Loading required package: Matrix
Attaching package: 'lmerTest'The following object is masked from 'package:lme4':
lmerThe following object is masked from 'package:stats':
stepLoading required package: Matrix
Attaching package: 'lmerTest'The following object is masked from 'package:lme4':
lmerThe following object is masked from 'package:stats':
stepReading
Basic Concepts
Models for non-normal data
Link functions
Logistic regression: Binary data
Probit regression: Binary data
Poisson regression: Count data
Consider the following multiple linear regression model: For
This is equivalent to
Reminder of the assumptions of the above model:
Therefore,
A generalized linear model (GLM) extends the linear regression to other distributions, where the response variable is generated from a distribution in the exponential family.
In the GLM,
Examples of distributions in the exponential family include: normal with known variance (link = identity), Bernoulli (link = logit or probit), binomial (link = logit or log), gamma, exponential (link = negative inverse), and others.

Let’s take a binary outcome where
where
We know that
The link function should have some desirable properties:
In general the logistic function is defined as :
The
There is no
It is really important to understand that the change in log-odds per one unit change in
#The Logit Function
p.i <- seq(0, 1, by = 0.001)
g_p.i <- log(p.i/(1-p.i))
plot(p.i, g_p.i, col = "red", xlab ="p", ylab = "Logit Function")
#The Logistic Function
p_i <- exp(g_p.i)/(1+exp(g_p.i))
plot(g_p.i, p_i, col = "blue", xlab = "x", ylab = "Logistic Function")
What are the effects of changing the baseline odds?
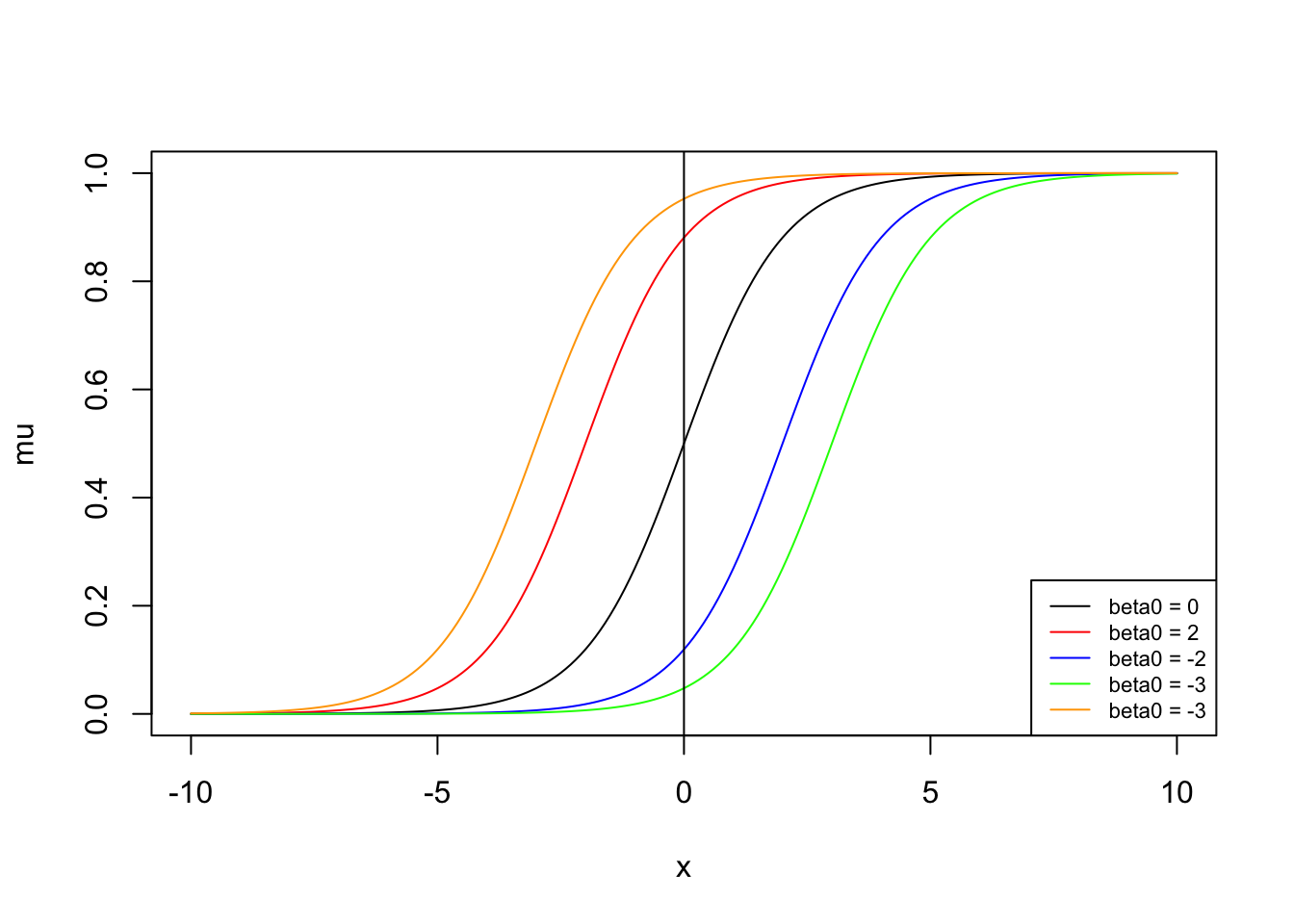
Note: The shape is maintained, and the baseline (intercept) probability changes.
What are the effects of the change in slope?
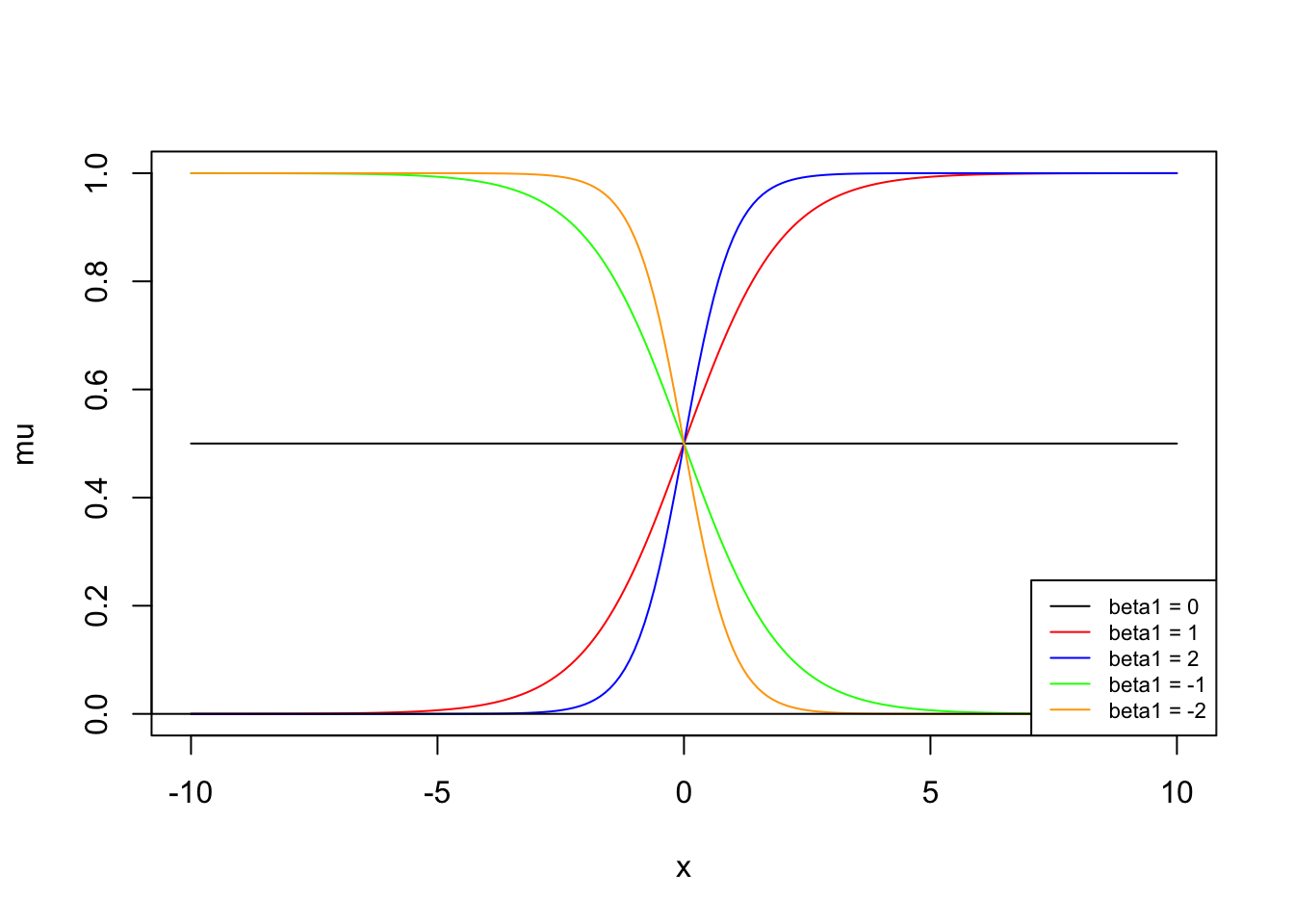
Note how the steepness changes with difference in slopes. Also note how the direction changes depending on the effect changes.
Fitting the GLM model in R is very similar to fitting a linear regression model. We need to specify the distribution as binomialand the link function as logit.
### The GLM Function
load("/Users/emilypeterson/Library/CloudStorage/OneDrive-EmoryUniversity/BIOS 526/BIOS526_Book/data/PTB.Rdata")
fit <- glm(formula = ptb ~ age + male + tobacco,
family = binomial(link = "logit"),
data = dat)
summary(fit)
Call:
glm(formula = ptb ~ age + male + tobacco, family = binomial(link = "logit"),
data = dat)
Deviance Residuals:
Min 1Q Median 3Q Max
-0.5160 -0.4236 -0.4103 -0.4088 2.2500
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.4212664 0.0631355 -38.350 < 2e-16 ***
age -0.0006295 0.0021596 -0.291 0.77068
maleM 0.0723659 0.0258672 2.798 0.00515 **
tobacco 0.4096495 0.0534627 7.662 1.83e-14 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 44908 on 77339 degrees of freedom
Residual deviance: 44846 on 77336 degrees of freedom
AIC: 44854
Number of Fisher Scoring iterations: 5Preterm delivery was significantly associated with male babies (p-value = 0.005) when controlling for age and mother’s smoking status. The odds ratio of a preterm birth for a male baby versus a female baby was 1.07 (95% CI: 1.02, 1.13).
How we get CIs:
NOTE: TRANSFORM THE INTERVALS. DO NOT TRANSFORM THE STANDARD ERRORS (this requires the delta method).
Preterm delivery was significantly associated with whether the mother smoked during pregnancy (p<0.001) when controlling for age and the baby’s sex.
The baseline proportion (female babies born to mother of age 25 who didn’t smoke) of preterm delivery was
We didn’t find an effect of mother’s age.
How can we test whether which model is better?
The likelihood for a binary logistic regression is given by:
The likelihood ratio test (LRT) is used to test the null hypothesis that any subset of the
The Likelihood Ratio Test is given by:
This gives the ratio of the likelihood of the null model evaluated at the MLEs to the likelihood of the full model evaluated at the MLEs.
In the normal case, this is the analogous to the F-test described in Chapter 1.
The likelihood ratio test statistic is given by:
The difference follows a Chi-square distribution, i.e.,
where
library(car)Loading required package: carDataRegistered S3 methods overwritten by 'car':
method from
influence.merMod lme4
cooks.distance.influence.merMod lme4
dfbeta.influence.merMod lme4
dfbetas.influence.merMod lme4Anova(fit)Analysis of Deviance Table (Type II tests)
Response: ptb
LR Chisq Df Pr(>Chisq)
age 0.085 1 0.770669
male 7.834 1 0.005126 **
tobacco 53.648 1 2.399e-13 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The Wald Test is the test of significance for individual regression coefficients in logistic regression. Wald test under regularity conditions and assuming asympotitc results,
We can write
In
For maximum likelihood estimates, the ratio
can be used to test
–> –> –>
–>
This is known as the population-averaged effect or marginal effect.
The two approaches are estimating different slopes!.
Note: the GLM likelihood assumes independence, resulting in incorrect SE.
We are modeling transformations of the expectations:
When
Now consider a unit change in
Covariate impacts are multiplicative rather than additive (apples to log models in general) on the count scale.
cancer <- readRDS(paste0("data/ohio_cancer.RDS"))
head(cancer) county sex race year death pop
1 1 1 1 1 6 8912
2 1 1 1 2 5 9139
3 1 1 1 3 8 9455
4 1 1 1 4 5 9876
5 1 1 1 5 8 10281
6 1 1 1 6 5 10876Consider a random-intercept Poisson model where we treat all stratified death counts within the same county as a group.
fit = lme4::glmer(death ~ factor(sex)*factor(race) + (1|county),
family = poisson, data =cancer)
summary(fit)Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: poisson ( log )
Formula: death ~ factor(sex) * factor(race) + (1 | county)
Data: cancer
AIC BIC logLik deviance df.resid
39383.0 39417.6 -19686.5 39373.0 7387
Scaled residuals:
Min 1Q Median 3Q Max
-7.4474 -1.0870 -0.6149 0.4008 14.8303
Random effects:
Groups Name Variance Std.Dev.
county (Intercept) 1.094 1.046
Number of obs: 7392, groups: county, 88
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 2.839314 0.111660 25.428 < 2e-16 ***
factor(sex)2 -1.014711 0.007458 -136.061 < 2e-16 ***
factor(race)2 -2.064818 0.011466 -180.088 < 2e-16 ***
factor(sex)2:factor(race)2 -0.165961 0.023499 -7.063 1.64e-12 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) fctr(s)2 fctr(r)2
factor(sx)2 -0.018
factor(rc)2 -0.012 0.173
fctr()2:()2 0.006 -0.317 -0.488 cancer$logpop = log(cancer$pop)
fit = lme4::glmer(death ~ offset(logpop) + factor(sex)*factor(race) + (1|county),
family = poisson,
data = cancer)
summary(fit)Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: poisson ( log )
Formula: death ~ offset(logpop) + factor(sex) * factor(race) + (1 | county)
Data: cancer
AIC BIC logLik deviance df.resid
31814.5 31849.0 -15902.2 31804.5 7387
Scaled residuals:
Min 1Q Median 3Q Max
-7.8261 -0.6352 -0.2335 0.4472 9.8153
Random effects:
Groups Name Variance Std.Dev.
county (Intercept) 0.03991 0.1998
Number of obs: 7392, groups: county, 88
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -7.367370 0.022071 -333.808 < 2e-16 ***
factor(sex)2 -1.077799 0.007458 -144.510 < 2e-16 ***
factor(race)2 0.034907 0.011735 2.974 0.00293 **
factor(sex)2:factor(race)2 -0.219521 0.023503 -9.340 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) fctr(s)2 fctr(r)2
factor(sx)2 -0.089
factor(rc)2 -0.030 0.171
fctr()2:()2 0.029 -0.317 -0.476 We are going to implement a more realistic Poisson regression model to explore the associations of sex and race on cancer incidence in Ohio.
Read in the Ohio cancer data from Canvas.
Create exploratory plots of the associations/relationships between
Run a Poisson regression in R using the following model:
Hint: the intercept can be broken down into two rand effects in the
5. What were the findings from the R output?
In Poisson, the assumption
One can add an additional overdispersion parameter also called a scale parameter.
One can adjust the parameter variance by this scale parameter. This is called the quais-poisson model.
Fitting a quasi-poisson in GLM
glm_quasi = glm(death ~ offset(logpop) + factor(sex)*factor(race) + (1|county),
family = "quasipoisson",
data = cancer)
summary(glm_quasi)
Call:
glm(formula = death ~ offset(logpop) + factor(sex) * factor(race) +
(1 | county), family = "quasipoisson", data = cancer)
Deviance Residuals:
Min 1Q Median 3Q Max
-7.1212 -0.9661 -0.3920 0.2381 10.6459
Coefficients: (1 not defined because of singularities)
Estimate Std. Error t value Pr(>|t|)
(Intercept) -7.280397 0.005707 -1275.600 < 2e-16 ***
factor(sex)2 -1.074482 0.011065 -97.106 < 2e-16 ***
factor(race)2 0.117448 0.017011 6.904 5.47e-12 ***
1 | countyTRUE NA NA NA NA
factor(sex)2:factor(race)2 -0.218442 0.034865 -6.265 3.93e-10 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasipoisson family taken to be 2.200393)
Null deviance: 43628 on 7391 degrees of freedom
Residual deviance: 15976 on 7388 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 5Concepts
Hierarchical linear models: three level random intercept model for Gaussain data ( a type of lmm).
Hierarchical generalized linear models (a type of glmm).
Hierarchical structure and covariance structures.
Reading
School data example adapted from Gelman and Hill.
Guatemalan data example: Rodriguez B and Goldman N (2001). Improved estimation procedures for multilevel models with binary response: a case study. Journal of Royal Statistical Society Series A 339-355.
Level 1: relating math scored to occasion (year) specific covariates:
Level 2: relating child-specific random intercepts to child. characteristics
Level 3: relating school-specific random intercepts to school characteristics.
Reparametrization of the multi-level model: The multilevel model can be combined to give:
Because
Every measurement and every child in school
male, non-Black, non-Hispanic
grade year at 3.5 not retained.


Hierarchical Specification of the multi-level model:
dat <- read.csv(paste0(getwd(), "/data/achievement.csv"))
dim(dat)[1] 7230 10dat[1:6,] year math retained female black hispanic size lowinc school child
1 0.5 1.146 0 0 0 1 380 40.3 2020 244
2 1.5 1.134 0 0 0 1 380 40.3 2020 244
3 2.5 2.300 0 0 0 1 380 40.3 2020 244
4 -1.5 -1.303 0 0 0 0 380 40.3 2020 248
5 -0.5 0.439 0 0 0 0 380 40.3 2020 248
6 0.5 2.430 0 0 0 0 380 40.3 2020 248We specify the multi-level model with two random intercept components. (Here child ID is unique, so the below code is equivalent to (1|school) + (1|school:child), see R code)
dat <- read.csv(paste0(getwd(), "/data/achievement.csv"))
fit <- lmer(math~ year + retained + female + black + hispanic + size + lowinc + (1|school) + (1|child), data = dat)
summary(fit)Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: math ~ year + retained + female + black + hispanic + size + lowinc +
(1 | school) + (1 | child)
Data: dat
REML criterion at convergence: 16706.4
Scaled residuals:
Min 1Q Median 3Q Max
-3.8439 -0.5837 -0.0267 0.5587 6.1702
Random effects:
Groups Name Variance Std.Dev.
child (Intercept) 0.6637 0.8147
school (Intercept) 0.0875 0.2958
Residual 0.3445 0.5870
Number of obs: 7230, groups: child, 1721; school, 60
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 2.398e-01 1.524e-01 6.406e+01 1.573 0.12064
year 7.483e-01 5.396e-03 5.744e+03 138.685 < 2e-16 ***
retained 1.481e-01 3.535e-02 5.802e+03 4.190 2.83e-05 ***
female -9.038e-05 4.223e-02 1.668e+03 -0.002 0.99829
black -5.182e-01 8.060e-02 1.154e+03 -6.429 1.88e-10 ***
hispanic -2.899e-01 8.910e-02 1.642e+03 -3.254 0.00116 **
size -1.028e-04 1.485e-04 5.719e+01 -0.692 0.49167
lowinc -8.002e-03 1.818e-03 6.900e+01 -4.401 3.84e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) year retand female black hispnc size
year -0.012
retained -0.009 0.086
female -0.141 -0.005 0.017
black -0.108 -0.006 -0.013 -0.006
hispanic -0.071 -0.010 -0.006 0.018 0.556
size -0.454 -0.009 0.008 -0.019 0.025 -0.079
lowinc -0.622 0.009 -0.009 0.015 -0.326 -0.188 -0.221Note that the random effects of child and school are assumed to be independent.
Note on nesting A child is nested within school because for a given child, school does not vary. Therefore, we can’t look at the interaction between child and school!. We can only look at interactions for crossed factors. There are many possible interactions here.
The higher number of interactions, the finer the level (the smaller the sample). So to keep things simple, we will ignore them here.
We cannot estimate whether a student from a disadvantaged background (low income, single family household, parents’ education, others) has lower scores.
In particular, race and ethnicity are correlated with other factors impacting on individual’s achievement scores.
The lower scores reflect products of systemic racism and shortcomings of the education system.
Results of this data set could be used to help prioritize education funds or policy.
Our 3-level model decomposes the total residual variation into three components:
Level 3: Between school variation
Level 2: Between child variation
Level 1: Between grade variation (within a child)
We can consider models that only include random intercepts for schools or for children.
Assume no between-child variation
# fit2 <- lmer (math ~ year + retained + female + black + hispanic + size + lowinc + (1|school) , data = dat)Assume no between-school variation
# fit3 <- lmer (math ~ year + retained + female + black + hispanic + size + lowinc + (1|child) , data = dat)Note: Likelihood Ratio Tests can be inaccurate due to issues of testing a null hypothesis on the boundary of the parameter space (generally, p-values too big). I suggest using a model that seems reasonable.
Level 1: (
Level 2: (
Level 3: (
dat <- read.csv(paste0(getwd(), "/data/guatemalan.csv"))
dim(dat)[1] 2159 10head(dat) mom cluster immun kid2p momEdPri momEdSec husEdPri husEdSec rural pcInd81
1 2 1 1 1 0 1 0 1 0 0.1075042
2 185 36 0 1 1 0 1 0 0 0.0437295
3 186 36 0 1 1 0 0 1 0 0.0437295
4 187 36 0 1 1 0 1 0 0 0.0437295
5 188 36 0 1 1 0 0 0 0 0.0437295
6 188 36 1 1 1 0 0 0 0 0.0437295length(unique(dat$mom)) #Total number of mothers[1] 1595length(unique(dat$cluster)) #Total number of communities[1] 161table(table(dat$mom)) #Number of children per mom
1 2 3
1063 500 32 Only two normal random effects.
We can think of

Recall the model is conditioned on the random effects:
set.seed(123)
fit <- glmer(immun ~ kid2p + momEdPri + momEdSec + husEdPri + husEdSec + rural + pcInd81 + (1|mom) + (1|cluster), family = binomial, data = dat)Warning in checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0260502 (tol = 0.002, component 1)summary(fit)Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: binomial ( logit )
Formula: immun ~ kid2p + momEdPri + momEdSec + husEdPri + husEdSec + rural +
pcInd81 + (1 | mom) + (1 | cluster)
Data: dat
AIC BIC logLik deviance df.resid
2739.8 2796.6 -1359.9 2719.8 2149
Scaled residuals:
Min 1Q Median 3Q Max
-1.5854 -0.6356 -0.3569 0.6972 2.4860
Random effects:
Groups Name Variance Std.Dev.
mom (Intercept) 1.2357 1.1116
cluster (Intercept) 0.5033 0.7094
Number of obs: 2159, groups: mom, 1595; cluster, 161
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.7610 0.2800 -2.718 0.00658 **
kid2p 1.2812 0.1581 8.106 5.24e-16 ***
momEdPri 0.2793 0.1470 1.900 0.05749 .
momEdSec 0.2858 0.3248 0.880 0.37884
husEdPri 0.3763 0.1457 2.583 0.00979 **
husEdSec 0.3262 0.2733 1.194 0.23257
rural -0.6690 0.2049 -3.266 0.00109 **
pcInd81 -0.9823 0.2486 -3.951 7.79e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) kid2p mmEdPr mmEdSc hsEdPr hsEdSc rural
kid2p -0.450
momEdPri -0.343 0.106
momEdSec -0.234 0.073 0.350
husEdPri -0.355 0.091 -0.154 -0.067
husEdSec -0.281 0.014 -0.172 -0.474 0.394
rural -0.542 -0.075 -0.003 0.109 0.052 0.197
pcInd81 -0.438 -0.108 0.195 0.105 0.024 0.046 0.050
optimizer (Nelder_Mead) convergence code: 0 (OK)
Model failed to converge with max|grad| = 0.0260502 (tol = 0.002, component 1)What warning is this??
Convergence issues can happen for several reasons:
Model complexity: The model may be too complex, with too many random effects or predictors that cause instability in the estimation process.
Insufficient data: You might not have enough data to estimate all parameters properly, especially for random effects.
Scaling issues: Poor scaling of variables can make the optimization problem numerically unstable.
Starting values: Sometimes, the optimizer doesn’t start close enough to a good solution.
Use the control argument in glmer to allow more iterations or to relax convergence criteria.
fit_opt <- glmer(immun ~ kid2p + momEdPri + momEdSec + husEdPri + husEdSec + rural + pcInd81 + (1|mom) + (1|cluster), family = binomial, data = dat, control = glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e5)))
summary(fit_opt)Generalized linear mixed model fit by maximum likelihood (Laplace
Approximation) [glmerMod]
Family: binomial ( logit )
Formula: immun ~ kid2p + momEdPri + momEdSec + husEdPri + husEdSec + rural +
pcInd81 + (1 | mom) + (1 | cluster)
Data: dat
Control: glmerControl(optimizer = "bobyqa", optCtrl = list(maxfun = 2e+05))
AIC BIC logLik deviance df.resid
2739.8 2796.6 -1359.9 2719.8 2149
Scaled residuals:
Min 1Q Median 3Q Max
-1.5874 -0.6355 -0.3568 0.6970 2.4865
Random effects:
Groups Name Variance Std.Dev.
mom (Intercept) 1.2373 1.1123
cluster (Intercept) 0.5038 0.7098
Number of obs: 2159, groups: mom, 1595; cluster, 161
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -0.7624 0.2801 -2.722 0.00649 **
kid2p 1.2815 0.1581 8.105 5.26e-16 ***
momEdPri 0.2793 0.1470 1.899 0.05753 .
momEdSec 0.2808 0.3248 0.864 0.38735
husEdPri 0.3771 0.1457 2.588 0.00966 **
husEdSec 0.3308 0.2734 1.210 0.22628
rural -0.6686 0.2049 -3.263 0.00110 **
pcInd81 -0.9824 0.2487 -3.950 7.83e-05 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr) kid2p mmEdPr mmEdSc hsEdPr hsEdSc rural
kid2p -0.450
momEdPri -0.343 0.106
momEdSec -0.234 0.072 0.349
husEdPri -0.355 0.091 -0.154 -0.067
husEdSec -0.281 0.015 -0.172 -0.474 0.394
rural -0.542 -0.075 -0.003 0.109 0.052 0.196
pcInd81 -0.438 -0.108 0.195 0.106 0.024 0.045 0.050