14 Day 14 (June 21)
14.3 Bootstrap confidence intervals
Google scholar demonstration
Motivation (see section 3.6 in Faraway (2014))
- Functions of parameters (i.e., “derived” quantities)
- Difficult to obtain the sampling distribution for non-linear functions of estimated parameters
- Further reading (Hesterberg 2015)
- Example: When do we expect the population to go extinct?
y <- c(63, 68, 61, 44, 103, 90, 107, 105, 76, 46, 60, 66, 58, 39, 64, 29, 37, 27, 38, 14, 38, 52, 84, 112, 112, 97, 131, 168, 70, 91, 52, 33, 33, 27, 18, 14, 5, 22, 31, 23, 14, 18, 23, 27, 44, 18, 19) year <- 1965:2011 df <- data.frame(y = y, year = year) plot(x = df$year, y = df$y, xlab = "Year", ylab = "Annual count", main = "", col = "brown", pch = 20, xlim = c(1965, 2040)) m1 <- lm(y~year) abline(m1)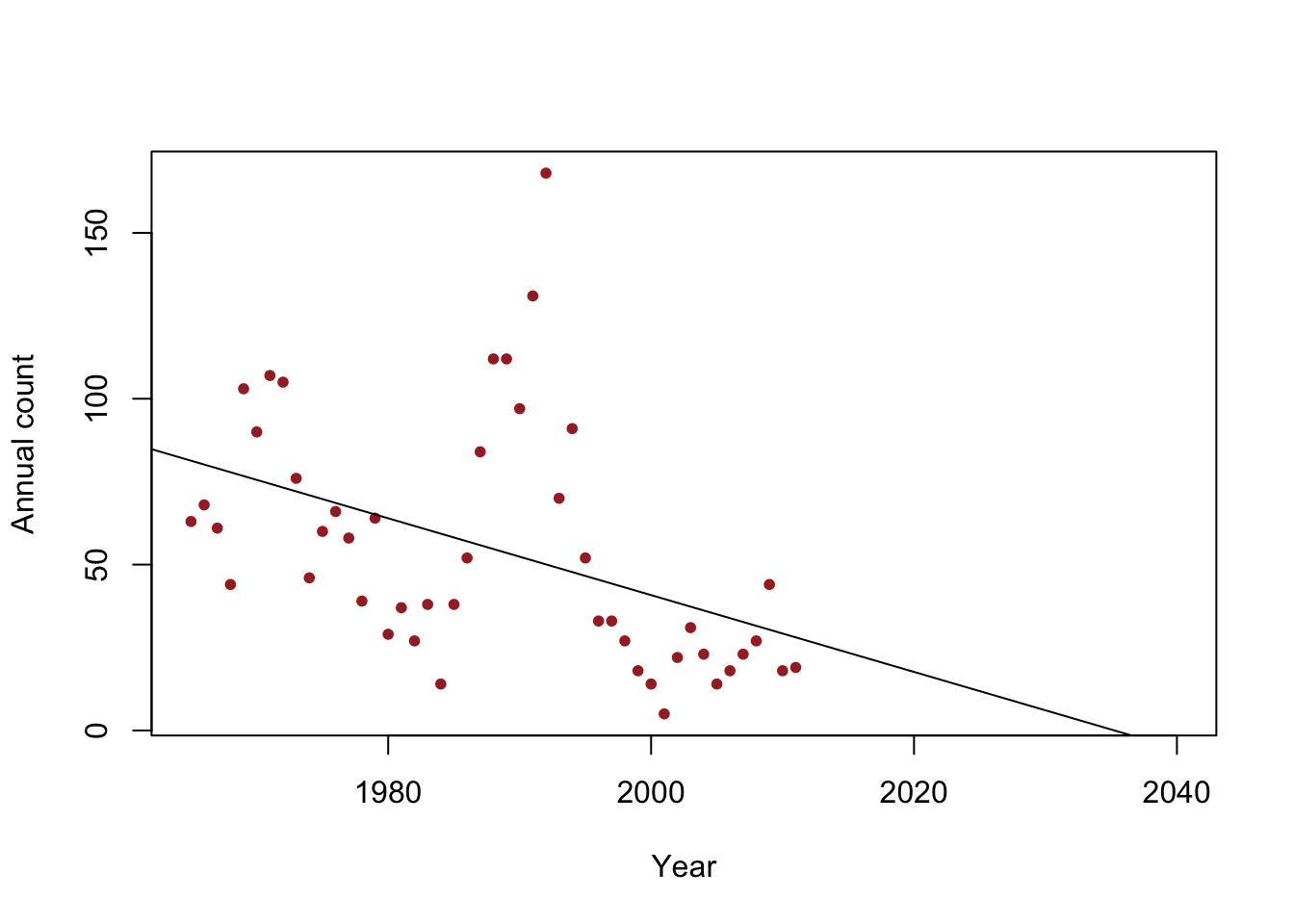
Non-parametric bootstrap (see Efron and Tibshirani (1994)).
- From a data set with n observations, take a sample of size n with replacement. For a linear model, the sample should include both the observed response (\(y_{i}\)) and covariates (\(x_{1}\),\(x_{2}\),…,\(x_{p}\)).
- Estimate the parameters for a statistical model using the sampled data from step 1.
- Save the estimates of interest. This could be parameters of interest (e.g., \(\boldsymbol{\beta}\)) or a a derived quantity (e.g., \(R^{2}\), \(\frac{1}{\boldsymbol{\beta}}\)).
- Repeats steps (1)-(3) \(m\) times.
Inference
- The \(m\) samples of the quantities of interest are samples from the empirical distribution.
- The empirical distribution can summarized using sample statistics (e.g., quantiles, mean, variance, etc). Conceptually, this is similar to Monte Carlo integration.
Example in R
library(latex2exp) # Number of bootstrap samples (m) m.boot <- 1000 # Create matrix to save empirical distribution of -beta2.hat/beta1.hat (expected time of extinction) ed.extinct.hat <- matrix(,m.boot,1) # Set random seed so results are the same if we run it again # Results would be different due to random resampling of data set.seed(1940) # Start for loop for non-parametric boostrap for(m in 1:m.boot){ # Sample data with replacement # boot.sample gives the rows of df that we use for estimation boot.sample <- sample(1:nrow(df),replace=TRUE) # Make temporary data frame that contains the resamples df.temp <- df[boot.sample,] # Estimate parameters for df.temp m1 <- lm(y~year,data=df.temp) # Save estimate of -beta0.hat/beta1.hat (expected time of extinction) ed.extinct.hat[m,] <- -coef(m1)[1]/coef(m1)[2] } par(mar=c(5,4,7,2)) hist(ed.extinct.hat,col="grey",xlab="Year",main=TeX('Empirical distribuiton of $$-$$\\hat{\\frac{$\\beta_0}{$\\beta_1}}'),freq=FALSE,breaks=20)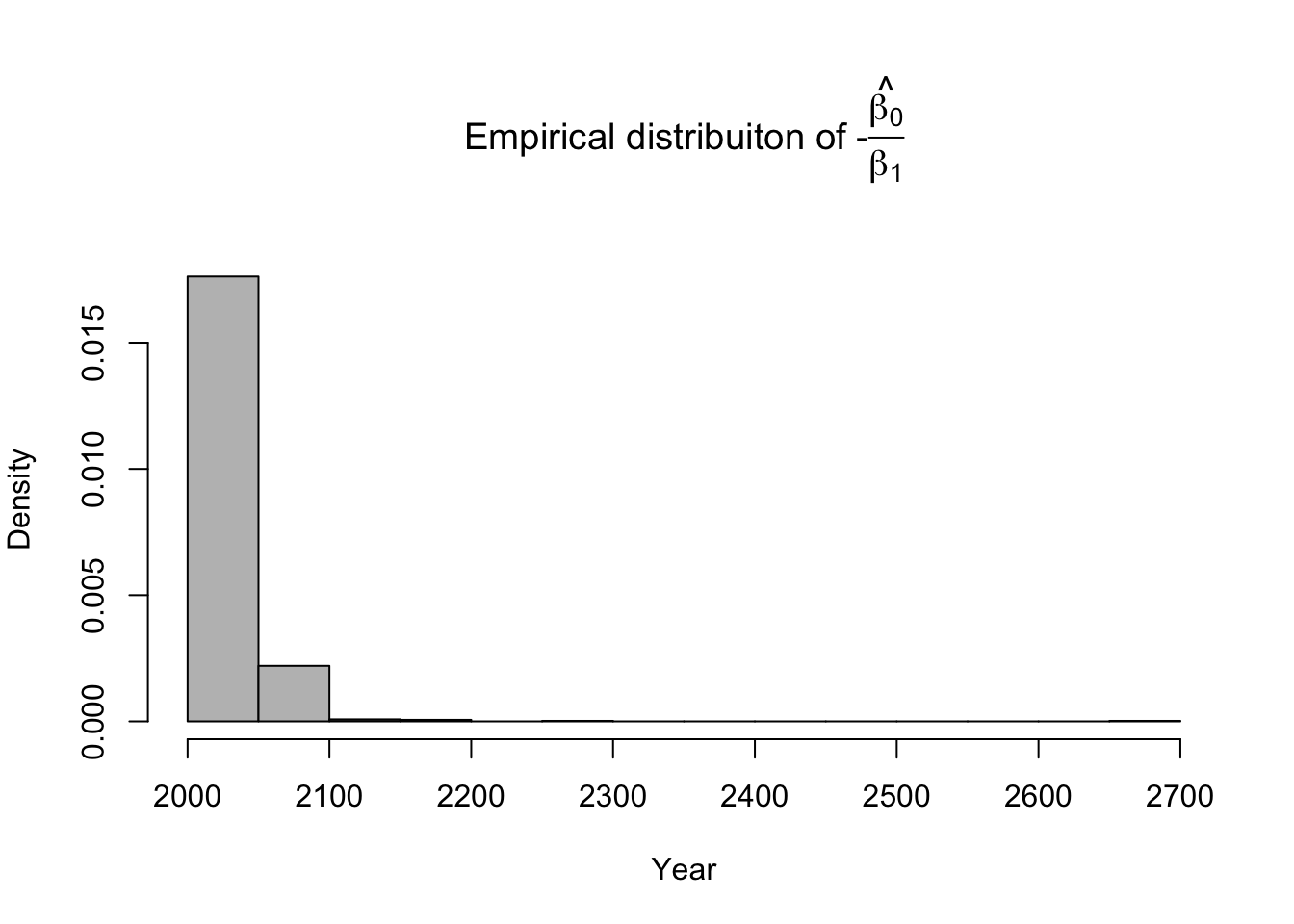
# 95% equal-tailed CI based on percentiles of the empirical distribution quantile(ed.extinct.hat,prob=c(0.025,0.975))## 2.5% 97.5% ## 2021.203 2077.168
Literature cited
Efron, Bradley, and Robert J Tibshirani. 1994. An Introduction to the Bootstrap. CRC press.
Faraway, J. J. 2014. Linear Models with r. CRC Press.
Hesterberg, Tim C. 2015. “What Teachers Should Know about the Bootstrap: Resampling in the Undergraduate Statistics Curriculum.” The American Statistician 69 (4): 371–86.