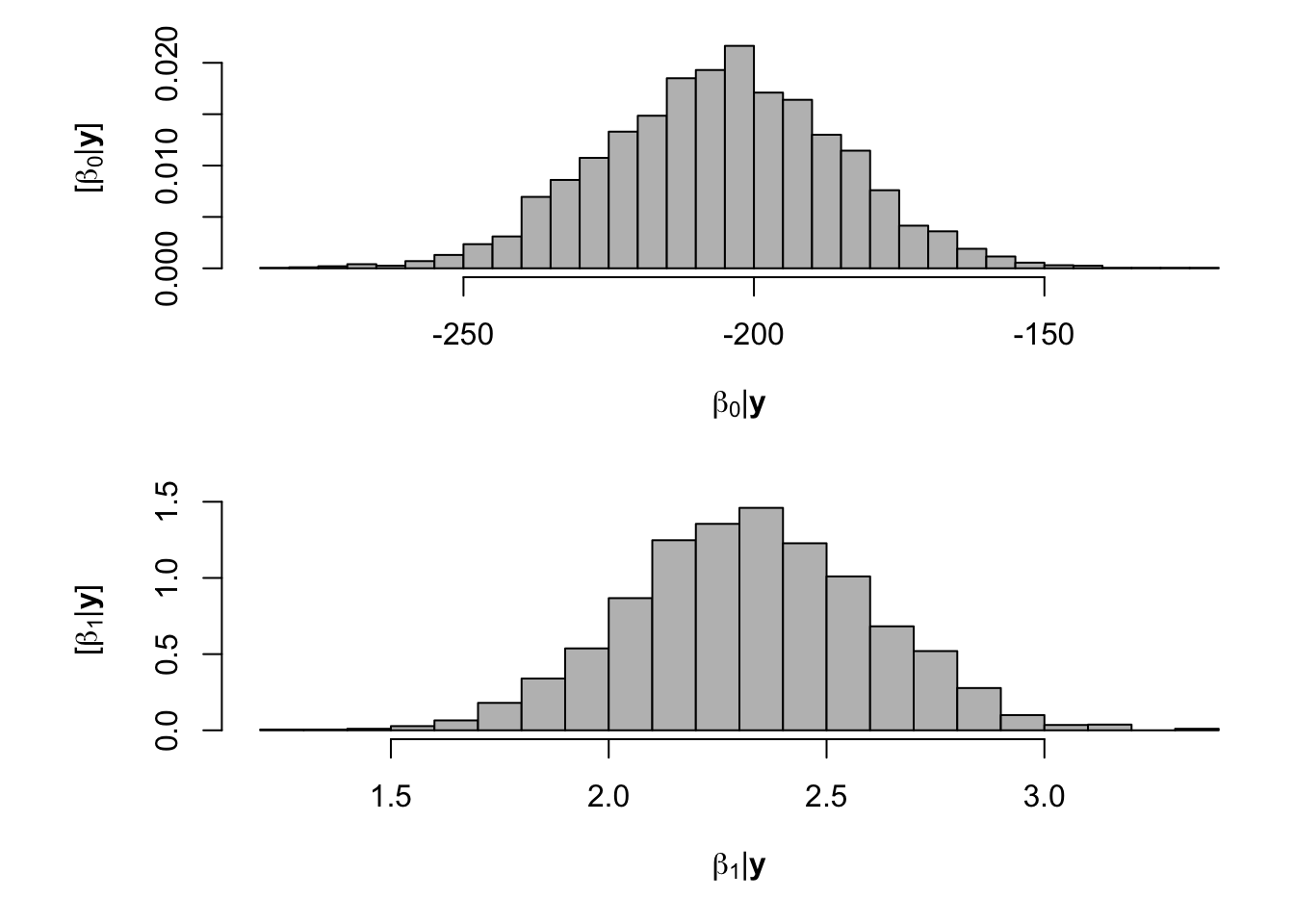
- What value of Ndfa is needed to achieve a neutral N balance?
hist(-samples$beta.save[1,-c(1:1000)]/samples$beta.save[2,-c(1:1000)],xlab=expression(theta*"|"*bold(y)),ylab=expression("["*theta*"|"*bold(y)*"]"),freq=FALSE,col="grey",main="",breaks=30)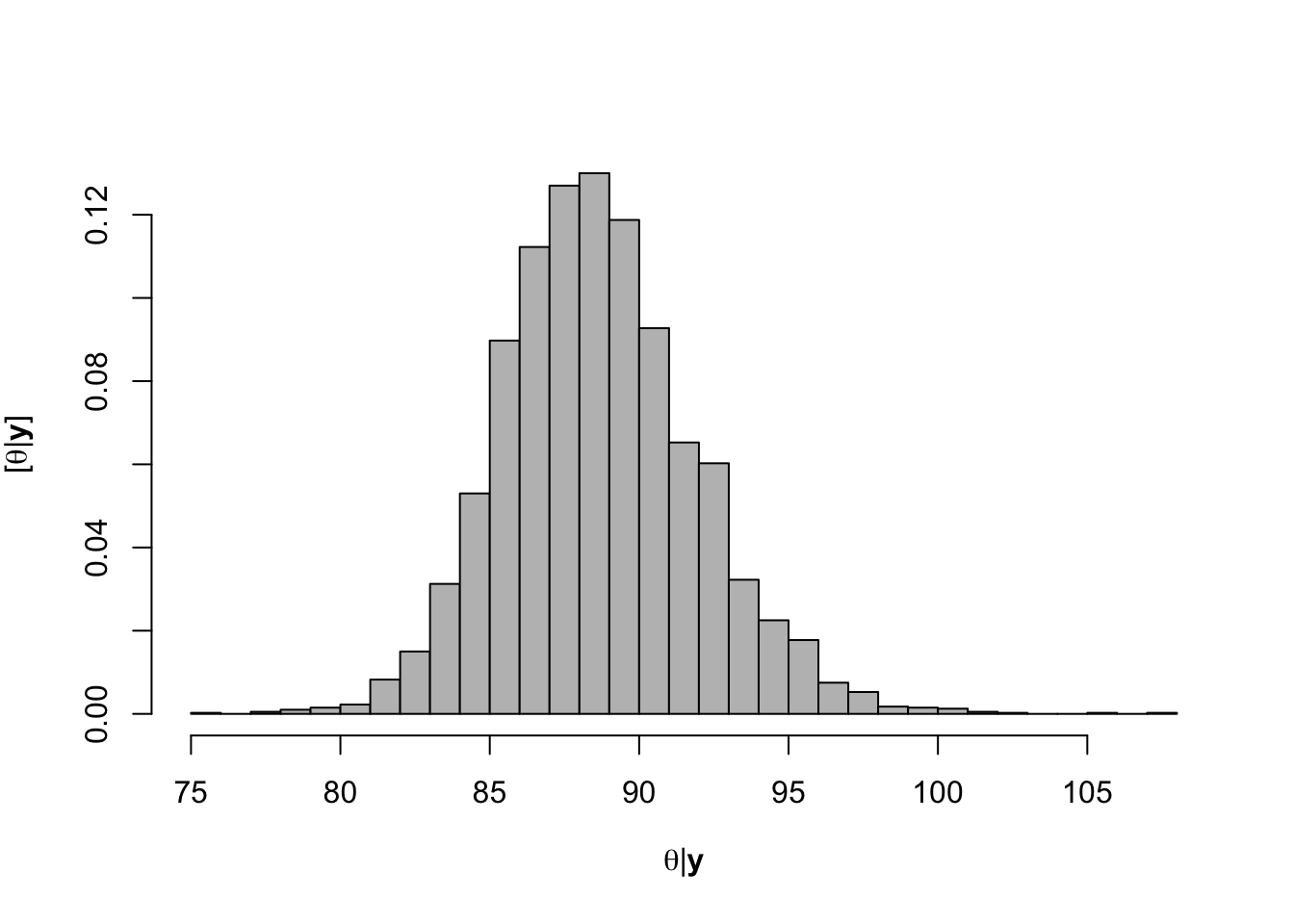
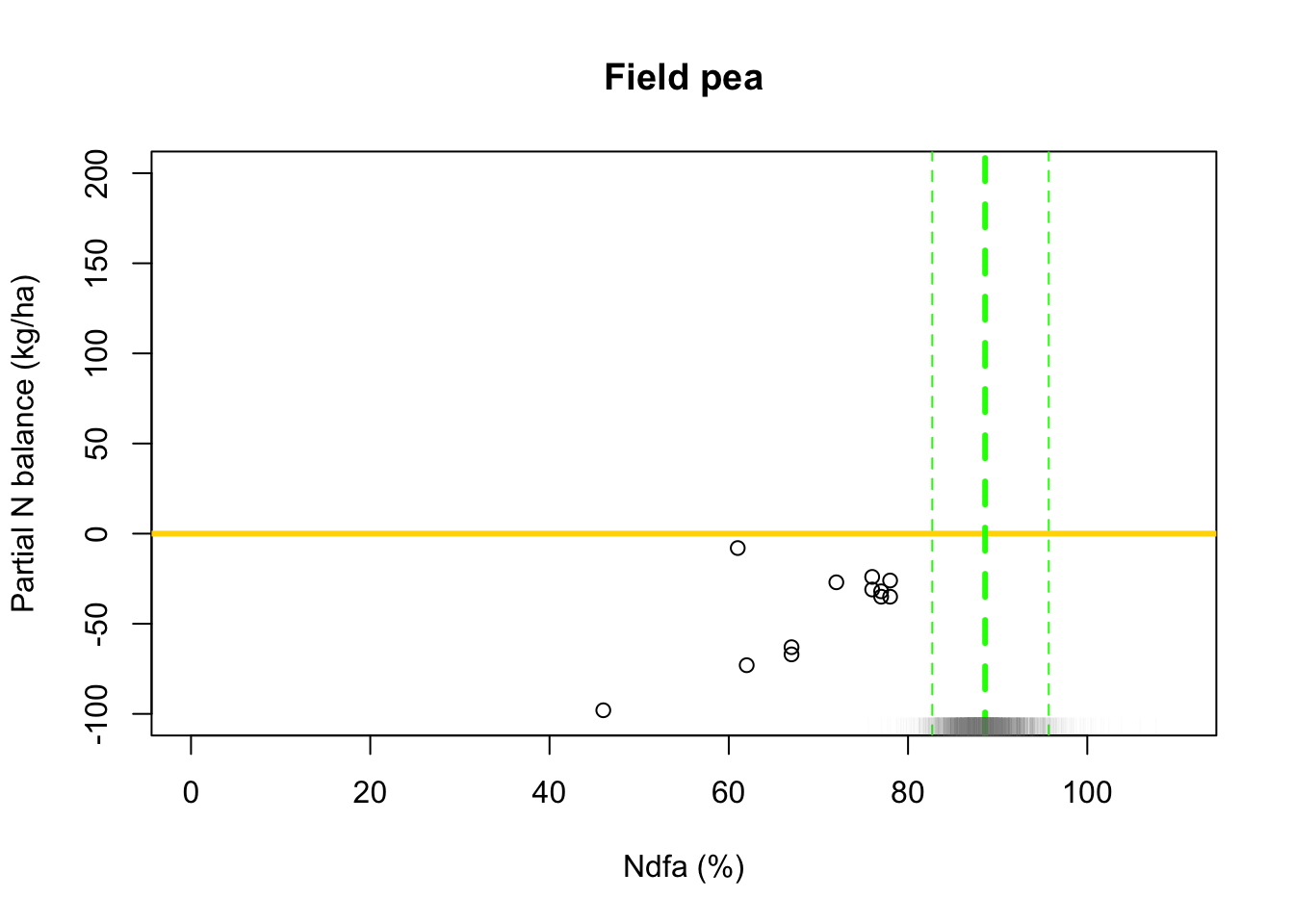
## [1] 88.59053# 95% credible intervals for theta quantile(-samples$beta.save[1,]/samples$beta.save[2,],prob=c(0.025,0.975))## 2.5% 97.5% ## 82.73209 95.53940# Plot data and theta plot(df.wl$Ndfa,df.wl$PartNbalance, xlab="Ndfa (%)",ylab="Partial N balance (kg/ha)", xlim=c(0,110),ylim=c(-100,200),main="Field pea") abline(a=0,b=0,col="gold",lwd=3) abline(v=88.6,lwd=3,lty=2,col="green") abline(v=82.7,lwd=1,lty=2,col="green") abline(v=95.7,lwd=1,lty=2,col="green") rug(-samples$beta.save[1,]/samples$beta.save[2,],col=gray(0.5,0.03))