ncm(otutab) -> ncm_res
plot(ncm_res)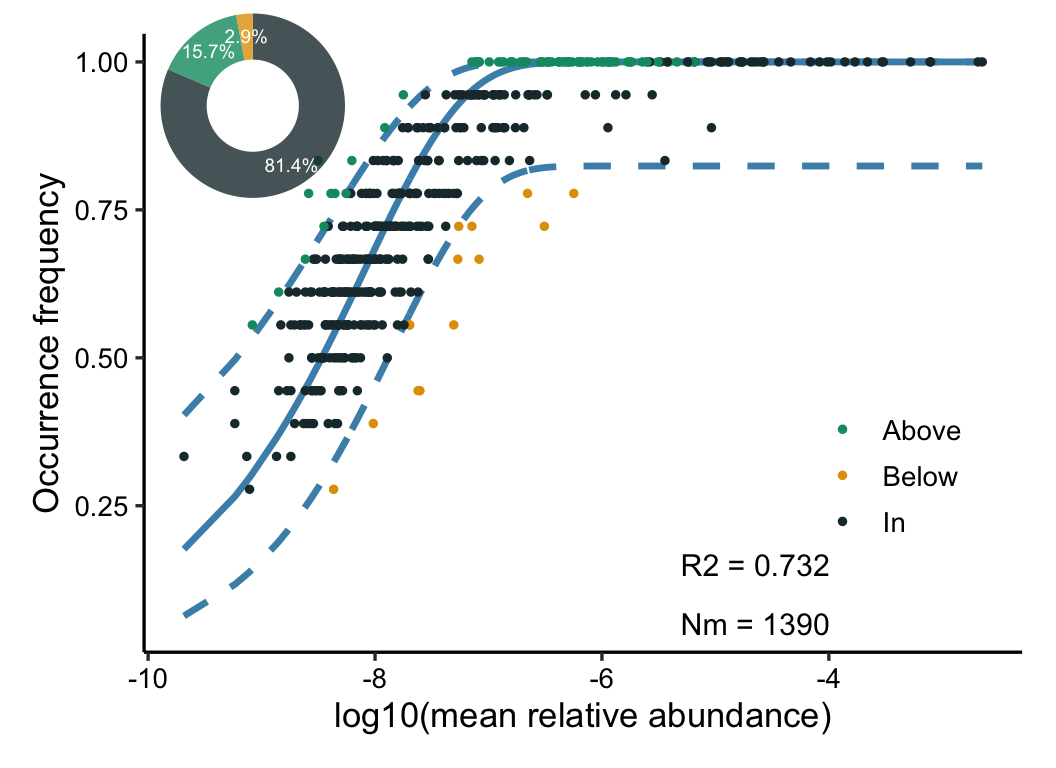
Community assembly in microbiome refers to the processes that shape the composition, diversity, and structure of microbial communities in a particular environment or host. Microbiome consist of diverse microbial populations that interact with each other and their surroundings, and understanding how these communities assemble is crucial for comprehending their ecological dynamics and functional implications.
For more details on diversity analysis, you can refer to Microbial community construction
It is only when species die or leave the system that community structure changes. At this point, the niche of the departing individual will become vacant, and other individuals will fill the niche through migration from outside the community or reproduction within the community. The dynamics of a community can therefore be described as a cycle of death - reproduction/diffusion - death.
ncm(otutab) -> ncm_res
plot(ncm_res)
R2 represents the overall goodness of fit of the neutral community model, and the higher the R2, the closer it is to the neutral model, that is, the more the community construction is affected by the stochastic process and the less the deterministic process.
df2tree(taxonomy) -> phylo
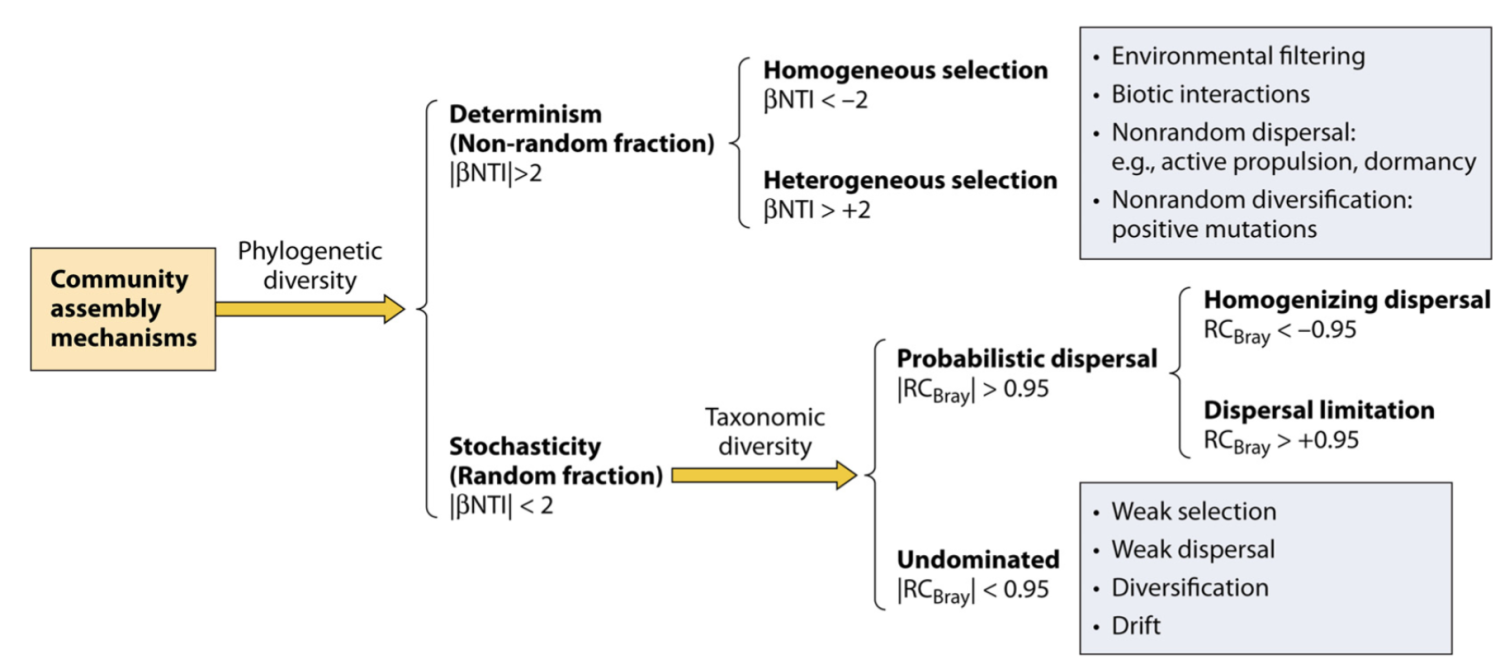
nti_rc(otutab, phylo, metadata["Group"], file = NULL) -> nti_res
plot(nti_res) + scale_fill_brewer(palette = "Set3")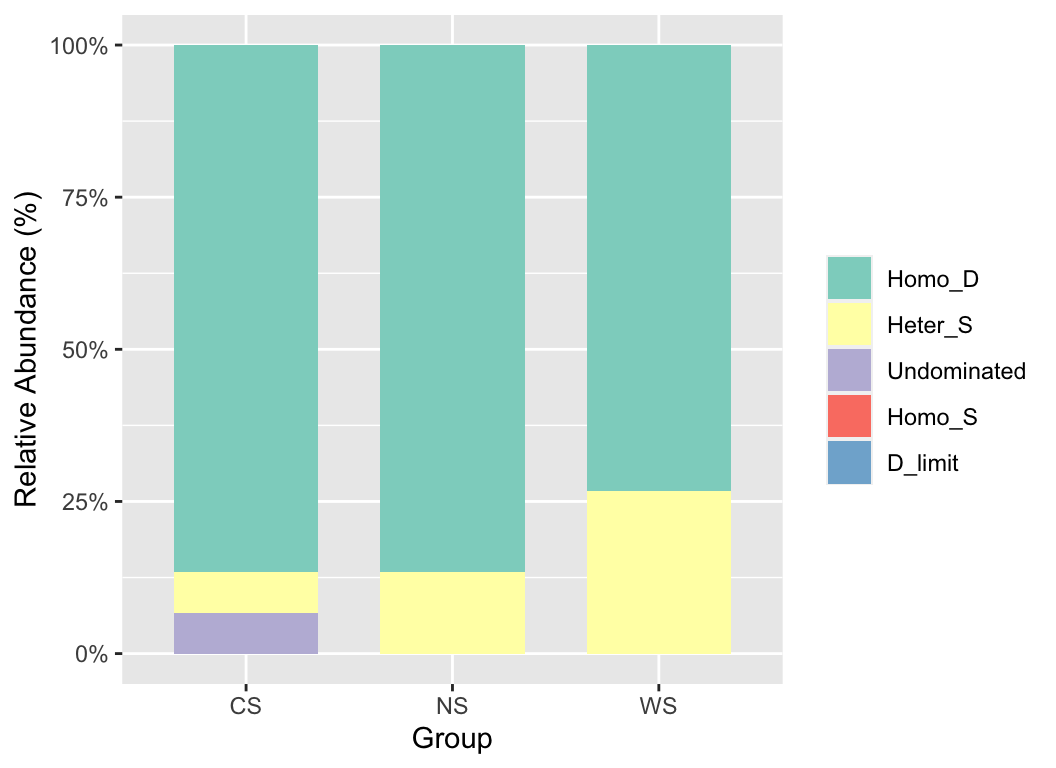
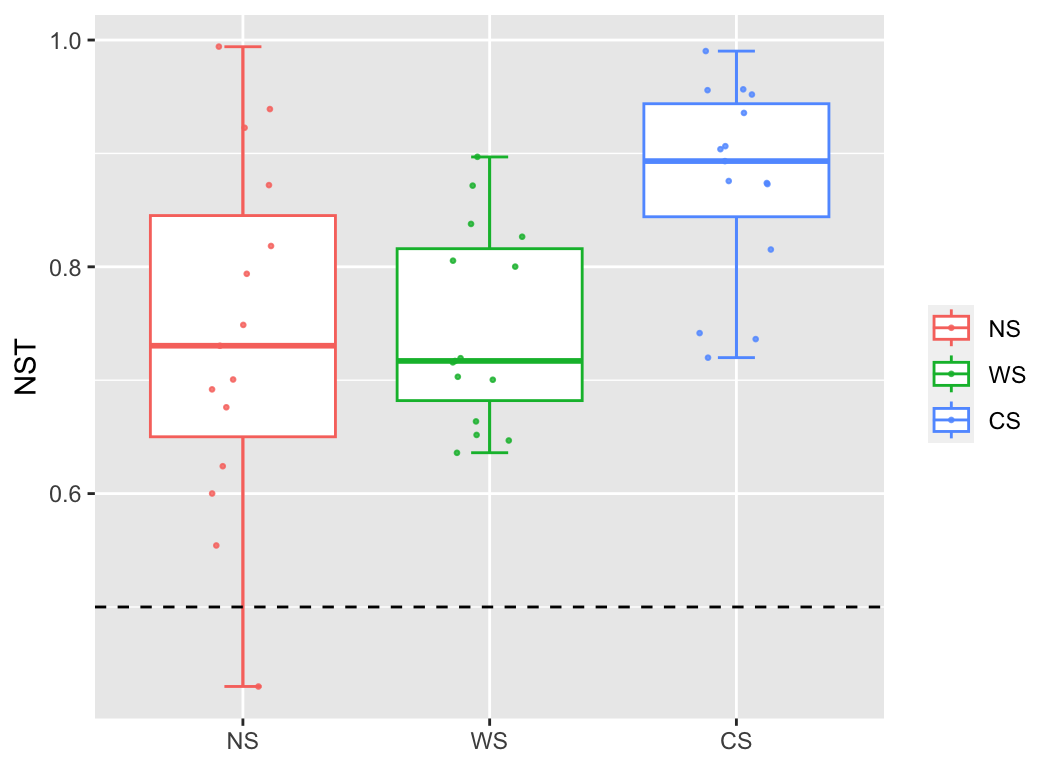
A new method is developed to quantify randomness in ecological processes. A new index, normalized stochasticity ratio (NST), is proposed as the boundary point between deterministic dominance (<50%) and stochastic dominance (>50%).
nst(otutab, metadata["Group"], file = NULL) -> nst_res
plot(nst_res, c_group = "intra") +
geom_hline(yintercept = 0.5, lty = 2) + ylab("NST")