Chapter 4 Scripting and Writing Functions in R
4.1 Overview
4.2 Introduction
[1] 4.3 7.1 6.3 5.2 3.2Tree is smallfor (i in 1:3) {
if (tree.heights[i] < 6) { cat('Tree',i,' is small\n') }
else { cat('Tree',i,' is large\n')} }Tree 1 is small
Tree 2 is large
Tree 3 is largeassess.tree.height <- function(tree.list, thresh)
{ for (i in 1:length(tree.list))
{ if(tree.list[i] < thresh) {cat('Tree',i, ' is small\n')}
else { cat('Tree',i,' is large\n')}
}
}
assess.tree.height(tree.heights, 6)Tree 1 is small
Tree 2 is large
Tree 3 is large
Tree 4 is small
Tree 5 is smallTree 1 is large
Tree 2 is large
Tree 3 is large
Tree 4 is small
Tree 5 is small4.3 Building blocks for Programs
4.3.1 Conditional Statements
x is negativex is negativex is positiveAll numbers are positiveSome numbers are positive[1] FALSE4.3.2 Code Blocks: \(\{\) and \(\}\)
x <- c(1,3,6,8,9,5)
if (all(x > 0)) {
cat("All numbers are positive\n")
total <- sum(x)
cat("Their sum is ",total) }All numbers are positive
Their sum is 32x <- c(1,3,6,8,9,-5)
if (all(x > 0)) {
cat("All numbers are positive\n")
total <- sum(x)
cat("Their sum is ",total) } else {
cat("Not all numbers are positive\n")
cat("This is probably an error as numbers are rainfall levels") }Not all numbers are positive
This is probably an error as numbers are rainfall levels4.3.3 Functions
mean.rainfall <- function(rf)
{ if (all(rf> 0)) #open Function
{ mean.value <- mean(rf) #open Consequent
cat("The mean is ",mean.value)
} else #close Consequent
{ cat("Warning: Not all values are positive\n") #open Alternative
} #close Alternative
} #close Function
mean.rainfall(c(8.5,9.3,6.5,9.3,9.4)) The mean is 8.6mean.rainfall2 <- function(rf) {
if (all(rf> 0)) {
return( mean(rf))} else {
return(NA)}
}
mr <- mean.rainfall2(c(8.5,9.3,6.5,9.3,9.4))
mr[1] 8.6[1] 8.6[1] "Tuesday"4.3.4 Loops and repetition
The cube of 1 is 1
The cube of 2 is 8
The cube of 3 is 27
The cube of 4 is 64
The cube of 5 is 125 for (val in seq(0,1,by=0.25)) {
val.squared <- val * val
cat("The square of",val,"is ",val.squared,"\n")}The square of 0 is 0
The square of 0.25 is 0.0625
The square of 0.5 is 0.25
The square of 0.75 is 0.5625
The square of 1 is 1 i <- 1; n <- 654
repeat{
i.squared <- i * i
if (i.squared > n) break
i <- i + 1}
cat("The first square number exceeding",n, "is ",i.squared,"\n")The first square number exceeding 654 is 676 first.bigger.square <- function(n) {
i <- 1
repeat{
i.squared <- i * i
if (i.squared > n) break
i <- i + 1 }
return(i.squared)}
first.bigger.square(76987)[1] 772844.3.5 Debugging
4.4 Writing Functions
4.4.1 Introduction
[1] 34.4.2 Data Checking
[1] NaNcube.root <- function(x) {
if (x >= 0) {
result <- x ^ (1/3) } else {
result <- -(-x) ^ (1/3) }
return(result)} [1] 1.44225[1] -1.442254.4.3 More Data Checking
4.4.4 Loops Revisited
4.4.4.1 Conditional Loops
4.4.4.2 Deterministic Loops
4.4.5 Further Activity
library(GISTools)
library(sf)
data(georgia)
# create an empty list for the results
adj.list <- list()
# convert georgia to sf
georgia_sf <- st_as_sf(georgia2)
# extract a single county
county.i <- georgia_sf[1,]
# determine the adjacent counties
# the [-1] removes Appling form its own list
adj.i <- unlist(st_intersects(county.i, georgia_sf))[-1]
# extract their names
adj.names.i <- georgia2$Name[adj.i]
# add to the list
adj.list[[1]] <- adj.i
# name the list elements
names(adj.list[[1]]) <- adj.names.i[[1]]
Bacon Jeff Davis Pierce Tattnall Toombs
3 80 113 132 138
Wayne
151 4.5 Spatial data structures
[1] "list" [,1] [,2]
[1,] 1292287 1075896
[2,] 1292654 1075919
[3,] 1292949 1075590
[4,] 1294045 1075841
[5,] 1294603 1075472
[6,] 1295467 1075621.
# plot Appling
plot(georgia.polys[[1]],asp=1,type='l',
xlab = "Easting", ylab = "Northing")
# plot adjacent county outlines
points(georgia.polys[[3]],asp=1,type='l', col = "red")
points(georgia.polys[[151]],asp=1,type='l', col = "blue", lty = 2)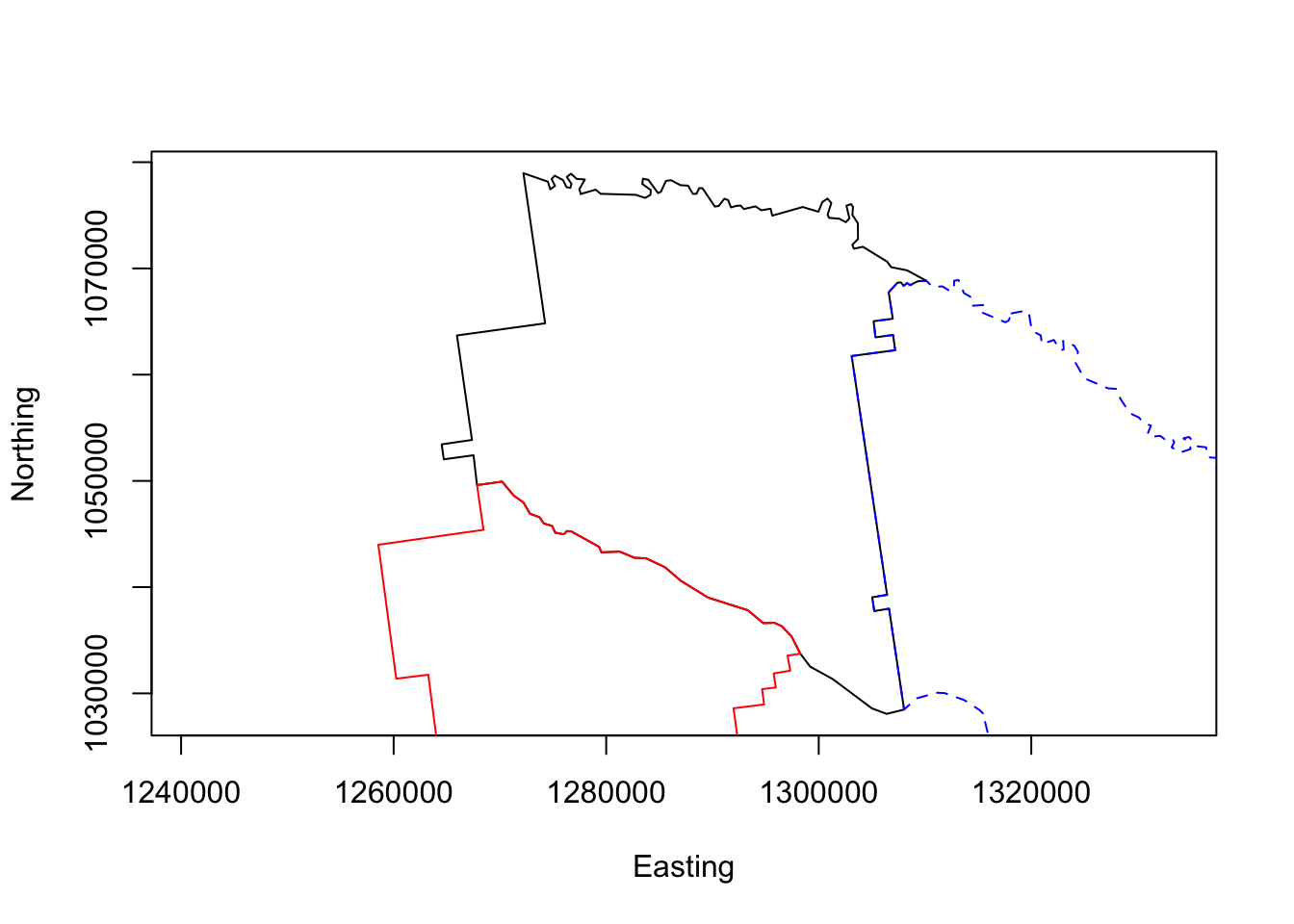
Figure 4.1: A simple plot of Appling County and 2 adjacent counties.
4.6 Apply functions
library(GISTools)
data(newhaven)
## the @data route
head(blocks@data[, 14:17])
## the data frame route
head(data.frame(blocks[, 14:17]))# set up vector to hold result
result.vector <- vector()
for (i in 1:nrow(blocks@data)){
# for each row determine which column has the max value
result.i <- which.max(data.frame(blocks[i,14:17]))
# put into the result vector
result.vector <- append(result.vector, result.i)
}res.vec <-apply(data.frame(blocks[,14:17]), 1, which.max)
# comapre the two results
identical(as.vector(res.vec), as.vector(result.vector))# Loop
t1 <- Sys.time()
result.vector <- vector()
for (i in 1:nrow(blocks@data)){
result.i <- which.max(data.frame(blocks[i,14:17]))
result.vector <- append(result.vector, result.i)
}
Sys.time() - t1
# Apply
t1 <- Sys.time()
res.vec <-apply(data.frame(blocks[,14:17]), 1, which.max)
Sys.time() - t1plot(bbox(georgia2)[1,], bbox(georgia2)[2,], asp = 1,
type='n',xlab='',ylab='',xaxt='n',yaxt='n',bty='n')
for (i in 1:length(georgia.polys)){
points(georgia.polys[[i]], type='l')
# small delay so that you can see the plotting
Sys.sleep(0.05)
}plot(bbox(georgia2)[1,], bbox(georgia2)[2,], asp = 1,
type='n',xlab='',ylab='',xaxt='n',yaxt='n',bty='n')
invisible(mapply(polygon,georgia.polys))# create a distance matrix
dMat <- as.matrix(dist(coordinates(georgia2)))
dim(dMat)
# create an empty vector
count.vec <- vector()
# create an empty list
names.list <- list()
# for each county...
for( i in 1:nrow(georgia_sf)) {
# which counties are within 50km
vec.i <- which(dMat[i,] <= 50000)
# add to the vector
count.vec <- append(count.vec, length(vec.i))
# find their names
names.i <- georgia_sf$Name[vec.i]
# add to the list
names.list[[i]] <- names.i
}
# have a look!
count.vec
names.list4.7 Manipulating data with dplyr
library(nycflights13)
library(tidyverse)
# select the variables
flights2 <- flights %>% select(origin, dest)
# remove Alaska and Hawaii
flights2 <- flights2[-grep("ANC", flights2$dest),]
flights2 <- flights2[-grep("HNL", flights2$dest),]
# group by destination
flights2 <- group_by(flights2, dest)
flights2 <- summarize(flights2, count = n())
# assign Lat and Lon for Origin
flights2$OrLat <- 40.6925
flights2$OrLon <- -74.16867
# have a look!
flights2# A tibble: 103 x 4
dest count OrLat OrLon
<chr> <int> <dbl> <dbl>
1 ABQ 254 40.7 -74.2
2 ACK 265 40.7 -74.2
3 ALB 439 40.7 -74.2
4 ATL 17215 40.7 -74.2
5 AUS 2439 40.7 -74.2
6 AVL 275 40.7 -74.2
7 BDL 443 40.7 -74.2
8 BGR 375 40.7 -74.2
9 BHM 297 40.7 -74.2
10 BNA 6333 40.7 -74.2
# ... with 93 more rowslibrary(maps)
library(geosphere)
# origin and desitnation examples
dest.eg <- matrix(c(77.1025, 28.7041), ncol = 2)
origin.eg <- matrix(c(-74.16867, 40.6925), ncol = 2)
# map the world from the maps package data
map("world", fill=TRUE, col="white", bg="lightblue")
# plot the points
points(dest.eg, col="red", pch=16, cex = 2)
points(origin.eg, col = "cyan", pch = 16, cex = 2)
# add the route
for (i in 1:nrow(dest.eg)) {
lines(gcIntermediate(dest.eg[i,], origin.eg[i,], n=50,
breakAtDateLine=FALSE, addStartEnd=FALSE,
sp=FALSE, sepNA), lwd = 2, lty = 2)
}4.8 Answers to self-test questions
Q1
cube.root.2 <- function(x)
{ if (is.numeric(x))
{ result <- sign(x)*abs(x)^(1/3)
return(result)
} else
{ cat("WARNING: Input must be numerical, not character\n")
return(NA) }
}Q2
gcd <- function(a,b)
{
divisor <- min(a,b) # line 1
dividend <- max(a,b) # line 1
repeat #line 5
{ remainder <- dividend %% divisor #line 2
dividend <- divisor # line 3
divisor <- remainder # line 4
if (remainder == 0) break #line 6
}
return(dividend)
}Q3 part i)
gcd.60 <- function(a)
{
for(i in 1:a)
{ divisor <- min(i,60)
dividend <- max(i,60)
repeat
{ remainder <- dividend %% divisor
dividend <- divisor
divisor <- remainder
if (remainder == 0) break
}
cat(dividend, "\n")
}
}Q3 part ii)
gcd.all <- function(x,y)
{ for(n1 in 1:x)
{ for (n2 in 1:y)
{ dividend <- gcd(n1, n2)
cat("when x is",n1,"& y is",n2,"dividend =",dividend,"\n")
}
}
}Q4
cube.root.table <- function(n)
{ for (x in seq(0.5, n, by = 0.5))
{ cat("The cube root of ",x," is",
sign(x)*abs(x)^(1/3),"\n") }
}cube.root.table <- function(n)
{ if (n < 0 ) by.val = 0.5
if (n < 0 ) by.val =-0.5
for (x in seq(0.5, n, by = by.val))
{ cat("The cube root of ",x," is",
sign(x)*abs(x)^(1/3),"\n") }
}Q5
# create an empty list for the results
adj.list <- list()
# convert georgia to sf
georgia_sf <- st_as_sf(georgia2)
for (i in 1:nrow(georgia_sf)) {
# extract a single county
county.i <- georgia_sf[i,]
# determine the adjacent counties
# the [-1] removes Appling form its own list
adj.i <- unlist(st_intersects(county.i, georgia_sf))[-1]
# extract their names
adj.names.i <- georgia2$Name[adj.i]
# add to the list
adj.list[[i]] <- adj.i
# name the list elements
names(adj.list[[i]]) <- adj.names.i
}Q6
return.adj <- function(sf.data){
# convert to sf regardless!
sf.data <- st_as_sf(sf.data)
adj.list <- list()
for (i in 1:nrow(sf.data)) {
# extract a single county
poly.i <- sf.data[i,]
# determine the adjacent counties
adj.i <- unlist(st_intersects(poly.i, sf.data))[-1]
# add to the list
adj.list[[i]] <- adj.i
}
return(adj.list)
}
# test it!
return.adj(georgia_sf)
return.adj(blocks)Q7
# number of counties within 50km
my.func1 <- function(x){
vec.i <- which(x <= 50000)[-i]
return(length(vec.i))
}
# their names
my.func2 <- function(x){
vec.i <- which(x <= 50000)
names.i <- georgia_sf$Name[vec.i]
return(names.i)
}
count.vec <- apply(dMat,1, my.func1)
names.list <- apply(dMat,1, my.func2)Q8
# Part 1: the join
flights2 <- flights2 %>% left_join(airports, c("dest" = "faa"))
flights2 <- flights2 %>% select(count,dest,OrLat,OrLon,
DestLat=lat,DestLon=lon)
# get rid of any NAs
flights2 <- flights2[!is.na(flights2$DestLat),]
flights2
# Part 2: the plot
# Using standard plots
dest.eg <- matrix(c(flights2$DestLon, flights2$DestLat), ncol = 2)
origin.eg <- matrix(c(flights2$OrLon, flights2$OrLat), ncol = 2)
map("usa", fill=TRUE, col="white", bg="lightblue")
points(dest.eg, col="red", pch=16, cex = 1)
points(origin.eg, col = "cyan", pch = 16, cex = 1)
for (i in 1:nrow(dest.eg)) {
lines(gcIntermediate(dest.eg[i,], origin.eg[i,], n=50,
breakAtDateLine=FALSE,
addStartEnd=FALSE, sp=FALSE, sepNA))
}
# using ggplot
all_states <- map_data("state")
dest.eg <- data.frame(DestLon = flights2$DestLon,
DestLat = flights2$DestLat)
origin.eg <- data.frame(OrLon = flights2$OrLon,
OrLat = flights2$OrLat)
library(GISTools)
# Figure 2 using ggplot
# create the main plot
mp <- ggplot() +
geom_polygon( data=all_states,
aes(x=long, y=lat, group = group),
colour="white", fill="grey20") +
coord_fixed() +
geom_point(aes(x = dest.eg$DestLon, y = dest.eg$DestLat),
color="#FB6A4A", size=2) +
theme(axis.title.x=element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank(),
axis.title.y=element_blank(),
axis.text.y=element_blank(),
axis.ticks.y=element_blank())
# create some transparent shading
cols = add.alpha(colorRampPalette(brewer.pal(9,"Reds"))(nrow(flights2)), 0.7)
# loop through the destinations
for (i in 1:nrow(flights2)) {
# line thickness related flights
lwd.i = 1+ (flights2$count[i]/max(flights2$count))
# a sequence of colours
cols.i = cols[i]
# create a dataset
link <- as.data.frame(gcIntermediate(dest.eg[i,], origin.eg[i,], n=50,
breakAtDateLine=FALSE, addStartEnd=FALSE, sp=FALSE, sepNA))
names(link) <- c("lon", "lat")
mp <- mp + geom_line(data=link, aes(x=lon, y=lat),
color= cols.i, size = lwd.i)
}
# plot!
mp