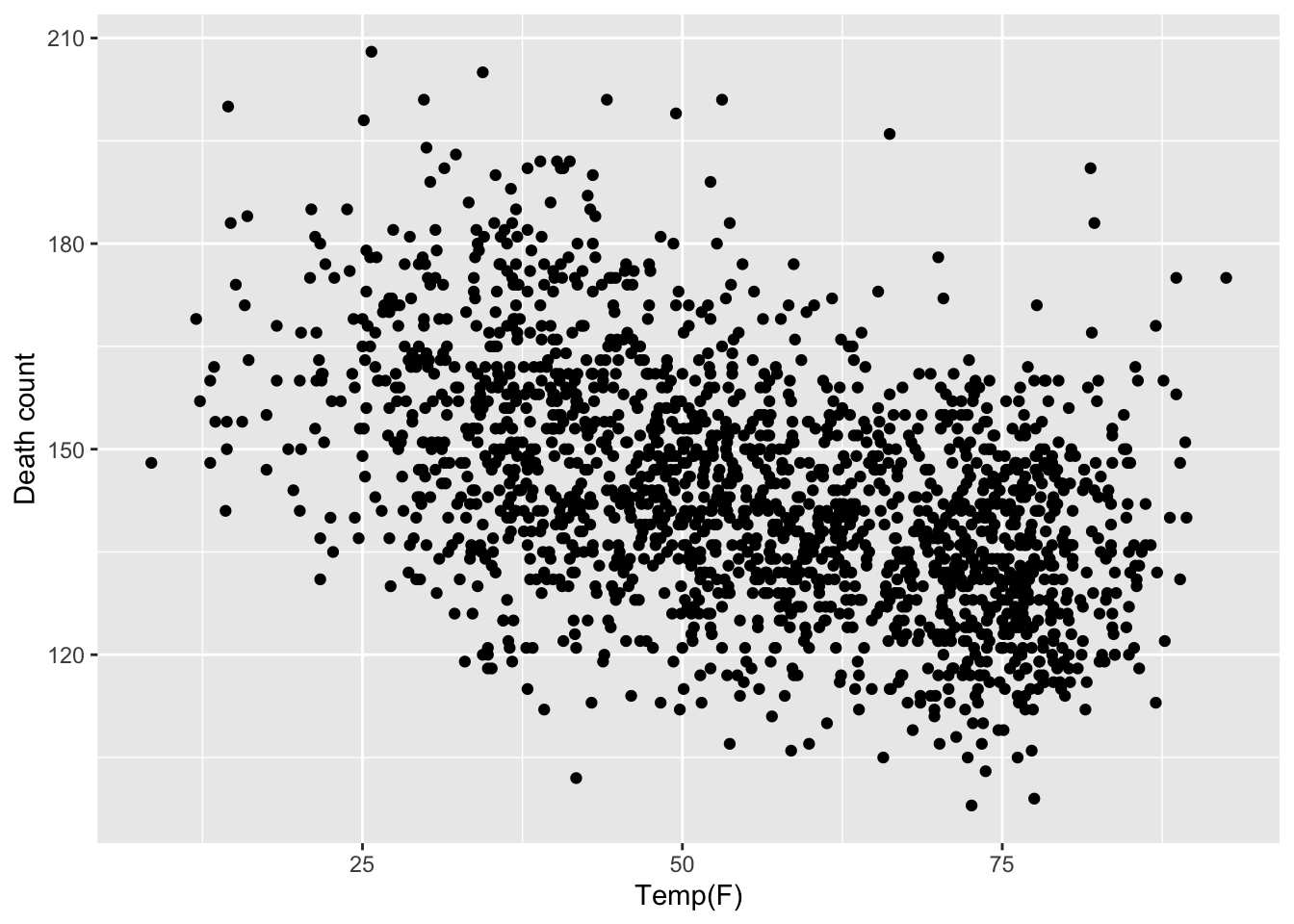
Module 5 Part 2: Penalized and Smoothing Splines
Reading
Chapter 5- Elements of Statistical Learning (Hastie)
Chapter 5- Introduction to Statistical Learning (James et al. )
Basic Concepts
- Constraints and penalized regression
- Smoothing matrix and smoothing parameter
- Generalized cross-validation to choose roughness penalty
- Mixed models to choose roughness penalty
Parametric Splines
Regression Problem
Let
We consider the nonparametric regression problem:
- We can approximate
e.g., linear spline with 9 equidistant interior knots,
Lets fit this to the data:
library(splines)
library(dplyr)
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionlibrary(tidyr)
# Define 9 evenly spaced knots
knots <- seq(min(health$Temp), max(health$Temp), length.out = 9)
# Create a linear spline basis using bs() function
spline_basis <- bs(health$Temp, knots = knots[-c(1, length(knots))], degree = 1) # Exclude boundary knots
# Fit a linear model with the spline basis
fit <- lm(alldeaths ~ spline_basis, data = health)
# Plot the results
plot(health$Temp, health$alldeaths, main = "Linear Spline with 9 Knots", col = "grey", pch = 16, xlab = "Temp", ylab = "Death Count")
lines(health$Temp, predict(fit), col = "blue", lwd = 2)
abline(v = knots, col = "red", lty = 2) # Show knot locations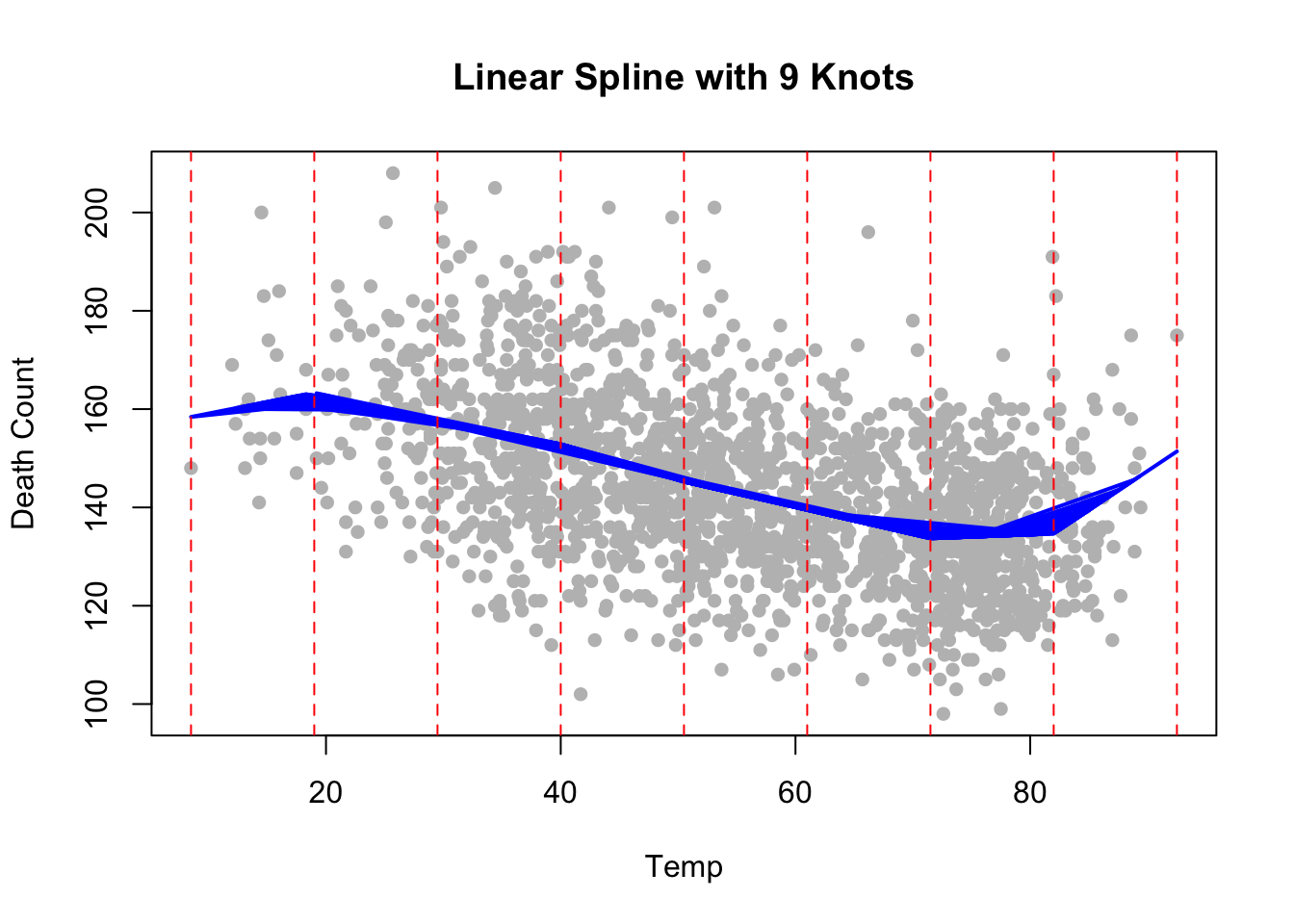
Automatic Knot Selection
What if we don’t know the number and locations of the knots?
Splines can overfit the data when number of knot increases. We want to avoid overfitting
Approach
- Start with a lot of knots. This ensures we will not miss important fine-scale behavior.
- Assume most of the knots are not useful and shrink their coefficients towards zero.
- Determine how much to shrink based on some criteria (e.g. CV, GCV, AIC).
Benefits
- Knot placement is not important if the number is dense enough.
- Shrinking most coefficients to zero will stabilize model estimation similar to performing variable selection.
Penalized Spline
Consider the basis expansion :
L2 Definition: The L2 norm, also called the Euclidean norm, is a measure of the magnitude (or length) of a vector in Euclidean space. It is defined as:
where
The L2 norm is the square root of the sum of the squared elements of the vector.
Properties:
1. Non-Negativity
- Scalability: If
- Triangle Inequality: For two vectors $ $ and
- Geometric Interpretation: The L2 norm corresponds to the Euclidean distance from the origin to the point
This is the standard distance formula in 2D geometry.
Penalties
Ridge regression
- Ridge shrinks coefficients of vectors in B-spline basis, but does not induce sparsity.
- Ridge is easy to solve- closed form solution!
Lasso = absolute value =
- Lasso tends to make some coefficients exactly zero.
- Trickier to solve. More on this later.
A small
Our goal: Convert the two problems: (1) number of knots, and (2) their location- into a single parameter that we can choose with GCV.
Matrix of
- For simplicity, consider a linear spline. Then evaluate the basis functions at each
where:
Writing
where
Residuals are given by:
where
Constrained Formulation
We define the objective function as:
The term
- Smaller values indicate a better fit.
- Want to minimize RSS, i.e., find
Constraint :
The constraint
- 1 in
- 0 in
- 1 in
Penalty Formulation
This problem can be equivalently formulated without constraint
There is a one-to-one mapping between
The smoothing parameter,
- Minimizing the residual sum of squares (fit to the data).
- Penalizing complexity of the model (regularization term
A larger value of
A smaller value of $$ allows more flexibility, potentially fitting the data more closely but risking overfitting.
Why This Matters
- Constraint-based and penalty-based—are equivalent in theory but have different practical uses:
- Constraint-based Formulation: Useful for explicitly enforcing a limit on model complexity (e.g., in constrained optimization problems).
- Penalty-based Formulation: Easier to solve using standard optimization techniques and more common in regularization methods like Ridge Regression or Smoothing Splines.
Closed-Form Solution
- The optimization problem is defined as:
- Differentiating with Respect to
To find the closed-form solution, differentiate the objective function with respect to
- Simplify:
- Rearranging terms:
- Final Solution: Solving for
for some positive number
- When
- When
- Note here
Come back to our mortality example
Consider the temperature and mortality analysis. Assume 40 equidistant knots and linear splines. The model is defined as:
where: -
We will penalize the coefficients
So
knots <- seq(range(health$Temp)[1], range(health$Temp)[2], length.out = 40+2)
#place knots evenly on interior of the range of x
knots = knots[c(2:(length(knots)-1))]
X = cbind(rep(1, length(health$Temp)), health$Temp)
head(X) [,1] [,2]
[1,] 1 27.6
[2,] 1 25.1
[3,] 1 25.3
[4,] 1 29.8
[5,] 1 29.8
[6,] 1 33.9for(i in 1:length(knots)){
X = cbind(X, (health$Temp - knots[i]) * (health$Temp>knots[i]))
}
#Penalty matrix for regularization, 42 = number of coeffs
B = diag(42)
#Only spline terms for 3rd coefficient will be penalized
B[1,1] = 0
B[2,2] =0
dim(X)[1] 1826 42dim(B)[1] 42 42We now search through different values of
Calculate penalized
Calculate
Calculate fitted values
Calculate the GCV using the matrix
We select the
Y = health$alldeaths
lambda <- seq(0, 50, by = 5)
calc_gvc <- function(lambda, X, B){
beta = solve(t(X) %*% X + lambda*B) %*% t(X) %*% Y
H = X %*% solve(t(X) %*% X + lambda*B) %*% t(X)
Yhat = X %*% beta
GCV = mean((Y- Yhat)^2)/ (1-mean(diag(H)))^2
C = t(beta) %*% B %*% beta
return(GCV)
}
GCV <- rep(NA, length(lambda))
for(i in 1:length(lambda)){
GCV[i] = calc_gvc(lambda = lambda[i], X, B)
}
GCV [1] 233.7743 231.8207 231.3576 231.1072 230.9433 230.8250 230.7344 230.6622
[9] 230.6028 230.5528 230.5099min(GCV)[1] 230.5099index = which(GCV == min(GCV))
lambda[index][1] 50Effect of Penalization
penalized_spline <- function(X, Y, lambda, B) {
# Create design matrix for linear splines
betahat = solve(t(X) %*% X + lambda*B) %*% t(X) %*% Y
H = X %*% solve(t(X) %*% X + lambda*B) %*% t(X)
Yhat = X %*% betahat
list_out <- list()
list_out[["Yhat"]] <- Yhat
list_out[["Betahat"]] <- betahat
return(list_out)
}
# Different lambda values to compare
lambdas <- seq(0, 50, by = 10)
# Create original data frame
original_data <- data.frame(x = health$Temp, y = health$alldeaths)
for(i in 1:length(lambdas)){
res <- penalized_spline(X=X, Y=Y, B=B, lambda = lambdas[i])
Yhat = res[["Yhat"]]
original_data[[paste0("lambda_", lambdas[i])]] <- Yhat
}
# Convert wide to long
long_data <- original_data %>%
pivot_longer(
cols = starts_with("lambda_"), # Select all lambda columns
names_to = "lambda", # Create a new column "lambda" for the column names
values_to = "prediction" # Create a new column "prediction" for the values
)
# Plot using ggplot2
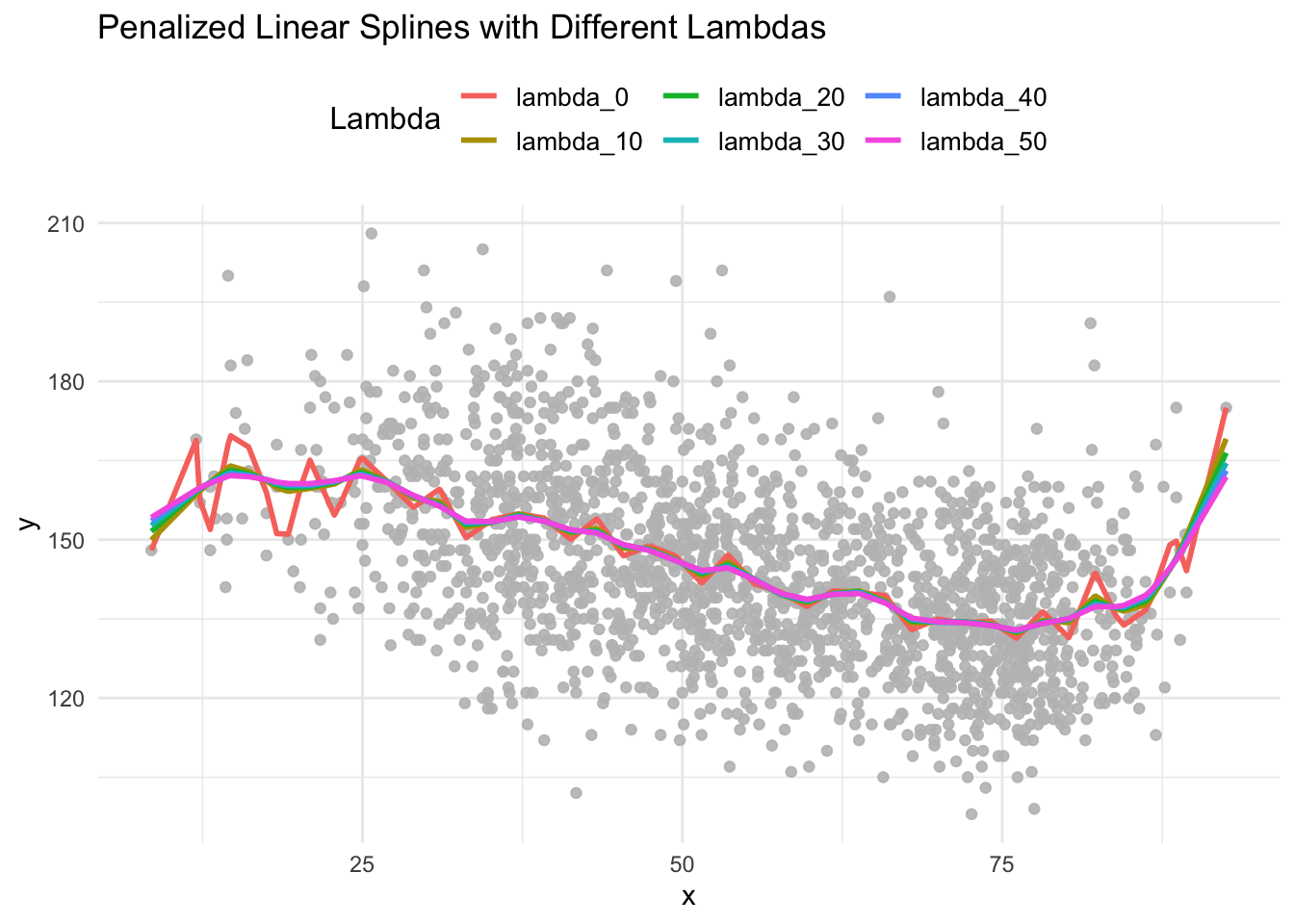
ggplot(long_data) +
geom_point(aes(x = x, y = y), color = "grey", alpha = 0.3) +
geom_line(aes(x = x, y = prediction[,1], color = lambda), linewidth = 1) +
labs(
title = "Penalized Linear Splines with Different Lambdas",
x = "x",
y = "y",
color = "Lambda"
) +
theme_minimal() +
theme(
legend.position = "top",
legend.title = element_text(size = 12),
legend.text = element_text(size = 10)
)
Betahat_df <- array(NA, c(42, length(lambdas)))
# Compute Betahat for each lambda
for (i in 1:length(lambdas)) {
res <- penalized_spline(X = X, Y = Y, B = B, lambda = lambdas[i])
Betahat_df[, i] <- res[["Betahat"]]
}
# Convert array to dataframe
Betahat_df <- as.data.frame(Betahat_df)
# Name the columns after the lambda values
colnames(Betahat_df) <- paste0("lambda_", lambdas)
# Add a column for the indices of the Betahat values
Betahat_df$Coefficient <- 1:42
Betahat_long <- Betahat_df %>%
pivot_longer(
cols = starts_with("lambda_"), # Select lambda columns
names_to = "Lambda", # Column for lambda values
values_to = "Betahat" # Column for Betahat values
) %>%
mutate(Lambda = as.numeric(sub("lambda_", "", Lambda))) # Clean lambda names
# Plot Betahat across different lambdas
ggplot(Betahat_long, aes(x = Coefficient, y = Betahat, color = as.factor(Lambda))) +
geom_line(size = 1) +
geom_point(size = 2, alpha = 0.8) +
labs(
title = "Penalized Linear Spline Coefficients Across Lambdas",
x = "Coefficient Index",
y = "Estimated Coefficient (Betahat)",
color = "Lambda"
) +
scale_color_brewer(palette = "Set1") + # Optional: nice color palette
theme_minimal() +
theme(
legend.position = "top",
legend.title = element_text(size = 12),
legend.text = element_text(size = 10),
axis.title = element_text(size = 14),
axis.text = element_text(size = 12)
)Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
ℹ Please use `linewidth` instead.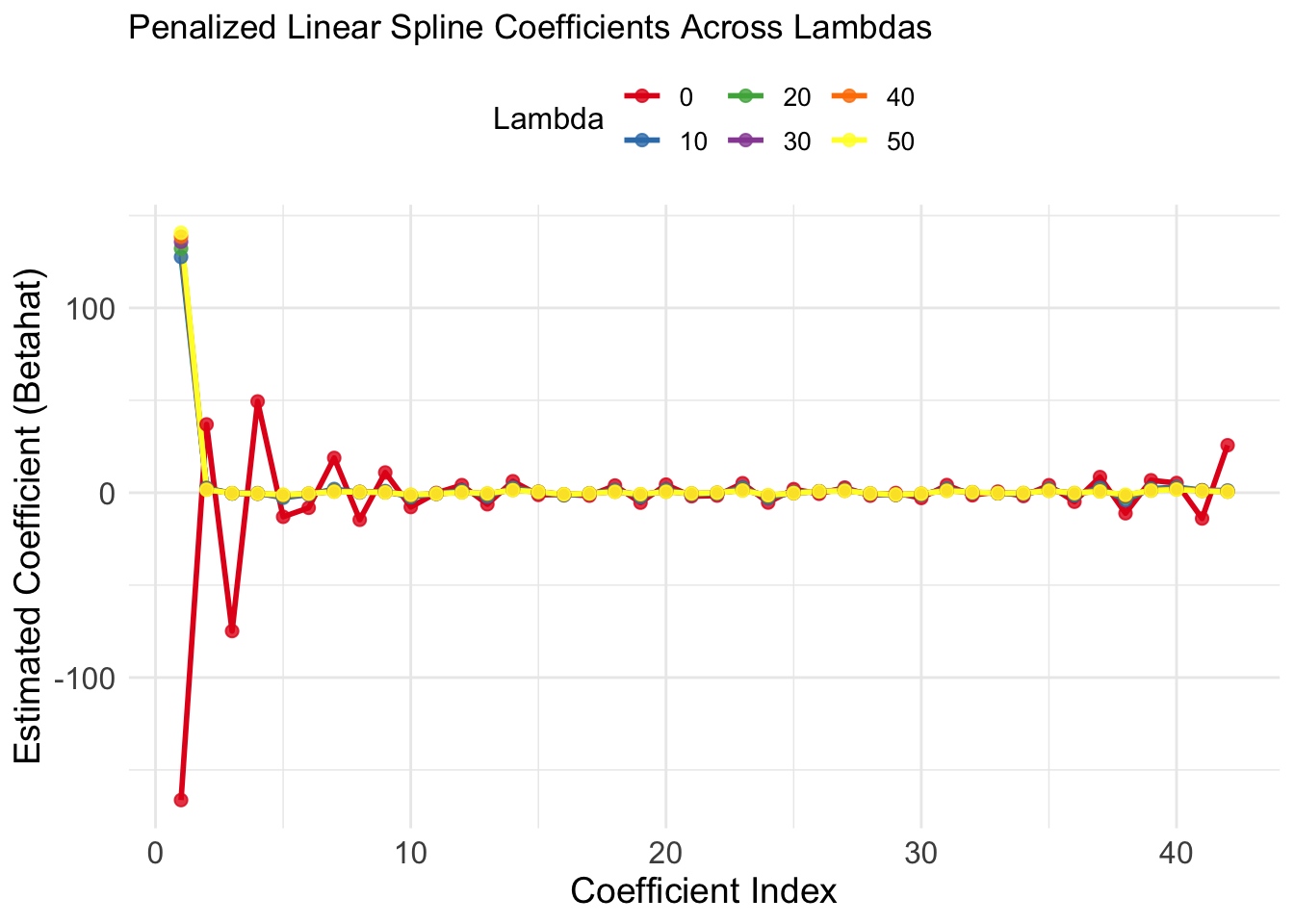
Key Takeaways from the Plot
As
Spline coefficients shrink, reducing their contribution to the model.
The model becomes smoother and less flexible, capturing broader trends but ignoring fine details.
The intercept and linear terms (not penalized) changes significantly across
The higher-index coefficients (spline terms) are the most affected, which is why you see their lines converge toward zero for larger
General Principle:
But how do we determine the level of shrinkage? i.e., how do we choose
Generalized Cross-Validation Error
An approximation of leave-one-out cross-validation. Generalized Cross Validation Error is defined as
Note that
Now we can apply GCV to any prediction of
where
Each element is shrunk towards zero. We can define effective degrees of freedom
Then GCV is defined as:
where
# Simulate data for illustration
set.seed(123)
lambda_values <- seq(0.1, 100, length.out = 100) # Range of lambda values
rss_values <- 500 / (lambda_values + 1) + rnorm(100, sd = 5) # Simulate RSS
df_values <- 20 - 18 * (lambda_values / (lambda_values + 10)) # Effective df
gcv_values <- rss_values / (1 - df_values / 20)^2 # Compute GCV
# Create a dataframe for plotting
gcv_data <- data.frame(
lambda = lambda_values,
RSS = rss_values,
df = df_values,
GCV = gcv_values
)
# Find optimal lambda
optimal_lambda <- lambda_values[which.min(gcv_values)]
optimal_lambda[1] 71.74545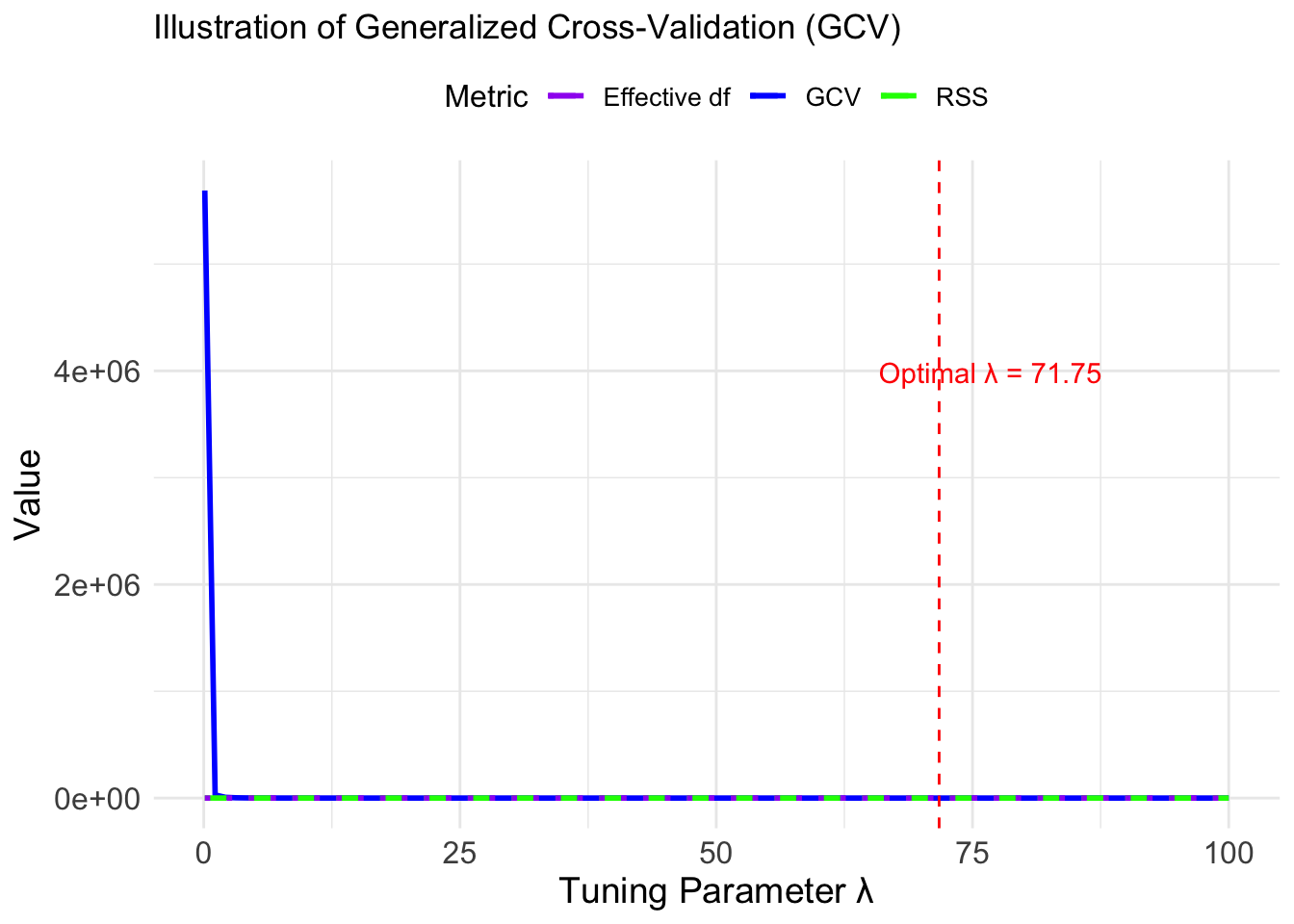
Residual Error Variance Estimate
We have two options:

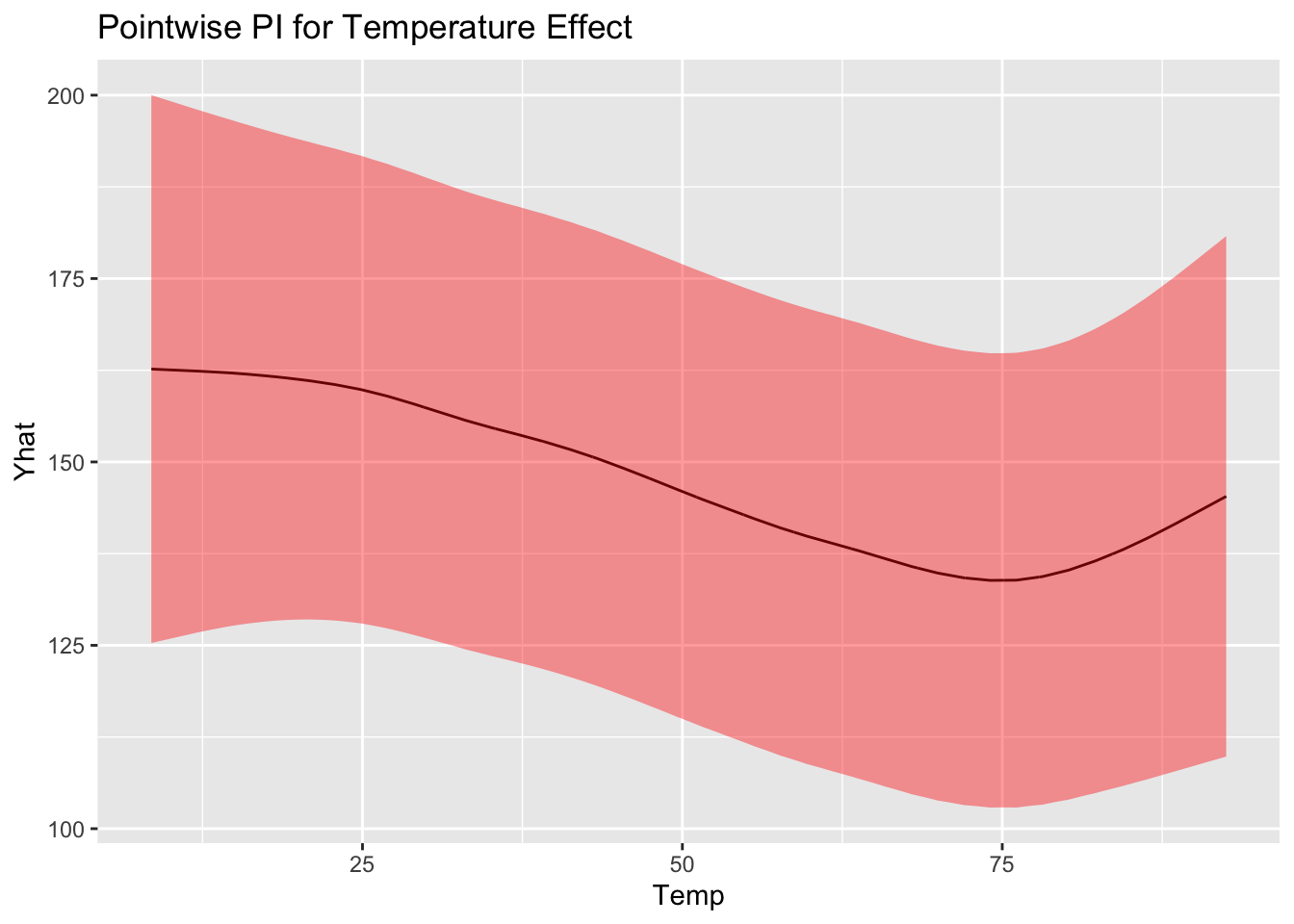
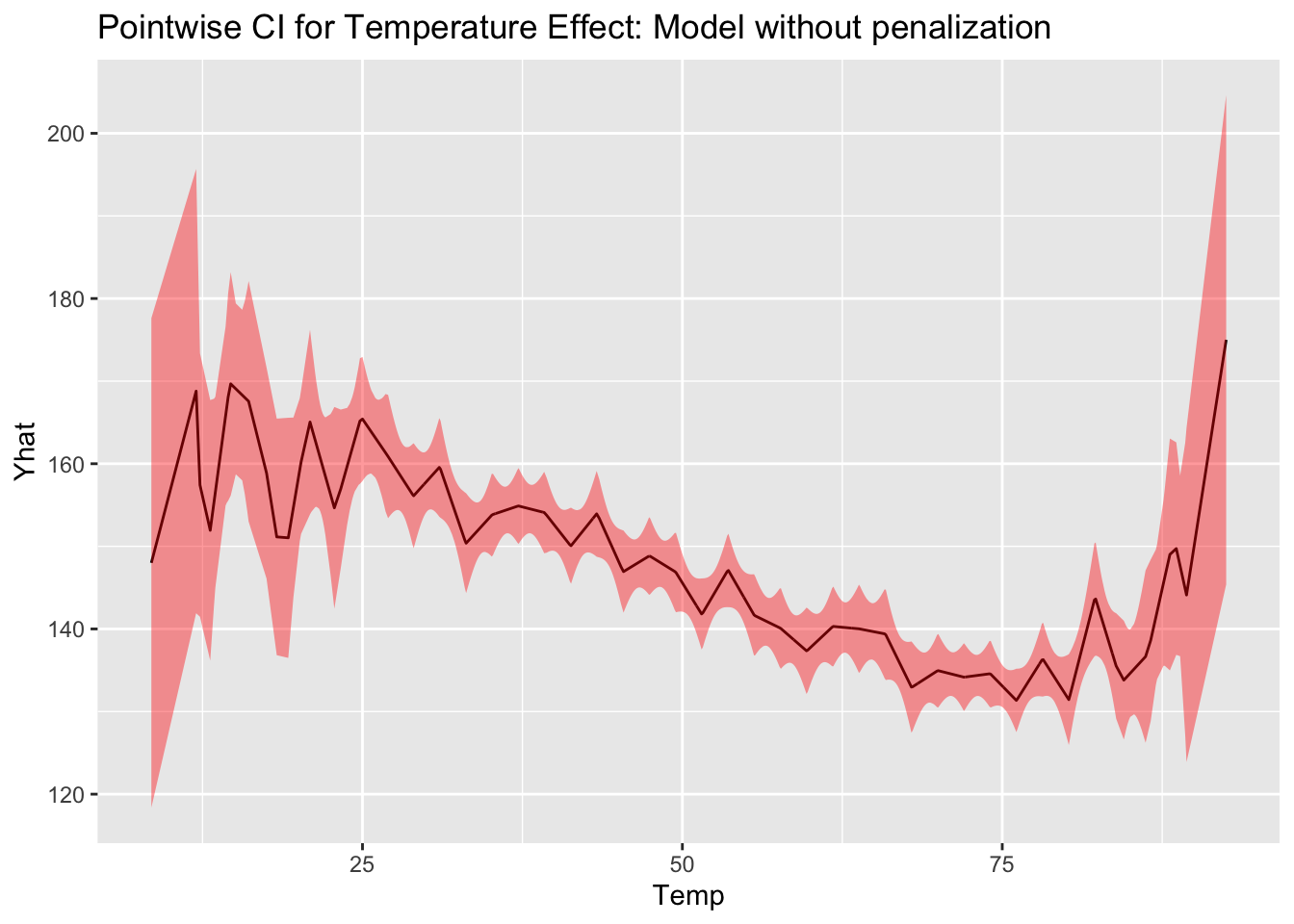
Confidence interval and prediction interval
Obtains point-wise confidence interval dervied from previous expression by plugging in
Similarly, the variance for an unobserved point
[1] 3600 user system elapsed
3.779 0.011 3.797 user system elapsed
0.044 0.013 0.056 [,1]
[1,] TRUE[1] 15.11997[1] 15.12668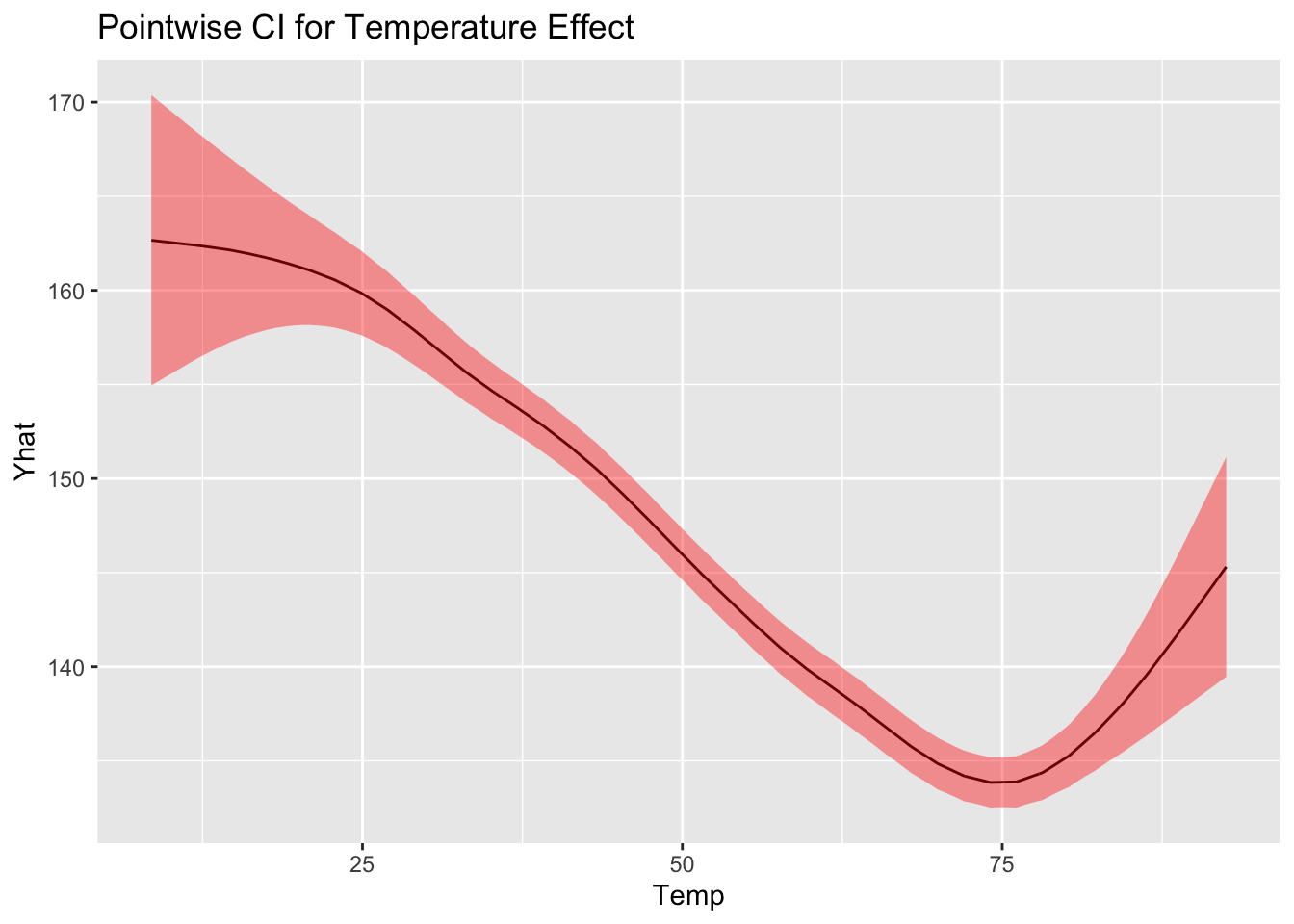
Smoothing Splines: other penalties
- A smoothing spline is a statistical tool used to fit a smooth curve to a set of data points.
- Goal is to approximate an underlying function that explains the relationship between variables without assuming a specific functional form (like a linear or polynomial relationship).
- Data Fitting: The smoothing spline minimizes the trade-off between model fit (through the sum of squared errors) and excessive curvature or smoothing.
- Smoothness: The curve generated by a smoothing spline is smooth (no discontinuities) while avoiding overfitting.
- Regularization: Smoothing splines are regularized, i.e., they include a penalty for too much curvature in the fitted curve, controlling the degree of smoothness. This penalty is determined by a smoothing parameter, often denoted by
- Note the 1st derivatives are not penalized
- 2nd part uses the squared 2nd-derivative that is a good measure of roughness.
- Shrinks coefficients in a cubic polynomial, causing function to change less quickly. (ie large penalties reduce wiggliness).
Smoothing Splines with mgcv in R
The mgcv (Mixed GAM Computation Vehicle) package in R provides the gam() function to fit a wide variety of smoothing splines with automatic smoothing parameter selection. Throughout this class, we will explore different options available in mgcv.
Below are some key default settings:
Default Options:
Basis Functions
Default: Thin plate regression spline.Basis Dimension
Default:- If
lm().
- If
Selection Methods
Default: Generalized Cross Validation (GCV).Family
Default: Gaussian.Standard Error Computation
Default: Bayesian.
Flexibility: GAMs can handle multiple predictors, each with potentially different non-linear relationships to the response. These relationships are modeled using smoothing splines, thin plate splines, or other basis functions.
Automatic Smoothing: GAM frameworks like
mgcvuse methods such as Generalized Cross-Validation (GCV) or Restricted Maximum Likelihood (REML) to automatically choose smoothing parameters for each term.
Comparing Smoothing Splines to GAMs
| Feature | Smoothing Splines | Generalized Additive Models (GAMs) |
|---|---|---|
| Scope | Single predictor | Multiple predictors |
| Additive Structure | No | Yes |
| Link Function | Not applicable (linear) | Supports different link functions (e.g., log, logit) |
| Smoothing | Single penalty term | Separate penalties for each predictor’s smooth term |
| Software | Standalone or integrated | Built into GAM frameworks like mgcv |
Relationship
- GAMs often use smoothing splines (or similar smoothers) as the functional form for each predictor in the additive model. For example, the
gam()function inmgcvallows specifying smooth terms likes(x)orte(x, z), which are internally fitted using basis functions, including smoothing splines.
Coming back to our example: Temperature Effect on Mortality
library (mgcv)Loading required package: nlme
Attaching package: 'nlme'The following object is masked from 'package:dplyr':
collapseThis is mgcv 1.8-42. For overview type 'help("mgcv-package")'.fit1 = gam(alldeaths~s(Temp), data= health)
summary(fit1)
Family: gaussian
Link function: identity
Formula:
alldeaths ~ s(Temp)
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 143.9168 0.3538 406.8 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(Temp) 6.026 7.2 80.62 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
R-sq.(adj) = 0.241 Deviance explained = 24.3%
GCV = 229.47 Scale est. = 228.58 n = 1826Checking GAMs
mgcv::gam.check(fit1)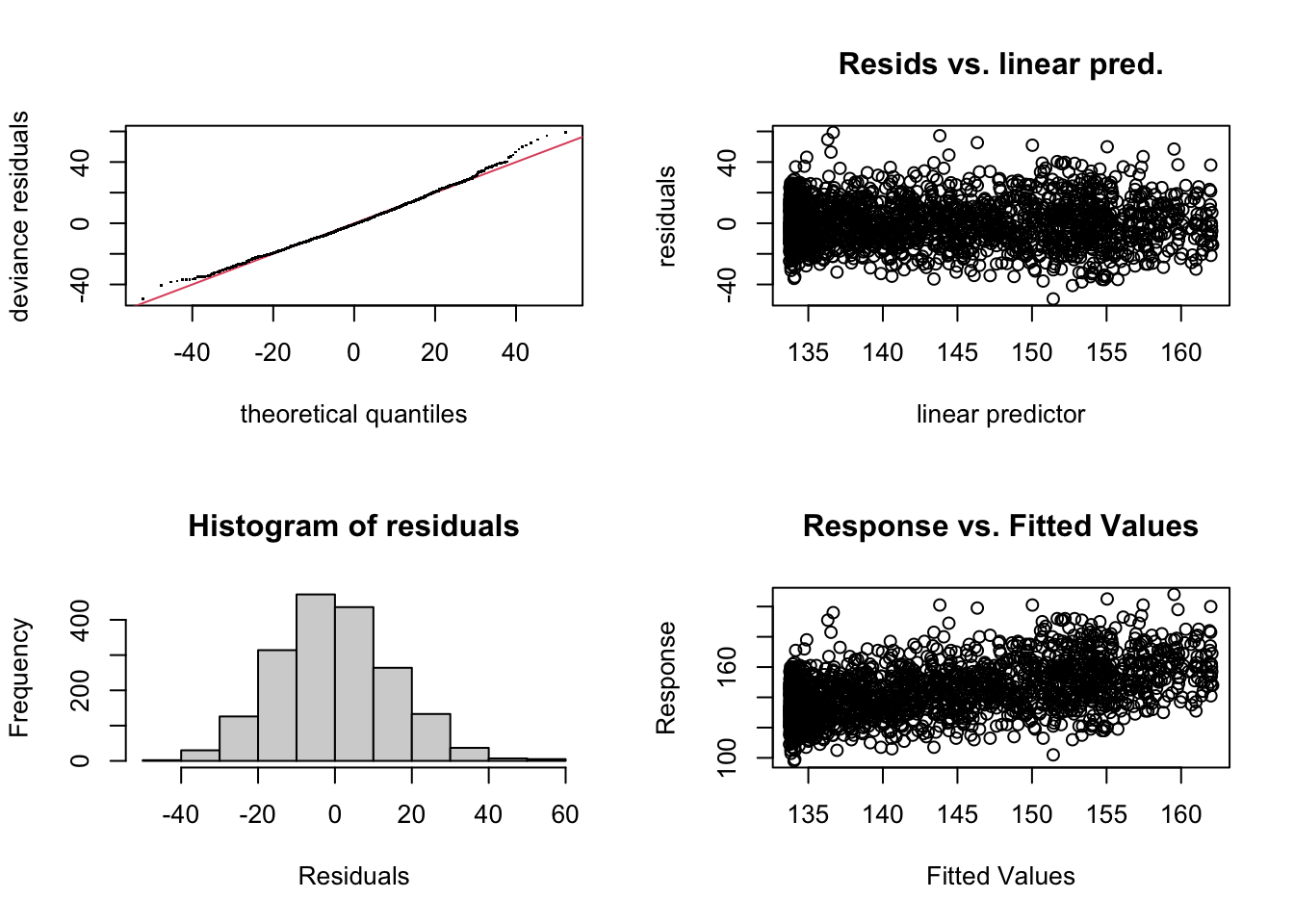
Method: GCV Optimizer: magic
Smoothing parameter selection converged after 5 iterations.
The RMS GCV score gradient at convergence was 7.241958e-05 .
The Hessian was positive definite.
Model rank = 10 / 10
Basis dimension (k) checking results. Low p-value (k-index<1) may
indicate that k is too low, especially if edf is close to k'.
k' edf k-index p-value
s(Temp) 9.00 6.03 1.02 0.86