Chapter 8 dyplyr
10월 15일 목요일, 202AIE17 송채은
What is dplyr?
dplyr is a grammar of data manipulation, he dplyr package provides a set of functions to transform or manipulate your data into the right form you need
- verb(a data frame, what to do with the data frame)
- filter() : picks cases based on their values
- select() : picks variables based on their names
- mutate() : adds new variables that are functions of existing variables
- arrange() : changes the ordering of the rows
- summarize() : reduces multiple values down to a single summary
These all combine naturally with group_by() which allows you to perform any operation “by group”
8.1 Tibbles
we often use a data frame to store our data. A tibble is the tidyverse version of the data frame in Base R
as_tibble() : convert a data frame to a tibble
## # A tibble: 150 x 5
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## <dbl> <dbl> <dbl> <dbl> <fct>
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
## # ... with 140 more rows8.2 Select columns with select()
select() subsets columns
## # A tibble: 336,776 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 1 1 517 515 2 830 819 11 UA 1545 N14228 EWR IAH
## 2 2013 1 1 533 529 4 850 830 20 UA 1714 N24211 LGA IAH
## 3 2013 1 1 542 540 2 923 850 33 AA 1141 N619AA JFK MIA
## 4 2013 1 1 544 545 -1 1004 1022 -18 B6 725 N804JB JFK BQN
## 5 2013 1 1 554 600 -6 812 837 -25 DL 461 N668DN LGA ATL
## 6 2013 1 1 554 558 -4 740 728 12 UA 1696 N39463 EWR ORD
## 7 2013 1 1 555 600 -5 913 854 19 B6 507 N516JB EWR FLL
## 8 2013 1 1 557 600 -3 709 723 -14 EV 5708 N829AS LGA IAD
## 9 2013 1 1 557 600 -3 838 846 -8 B6 79 N593JB JFK MCO
## 10 2013 1 1 558 600 -2 753 745 8 AA 301 N3ALAA LGA ORD
## # ... with 336,766 more rows, and 5 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>names() displays column names
## [1] "year" "month" "day" "dep_time" "sched_dep_time" "dep_delay" "arr_time"
## [8] "sched_arr_time" "arr_delay" "carrier" "flight" "tailnum" "origin" "dest"
## [15] "air_time" "distance" "hour" "minute" "time_hour"## # A tibble: 336,776 x 3
## year month day
## <int> <int> <int>
## 1 2013 1 1
## 2 2013 1 1
## 3 2013 1 1
## 4 2013 1 1
## 5 2013 1 1
## 6 2013 1 1
## 7 2013 1 1
## 8 2013 1 1
## 9 2013 1 1
## 10 2013 1 1
## # ... with 336,766 more rows: for selecting a range of consecutive variables
## # A tibble: 336,776 x 3
## year month day
## <int> <int> <int>
## 1 2013 1 1
## 2 2013 1 1
## 3 2013 1 1
## 4 2013 1 1
## 5 2013 1 1
## 6 2013 1 1
## 7 2013 1 1
## 8 2013 1 1
## 9 2013 1 1
## 10 2013 1 1
## # ... with 336,766 more rows! for taking the complement of a set of variables
## # A tibble: 336,776 x 16
## dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest air_time distance hour
## <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 517 515 2 830 819 11 UA 1545 N14228 EWR IAH 227 1400 5
## 2 533 529 4 850 830 20 UA 1714 N24211 LGA IAH 227 1416 5
## 3 542 540 2 923 850 33 AA 1141 N619AA JFK MIA 160 1089 5
## 4 544 545 -1 1004 1022 -18 B6 725 N804JB JFK BQN 183 1576 5
## 5 554 600 -6 812 837 -25 DL 461 N668DN LGA ATL 116 762 6
## 6 554 558 -4 740 728 12 UA 1696 N39463 EWR ORD 150 719 5
## 7 555 600 -5 913 854 19 B6 507 N516JB EWR FLL 158 1065 6
## 8 557 600 -3 709 723 -14 EV 5708 N829AS LGA IAD 53 229 6
## 9 557 600 -3 838 846 -8 B6 79 N593JB JFK MCO 140 944 6
## 10 558 600 -2 753 745 8 AA 301 N3ALAA LGA ORD 138 733 6
## # ... with 336,766 more rows, and 2 more variables: minute <dbl>, time_hour <dttm>- Selection helper functions
- starts_with() : Starts with a prefix
- ends_with() : Ends with a suffix
- contains() : Contains a literal string
- matches() : Matches a regular expression
- num_range() : Matches a numerical range like x01, x02, x03
select all columns starts with “dep”
## # A tibble: 336,776 x 2
## dep_time dep_delay
## <int> <dbl>
## 1 517 2
## 2 533 4
## 3 542 2
## 4 544 -1
## 5 554 -6
## 6 554 -4
## 7 555 -5
## 8 557 -3
## 9 557 -3
## 10 558 -2
## # ... with 336,766 more rowsselect all columns ends with “time”
## # A tibble: 336,776 x 5
## dep_time sched_dep_time arr_time sched_arr_time air_time
## <int> <int> <int> <int> <dbl>
## 1 517 515 830 819 227
## 2 533 529 850 830 227
## 3 542 540 923 850 160
## 4 544 545 1004 1022 183
## 5 554 600 812 837 116
## 6 554 558 740 728 150
## 7 555 600 913 854 158
## 8 557 600 709 723 53
## 9 557 600 838 846 140
## 10 558 600 753 745 138
## # ... with 336,766 more rowsselect all columns contains “time”
## # A tibble: 336,776 x 6
## dep_time sched_dep_time arr_time sched_arr_time air_time time_hour
## <int> <int> <int> <int> <dbl> <dttm>
## 1 517 515 830 819 227 2013-01-01 05:00:00
## 2 533 529 850 830 227 2013-01-01 05:00:00
## 3 542 540 923 850 160 2013-01-01 05:00:00
## 4 544 545 1004 1022 183 2013-01-01 05:00:00
## 5 554 600 812 837 116 2013-01-01 06:00:00
## 6 554 558 740 728 150 2013-01-01 05:00:00
## 7 555 600 913 854 158 2013-01-01 06:00:00
## 8 557 600 709 723 53 2013-01-01 06:00:00
## 9 557 600 838 846 140 2013-01-01 06:00:00
## 10 558 600 753 745 138 2013-01-01 06:00:00
## # ... with 336,766 more rowsselect all columns starts with “dep”, contains “time”, and between year and day (inclusive)
## # A tibble: 336,776 x 10
## dep_time dep_delay sched_dep_time arr_time sched_arr_time air_time time_hour year month day
## <int> <dbl> <int> <int> <int> <dbl> <dttm> <int> <int> <int>
## 1 517 2 515 830 819 227 2013-01-01 05:00:00 2013 1 1
## 2 533 4 529 850 830 227 2013-01-01 05:00:00 2013 1 1
## 3 542 2 540 923 850 160 2013-01-01 05:00:00 2013 1 1
## 4 544 -1 545 1004 1022 183 2013-01-01 05:00:00 2013 1 1
## 5 554 -6 600 812 837 116 2013-01-01 06:00:00 2013 1 1
## 6 554 -4 558 740 728 150 2013-01-01 05:00:00 2013 1 1
## 7 555 -5 600 913 854 158 2013-01-01 06:00:00 2013 1 1
## 8 557 -3 600 709 723 53 2013-01-01 06:00:00 2013 1 1
## 9 557 -3 600 838 846 140 2013-01-01 06:00:00 2013 1 1
## 10 558 -2 600 753 745 138 2013-01-01 06:00:00 2013 1 1
## # ... with 336,766 more rowsselect columns 1,3,5
## # A tibble: 336,776 x 3
## year day sched_dep_time
## <int> <int> <int>
## 1 2013 1 515
## 2 2013 1 529
## 3 2013 1 540
## 4 2013 1 545
## 5 2013 1 600
## 6 2013 1 558
## 7 2013 1 600
## 8 2013 1 600
## 9 2013 1 600
## 10 2013 1 600
## # ... with 336,766 more rows## # A tibble: 317 x 79
## artist track date.entered wk1 wk2 wk3 wk4 wk5 wk6 wk7 wk8 wk9 wk10 wk11 wk12 wk13 wk14 wk15 wk16 wk17
## <chr> <chr> <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2 Pac Baby~ 2000-02-26 87 82 72 77 87 94 99 NA NA NA NA NA NA NA NA NA NA
## 2 2Ge+h~ The ~ 2000-09-02 91 87 92 NA NA NA NA NA NA NA NA NA NA NA NA NA NA
## 3 3 Doo~ Kryp~ 2000-04-08 81 70 68 67 66 57 54 53 51 51 51 51 47 44 38 28 22
## 4 3 Doo~ Loser 2000-10-21 76 76 72 69 67 65 55 59 62 61 61 59 61 66 72 76 75
## 5 504 B~ Wobb~ 2000-04-15 57 34 25 17 17 31 36 49 53 57 64 70 75 76 78 85 92
## 6 98^0 Give~ 2000-08-19 51 39 34 26 26 19 2 2 3 6 7 22 29 36 47 67 66
## 7 A*Tee~ Danc~ 2000-07-08 97 97 96 95 100 NA NA NA NA NA NA NA NA NA NA NA NA
## 8 Aaliy~ I Do~ 2000-01-29 84 62 51 41 38 35 35 38 38 36 37 37 38 49 61 63 62
## 9 Aaliy~ Try ~ 2000-03-18 59 53 38 28 21 18 16 14 12 10 9 8 6 1 2 2 2
## 10 Adams~ Open~ 2000-08-26 76 76 74 69 68 67 61 58 57 59 66 68 61 67 59 63 67
## # ... with 307 more rows, and 59 more variables: wk18 <dbl>, wk19 <dbl>, wk20 <dbl>, wk21 <dbl>, wk22 <dbl>, wk23 <dbl>,
## # wk24 <dbl>, wk25 <dbl>, wk26 <dbl>, wk27 <dbl>, wk28 <dbl>, wk29 <dbl>, wk30 <dbl>, wk31 <dbl>, wk32 <dbl>, wk33 <dbl>,
## # wk34 <dbl>, wk35 <dbl>, wk36 <dbl>, wk37 <dbl>, wk38 <dbl>, wk39 <dbl>, wk40 <dbl>, wk41 <dbl>, wk42 <dbl>, wk43 <dbl>,
## # wk44 <dbl>, wk45 <dbl>, wk46 <dbl>, wk47 <dbl>, wk48 <dbl>, wk49 <dbl>, wk50 <dbl>, wk51 <dbl>, wk52 <dbl>, wk53 <dbl>,
## # wk54 <dbl>, wk55 <dbl>, wk56 <dbl>, wk57 <dbl>, wk58 <dbl>, wk59 <dbl>, wk60 <dbl>, wk61 <dbl>, wk62 <dbl>, wk63 <dbl>,
## # wk64 <dbl>, wk65 <dbl>, wk66 <lgl>, wk67 <lgl>, wk68 <lgl>, wk69 <lgl>, wk70 <lgl>, wk71 <lgl>, wk72 <lgl>, wk73 <lgl>,
## # wk74 <lgl>, wk75 <lgl>, wk76 <lgl>names() returns columns of a dataset
## [1] "artist" "track" "date.entered" "wk1" "wk2" "wk3" "wk4" "wk5"
## [9] "wk6" "wk7" "wk8" "wk9" "wk10" "wk11" "wk12" "wk13"
## [17] "wk14" "wk15" "wk16" "wk17" "wk18" "wk19" "wk20" "wk21"
## [25] "wk22" "wk23" "wk24" "wk25" "wk26" "wk27" "wk28" "wk29"
## [33] "wk30" "wk31" "wk32" "wk33" "wk34" "wk35" "wk36" "wk37"
## [41] "wk38" "wk39" "wk40" "wk41" "wk42" "wk43" "wk44" "wk45"
## [49] "wk46" "wk47" "wk48" "wk49" "wk50" "wk51" "wk52" "wk53"
## [57] "wk54" "wk55" "wk56" "wk57" "wk58" "wk59" "wk60" "wk61"
## [65] "wk62" "wk63" "wk64" "wk65" "wk66" "wk67" "wk68" "wk69"
## [73] "wk70" "wk71" "wk72" "wk73" "wk74" "wk75" "wk76"## # A tibble: 317 x 4
## wk20 wk21 wk22 wk23
## <dbl> <dbl> <dbl> <dbl>
## 1 NA NA NA NA
## 2 NA NA NA NA
## 3 14 12 7 6
## 4 70 NA NA NA
## 5 NA NA NA NA
## 6 94 NA NA NA
## 7 NA NA NA NA
## 8 86 NA NA NA
## 9 4 5 5 6
## 10 89 NA NA NA
## # ... with 307 more rows8.3 Filter rows with filter()
filter() subsets observations based on their values
filter the flights departed on Jan 1
## # A tibble: 842 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 1 1 517 515 2 830 819 11 UA 1545 N14228 EWR IAH
## 2 2013 1 1 533 529 4 850 830 20 UA 1714 N24211 LGA IAH
## 3 2013 1 1 542 540 2 923 850 33 AA 1141 N619AA JFK MIA
## 4 2013 1 1 544 545 -1 1004 1022 -18 B6 725 N804JB JFK BQN
## 5 2013 1 1 554 600 -6 812 837 -25 DL 461 N668DN LGA ATL
## 6 2013 1 1 554 558 -4 740 728 12 UA 1696 N39463 EWR ORD
## 7 2013 1 1 555 600 -5 913 854 19 B6 507 N516JB EWR FLL
## 8 2013 1 1 557 600 -3 709 723 -14 EV 5708 N829AS LGA IAD
## 9 2013 1 1 557 600 -3 838 846 -8 B6 79 N593JB JFK MCO
## 10 2013 1 1 558 600 -2 753 745 8 AA 301 N3ALAA LGA ORD
## # ... with 832 more rows, and 5 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>filter the flights departed on Nov or Dec
## # A tibble: 55,403 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 11 1 5 2359 6 352 345 7 B6 745 N568JB JFK PSE
## 2 2013 11 1 35 2250 105 123 2356 87 B6 1816 N353JB JFK SYR
## 3 2013 11 1 455 500 -5 641 651 -10 US 1895 N192UW EWR CLT
## 4 2013 11 1 539 545 -6 856 827 29 UA 1714 N38727 LGA IAH
## 5 2013 11 1 542 545 -3 831 855 -24 AA 2243 N5CLAA JFK MIA
## 6 2013 11 1 549 600 -11 912 923 -11 UA 303 N595UA JFK SFO
## 7 2013 11 1 550 600 -10 705 659 6 US 2167 N748UW LGA DCA
## 8 2013 11 1 554 600 -6 659 701 -2 US 2134 N742PS LGA BOS
## 9 2013 11 1 554 600 -6 826 827 -1 DL 563 N912DE LGA ATL
## 10 2013 11 1 554 600 -6 749 751 -2 DL 731 N315NB LGA DTW
## # ... with 55,393 more rows, and 5 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>filter the flights to IAH
## # A tibble: 7,198 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 1 1 517 515 2 830 819 11 UA 1545 N14228 EWR IAH
## 2 2013 1 1 533 529 4 850 830 20 UA 1714 N24211 LGA IAH
## 3 2013 1 1 623 627 -4 933 932 1 UA 496 N459UA LGA IAH
## 4 2013 1 1 728 732 -4 1041 1038 3 UA 473 N488UA LGA IAH
## 5 2013 1 1 739 739 0 1104 1038 26 UA 1479 N37408 EWR IAH
## 6 2013 1 1 908 908 0 1228 1219 9 UA 1220 N12216 EWR IAH
## 7 2013 1 1 1028 1026 2 1350 1339 11 UA 1004 N76508 LGA IAH
## 8 2013 1 1 1044 1045 -1 1352 1351 1 UA 455 N667UA EWR IAH
## 9 2013 1 1 1114 900 134 1447 1222 145 UA 1086 N76502 LGA IAH
## 10 2013 1 1 1205 1200 5 1503 1505 -2 UA 1461 N39418 EWR IAH
## # ... with 7,188 more rows, and 5 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>filter the flights with arr_delay <= 120 or dep_delay > 120
## # A tibble: 325,773 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 1 1 517 515 2 830 819 11 UA 1545 N14228 EWR IAH
## 2 2013 1 1 533 529 4 850 830 20 UA 1714 N24211 LGA IAH
## 3 2013 1 1 542 540 2 923 850 33 AA 1141 N619AA JFK MIA
## 4 2013 1 1 544 545 -1 1004 1022 -18 B6 725 N804JB JFK BQN
## 5 2013 1 1 554 600 -6 812 837 -25 DL 461 N668DN LGA ATL
## 6 2013 1 1 554 558 -4 740 728 12 UA 1696 N39463 EWR ORD
## 7 2013 1 1 555 600 -5 913 854 19 B6 507 N516JB EWR FLL
## 8 2013 1 1 557 600 -3 709 723 -14 EV 5708 N829AS LGA IAD
## 9 2013 1 1 557 600 -3 838 846 -8 B6 79 N593JB JFK MCO
## 10 2013 1 1 558 600 -2 753 745 8 AA 301 N3ALAA LGA ORD
## # ... with 325,763 more rows, and 5 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>8.4 Add new variables with mutate()
mutate() create a new variable
mutate() executes the transformations iteratively so that later transformations can use the columns created by earlier transformations
## # A tibble: 336,776 x 22
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 1 1 517 515 2 830 819 11 UA 1545 N14228 EWR IAH
## 2 2013 1 1 533 529 4 850 830 20 UA 1714 N24211 LGA IAH
## 3 2013 1 1 542 540 2 923 850 33 AA 1141 N619AA JFK MIA
## 4 2013 1 1 544 545 -1 1004 1022 -18 B6 725 N804JB JFK BQN
## 5 2013 1 1 554 600 -6 812 837 -25 DL 461 N668DN LGA ATL
## 6 2013 1 1 554 558 -4 740 728 12 UA 1696 N39463 EWR ORD
## 7 2013 1 1 555 600 -5 913 854 19 B6 507 N516JB EWR FLL
## 8 2013 1 1 557 600 -3 709 723 -14 EV 5708 N829AS LGA IAD
## 9 2013 1 1 557 600 -3 838 846 -8 B6 79 N593JB JFK MCO
## 10 2013 1 1 558 600 -2 753 745 8 AA 301 N3ALAA LGA ORD
## # ... with 336,766 more rows, and 8 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>,
## # gain <dbl>, hours <dbl>, gain_per_hour <dbl>transmute() creates a new variable but drops others
transmute(flights, gain = arr_delay - dep_delay, hours = air_time / 60, gain_per_hour = gain / hours)## # A tibble: 336,776 x 3
## gain hours gain_per_hour
## <dbl> <dbl> <dbl>
## 1 9 3.78 2.38
## 2 16 3.78 4.23
## 3 31 2.67 11.6
## 4 -17 3.05 -5.57
## 5 -19 1.93 -9.83
## 6 16 2.5 6.4
## 7 24 2.63 9.11
## 8 -11 0.883 -12.5
## 9 -5 2.33 -2.14
## 10 10 2.3 4.35
## # ... with 336,766 more rowsThe scoped variant of mutate() and transmute() help us to apply the same transformation to multiple variables - mutate_all() and transmute_all() affect every variable - mutate_at() and transmute_at() affect variables selected with a character vector - mutate_if() and transmute_if() affect variables selected with a predicate function (a function that returns TRUE or FALSE)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5.0 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
## 11 5.4 3.7 1.5 0.2 setosa
## 12 4.8 3.4 1.6 0.2 setosa
## 13 4.8 3.0 1.4 0.1 setosa
## 14 4.3 3.0 1.1 0.1 setosa
## 15 5.8 4.0 1.2 0.2 setosa
## 16 5.7 4.4 1.5 0.4 setosa
## 17 5.4 3.9 1.3 0.4 setosa
## 18 5.1 3.5 1.4 0.3 setosa
## 19 5.7 3.8 1.7 0.3 setosa
## 20 5.1 3.8 1.5 0.3 setosa
## 21 5.4 3.4 1.7 0.2 setosa
## 22 5.1 3.7 1.5 0.4 setosa
## 23 4.6 3.6 1.0 0.2 setosa
## 24 5.1 3.3 1.7 0.5 setosa
## 25 4.8 3.4 1.9 0.2 setosa
## 26 5.0 3.0 1.6 0.2 setosa
## 27 5.0 3.4 1.6 0.4 setosa
## 28 5.2 3.5 1.5 0.2 setosa
## 29 5.2 3.4 1.4 0.2 setosa
## 30 4.7 3.2 1.6 0.2 setosa
## 31 4.8 3.1 1.6 0.2 setosa
## 32 5.4 3.4 1.5 0.4 setosa
## 33 5.2 4.1 1.5 0.1 setosa
## 34 5.5 4.2 1.4 0.2 setosa
## 35 4.9 3.1 1.5 0.2 setosa
## 36 5.0 3.2 1.2 0.2 setosa
## 37 5.5 3.5 1.3 0.2 setosa
## 38 4.9 3.6 1.4 0.1 setosa
## 39 4.4 3.0 1.3 0.2 setosa
## 40 5.1 3.4 1.5 0.2 setosa
## 41 5.0 3.5 1.3 0.3 setosa
## 42 4.5 2.3 1.3 0.3 setosa
## 43 4.4 3.2 1.3 0.2 setosa
## 44 5.0 3.5 1.6 0.6 setosa
## 45 5.1 3.8 1.9 0.4 setosa
## 46 4.8 3.0 1.4 0.3 setosa
## 47 5.1 3.8 1.6 0.2 setosa
## 48 4.6 3.2 1.4 0.2 setosa
## 49 5.3 3.7 1.5 0.2 setosa
## 50 5.0 3.3 1.4 0.2 setosa
## 51 7.0 3.2 4.7 1.4 versicolor
## 52 6.4 3.2 4.5 1.5 versicolor
## 53 6.9 3.1 4.9 1.5 versicolor
## 54 5.5 2.3 4.0 1.3 versicolor
## 55 6.5 2.8 4.6 1.5 versicolor
## 56 5.7 2.8 4.5 1.3 versicolor
## 57 6.3 3.3 4.7 1.6 versicolor
## 58 4.9 2.4 3.3 1.0 versicolor
## 59 6.6 2.9 4.6 1.3 versicolor
## 60 5.2 2.7 3.9 1.4 versicolor
## 61 5.0 2.0 3.5 1.0 versicolor
## 62 5.9 3.0 4.2 1.5 versicolor
## 63 6.0 2.2 4.0 1.0 versicolor
## 64 6.1 2.9 4.7 1.4 versicolor
## 65 5.6 2.9 3.6 1.3 versicolor
## 66 6.7 3.1 4.4 1.4 versicolor
## 67 5.6 3.0 4.5 1.5 versicolor
## 68 5.8 2.7 4.1 1.0 versicolor
## 69 6.2 2.2 4.5 1.5 versicolor
## 70 5.6 2.5 3.9 1.1 versicolor
## 71 5.9 3.2 4.8 1.8 versicolor
## 72 6.1 2.8 4.0 1.3 versicolor
## 73 6.3 2.5 4.9 1.5 versicolor
## 74 6.1 2.8 4.7 1.2 versicolor
## 75 6.4 2.9 4.3 1.3 versicolor
## 76 6.6 3.0 4.4 1.4 versicolor
## 77 6.8 2.8 4.8 1.4 versicolor
## 78 6.7 3.0 5.0 1.7 versicolor
## 79 6.0 2.9 4.5 1.5 versicolor
## 80 5.7 2.6 3.5 1.0 versicolor
## 81 5.5 2.4 3.8 1.1 versicolor
## 82 5.5 2.4 3.7 1.0 versicolor
## 83 5.8 2.7 3.9 1.2 versicolor
## 84 6.0 2.7 5.1 1.6 versicolor
## 85 5.4 3.0 4.5 1.5 versicolor
## 86 6.0 3.4 4.5 1.6 versicolor
## 87 6.7 3.1 4.7 1.5 versicolor
## 88 6.3 2.3 4.4 1.3 versicolor
## 89 5.6 3.0 4.1 1.3 versicolor
## 90 5.5 2.5 4.0 1.3 versicolor
## 91 5.5 2.6 4.4 1.2 versicolor
## 92 6.1 3.0 4.6 1.4 versicolor
## 93 5.8 2.6 4.0 1.2 versicolor
## 94 5.0 2.3 3.3 1.0 versicolor
## 95 5.6 2.7 4.2 1.3 versicolor
## 96 5.7 3.0 4.2 1.2 versicolor
## 97 5.7 2.9 4.2 1.3 versicolor
## 98 6.2 2.9 4.3 1.3 versicolor
## 99 5.1 2.5 3.0 1.1 versicolor
## 100 5.7 2.8 4.1 1.3 versicolor
## 101 6.3 3.3 6.0 2.5 virginica
## 102 5.8 2.7 5.1 1.9 virginica
## 103 7.1 3.0 5.9 2.1 virginica
## 104 6.3 2.9 5.6 1.8 virginica
## 105 6.5 3.0 5.8 2.2 virginica
## 106 7.6 3.0 6.6 2.1 virginica
## 107 4.9 2.5 4.5 1.7 virginica
## 108 7.3 2.9 6.3 1.8 virginica
## 109 6.7 2.5 5.8 1.8 virginica
## 110 7.2 3.6 6.1 2.5 virginica
## 111 6.5 3.2 5.1 2.0 virginica
## 112 6.4 2.7 5.3 1.9 virginica
## 113 6.8 3.0 5.5 2.1 virginica
## 114 5.7 2.5 5.0 2.0 virginica
## 115 5.8 2.8 5.1 2.4 virginica
## 116 6.4 3.2 5.3 2.3 virginica
## 117 6.5 3.0 5.5 1.8 virginica
## 118 7.7 3.8 6.7 2.2 virginica
## 119 7.7 2.6 6.9 2.3 virginica
## 120 6.0 2.2 5.0 1.5 virginica
## 121 6.9 3.2 5.7 2.3 virginica
## 122 5.6 2.8 4.9 2.0 virginica
## 123 7.7 2.8 6.7 2.0 virginica
## 124 6.3 2.7 4.9 1.8 virginica
## 125 6.7 3.3 5.7 2.1 virginica
## 126 7.2 3.2 6.0 1.8 virginica
## 127 6.2 2.8 4.8 1.8 virginica
## 128 6.1 3.0 4.9 1.8 virginica
## 129 6.4 2.8 5.6 2.1 virginica
## 130 7.2 3.0 5.8 1.6 virginica
## 131 7.4 2.8 6.1 1.9 virginica
## 132 7.9 3.8 6.4 2.0 virginica
## 133 6.4 2.8 5.6 2.2 virginica
## 134 6.3 2.8 5.1 1.5 virginica
## 135 6.1 2.6 5.6 1.4 virginica
## 136 7.7 3.0 6.1 2.3 virginica
## 137 6.3 3.4 5.6 2.4 virginica
## 138 6.4 3.1 5.5 1.8 virginica
## 139 6.0 3.0 4.8 1.8 virginica
## 140 6.9 3.1 5.4 2.1 virginica
## 141 6.7 3.1 5.6 2.4 virginica
## 142 6.9 3.1 5.1 2.3 virginica
## 143 5.8 2.7 5.1 1.9 virginica
## 144 6.8 3.2 5.9 2.3 virginica
## 145 6.7 3.3 5.7 2.5 virginica
## 146 6.7 3.0 5.2 2.3 virginica
## 147 6.3 2.5 5.0 1.9 virginica
## 148 6.5 3.0 5.2 2.0 virginica
## 149 6.2 3.4 5.4 2.3 virginica
## 150 5.9 3.0 5.1 1.8 virginicaapply log() to the columns that contain “Sepal”
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 1.629241 1.252763 1.4 0.2 setosa
## 2 1.589235 1.098612 1.4 0.2 setosa
## 3 1.547563 1.163151 1.3 0.2 setosa
## 4 1.526056 1.131402 1.5 0.2 setosa
## 5 1.609438 1.280934 1.4 0.2 setosa
## 6 1.686399 1.360977 1.7 0.4 setosaapply log() to the numeric columns
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 1.629241 1.252763 0.3364722 -1.6094379 setosa
## 2 1.589235 1.098612 0.3364722 -1.6094379 setosa
## 3 1.547563 1.163151 0.2623643 -1.6094379 setosa
## 4 1.526056 1.131402 0.4054651 -1.6094379 setosa
## 5 1.609438 1.280934 0.3364722 -1.6094379 setosa
## 6 1.686399 1.360977 0.5306283 -0.9162907 setosa8.5 Arrange rows with arrange()
arrange() reorders rows in ascending order by default (use desc() to order in descending order)
## # A tibble: 336,776 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 1 1 517 515 2 830 819 11 UA 1545 N14228 EWR IAH
## 2 2013 1 1 533 529 4 850 830 20 UA 1714 N24211 LGA IAH
## 3 2013 1 1 542 540 2 923 850 33 AA 1141 N619AA JFK MIA
## 4 2013 1 1 544 545 -1 1004 1022 -18 B6 725 N804JB JFK BQN
## 5 2013 1 1 554 600 -6 812 837 -25 DL 461 N668DN LGA ATL
## 6 2013 1 1 554 558 -4 740 728 12 UA 1696 N39463 EWR ORD
## 7 2013 1 1 555 600 -5 913 854 19 B6 507 N516JB EWR FLL
## 8 2013 1 1 557 600 -3 709 723 -14 EV 5708 N829AS LGA IAD
## 9 2013 1 1 557 600 -3 838 846 -8 B6 79 N593JB JFK MCO
## 10 2013 1 1 558 600 -2 753 745 8 AA 301 N3ALAA LGA ORD
## # ... with 336,766 more rows, and 5 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>## # A tibble: 336,776 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 12 1 13 2359 14 446 445 1 B6 745 N715JB JFK PSE
## 2 2013 12 1 17 2359 18 443 437 6 B6 839 N593JB JFK BQN
## 3 2013 12 1 453 500 -7 636 651 -15 US 1895 N197UW EWR CLT
## 4 2013 12 1 520 515 5 749 808 -19 UA 1487 N69804 EWR IAH
## 5 2013 12 1 536 540 -4 845 850 -5 AA 2243 N634AA JFK MIA
## 6 2013 12 1 540 550 -10 1005 1027 -22 B6 939 N821JB JFK BQN
## 7 2013 12 1 541 545 -4 734 755 -21 EV 3819 N13968 EWR CVG
## 8 2013 12 1 546 545 1 826 835 -9 UA 1441 N23708 LGA IAH
## 9 2013 12 1 549 600 -11 648 659 -11 US 2167 N945UW LGA DCA
## 10 2013 12 1 550 600 -10 825 854 -29 B6 605 N706JB EWR FLL
## # ... with 336,766 more rows, and 5 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>8.6 Grouped summaries with summarize()
summarize() collapses a tibble to a single row for summary (imagine the sum and average function in Excel)
Summary functions takes a vector as an input and return a single value as an output : Counts, Locations, Position
summarize(flights, delay_mean = mean(dep_delay, na.rm = TRUE), delay_sd = sd(dep_delay, na.rm = TRUE))## # A tibble: 1 x 2
## delay_mean delay_sd
## <dbl> <dbl>
## 1 12.6 40.2Together __group_by()__, __summarize()__ will produce one row for each group
## # A tibble: 365 x 4
## # Groups: year, month [12]
## year month day delay
## <int> <int> <int> <dbl>
## 1 2013 1 1 11.5
## 2 2013 1 2 13.9
## 3 2013 1 3 11.0
## 4 2013 1 4 8.95
## 5 2013 1 5 5.73
## 6 2013 1 6 7.15
## 7 2013 1 7 5.42
## 8 2013 1 8 2.55
## 9 2013 1 9 2.28
## 10 2013 1 10 2.84
## # ... with 355 more rows## # A tibble: 150 x 5
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## <dbl> <dbl> <dbl> <dbl> <fct>
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
## # ... with 140 more rowswithout the pipe operator %>%
## # A tibble: 3 x 5
## Species Sepal.Length Sepal.Width Petal.Length Petal.Width
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 setosa 5.01 3.43 1.46 0.246
## 2 versicolor 5.94 2.77 4.26 1.33
## 3 virginica 6.59 2.97 5.55 2.03with the pipe operator %>%
## # A tibble: 3 x 5
## Species Sepal.Length Sepal.Width Petal.Length Petal.Width
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 setosa 5.01 3.43 1.46 0.246
## 2 versicolor 5.94 2.77 4.26 1.33
## 3 virginica 6.59 2.97 5.55 2.03## # A tibble: 3 x 3
## Species Sepal.Length Sepal.Width
## <fct> <dbl> <dbl>
## 1 setosa 5.01 3.43
## 2 versicolor 5.94 2.77
## 3 virginica 6.59 2.97data() loads specific data sets
## # A tibble: 600 x 15
## id gender race ses sch prog locus concept mot career read write math sci ss
## <dbl> <fct> <fct> <fct> <fct> <fct> <dbl> <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 55 female hispanic low public general -1.78 0.560 1 prof1 28.3 46.3 42.8 44.4 50.6
## 2 114 male african-amer middle public academic 0.240 -0.350 1 operative 30.5 35.9 36.9 33.6 40.6
## 3 490 male white middle public vocation -1.28 0.340 0.330 prof1 31 35.9 46.1 39 45.6
## 4 44 female hispanic low public vocation 0.220 -0.760 1 service 31 41.1 49.2 33.6 35.6
## 5 26 female hispanic middle public academic 1.12 -0.740 0.670 service 31 41.1 36 36.9 45.6
## 6 510 male white middle public vocation -0.860 1.19 0.330 operative 33.6 28.1 31.8 39.6 35.6
## 7 133 female african-amer low public vocation -0.230 0.440 0.330 school 33.6 35.2 40.9 29.3 25.7
## 8 213 female white low public general 0.0400 -0.470 0.670 clerical 33.6 59.3 44.7 47.1 45.6
## 9 548 female white middle private academic 0.470 0.340 0.670 prof2 33.6 43 41 49.8 35.6
## 10 309 female white high public general 0.320 0.900 0.670 prof2 33.6 51.5 41.9 49.8 50.6
## # ... with 590 more rowsnames() displays the column names of the HSB data
## [1] "id" "gender" "race" "ses" "sch" "prog" "locus" "concept" "mot" "career" "read" "write"
## [13] "math" "sci" "ss"## tibble [600 x 15] (S3: tbl_df/tbl/data.frame)
## $ id : num [1:600] 55 114 490 44 26 510 133 213 548 309 ...
## $ gender : Factor w/ 2 levels "male","female": 2 1 1 2 2 1 2 2 2 2 ...
## $ race : Factor w/ 4 levels "hispanic","asian",..: 1 3 4 1 1 4 3 4 4 4 ...
## $ ses : Factor w/ 3 levels "low","middle",..: 1 2 2 1 2 2 1 1 2 3 ...
## $ sch : Factor w/ 2 levels "public","private": 1 1 1 1 1 1 1 1 2 1 ...
## $ prog : Factor w/ 3 levels "general","academic",..: 1 2 3 3 2 3 3 1 2 1 ...
## $ locus : num [1:600] -1.78 0.24 -1.28 0.22 1.12 ...
## $ concept: num [1:600] 0.56 -0.35 0.34 -0.76 -0.74 ...
## $ mot : num [1:600] 1 1 0.33 1 0.67 ...
## $ career : Factor w/ 17 levels "clerical","craftsman",..: 9 8 9 15 15 8 14 1 10 10 ...
## $ read : num [1:600] 28.3 30.5 31 31 31 ...
## $ write : num [1:600] 46.3 35.9 35.9 41.1 41.1 ...
## $ math : num [1:600] 42.8 36.9 46.1 49.2 36 ...
## $ sci : num [1:600] 44.4 33.6 39 33.6 36.9 ...
## $ ss : num [1:600] 50.6 40.6 45.6 35.6 45.6 ...n() gives the current group size
## # A tibble: 4 x 2
## race n
## <fct> <int>
## 1 hispanic 71
## 2 asian 34
## 3 african-amer 58
## 4 white 437count() counts the unique values of variables
## # A tibble: 4 x 2
## race n
## <fct> <int>
## 1 hispanic 71
## 2 asian 34
## 3 african-amer 58
## 4 white 437## # A tibble: 8 x 3
## race gender n
## <fct> <fct> <int>
## 1 hispanic male 37
## 2 hispanic female 34
## 3 asian male 15
## 4 asian female 19
## 5 african-amer male 24
## 6 african-amer female 34
## 7 white male 197
## 8 white female 240the means of reading, writing, math, and social science scores
## # A tibble: 1 x 4
## readm writem mathm ssm
## <dbl> <dbl> <dbl> <dbl>
## 1 51.9 52.4 51.8 52.0the means of reading, writing, math, and social science scores by gender
HSB_tbl %>%
group_by(gender) %>%
summarise(readm = mean(read), writem = mean(write), mathm = mean(math), ssm = mean(ss))## # A tibble: 2 x 5
## gender readm writem mathm ssm
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 male 52.4 49.8 52.3 51.4
## 2 female 51.5 54.6 51.4 52.6the means of all numeric variables by gender
## # A tibble: 2 x 10
## gender id locus concept mot read write math sci ss
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 male 305. 0.0134 0.102 0.624 52.4 49.8 52.3 53.2 51.4
## 2 female 297. 0.166 -0.0762 0.692 51.5 54.6 51.4 50.5 52.6the means of all numeric variables by ses for only asian and hispanic
HSB_tbl %>%
filter(race %in% c("asian", "hispanic")) %>%
group_by(ses) %>%
summarise(across(where(is.numeric), mean, na.rm = TRUE))## # A tibble: 3 x 10
## ses id locus concept mot read write math sci ss
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 low 44.8 -0.142 -0.189 0.537 44.7 45.9 45.8 43.0 45.5
## 2 middle 50.8 -0.122 0.0525 0.584 47.3 48.6 48.3 47.5 47.9
## 3 high 66.9 0.118 0.0619 0.705 55.6 55.9 56.6 54.0 55.08.7 The pipe operator %>%
x %>% f(y) turns into f(x, y)
x %>% f(y) %>% g(z) turns into g(f(x,y), z)
## # A tibble: 336,776 x 19
## # Groups: dest [105]
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 1 1 517 515 2 830 819 11 UA 1545 N14228 EWR IAH
## 2 2013 1 1 533 529 4 850 830 20 UA 1714 N24211 LGA IAH
## 3 2013 1 1 542 540 2 923 850 33 AA 1141 N619AA JFK MIA
## 4 2013 1 1 544 545 -1 1004 1022 -18 B6 725 N804JB JFK BQN
## 5 2013 1 1 554 600 -6 812 837 -25 DL 461 N668DN LGA ATL
## 6 2013 1 1 554 558 -4 740 728 12 UA 1696 N39463 EWR ORD
## 7 2013 1 1 555 600 -5 913 854 19 B6 507 N516JB EWR FLL
## 8 2013 1 1 557 600 -3 709 723 -14 EV 5708 N829AS LGA IAD
## 9 2013 1 1 557 600 -3 838 846 -8 B6 79 N593JB JFK MCO
## 10 2013 1 1 558 600 -2 753 745 8 AA 301 N3ALAA LGA ORD
## # ... with 336,766 more rows, and 5 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>delay <- summarize(by_dest, count = n(), dist = mean(distance, na.rm = TRUE), delay = mean(arr_delay, na.rm = TRUE)) ## # A tibble: 96 x 4
## dest count dist delay
## <chr> <int> <dbl> <dbl>
## 1 ABQ 254 1826 4.38
## 2 ACK 265 199 4.85
## 3 ALB 439 143 14.4
## 4 ATL 17215 757. 11.3
## 5 AUS 2439 1514. 6.02
## 6 AVL 275 584. 8.00
## 7 BDL 443 116 7.05
## 8 BGR 375 378 8.03
## 9 BHM 297 866. 16.9
## 10 BNA 6333 758. 11.8
## # ... with 86 more rowsggplot(delay, aes(x = dist, y = delay)) +
geom_point(aes(size = count), alpha = 1/3) +
geom_smooth(se = FALSE)## `geom_smooth()` using method = 'loess' and formula 'y ~ x'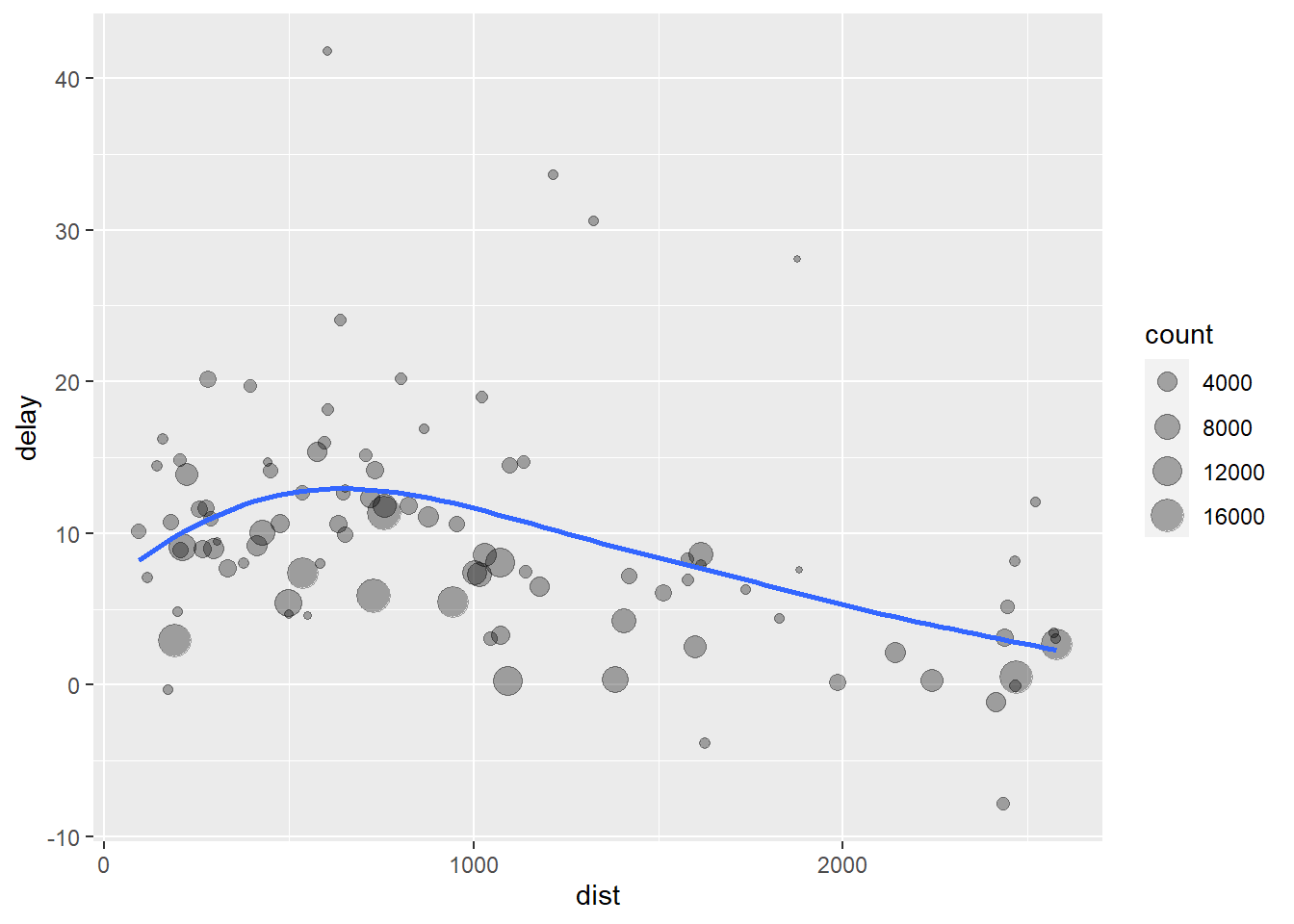
flights %>%
group_by(dest) %>%
summarize(count = n(), dist = mean(distance, na.rm = TRUE), delay = mean(arr_delay, na.rm = TRUE)) %>%
filter(count > 20, dest != "HNL") %>%
ggplot(aes(dist, delay)) +
geom_point(aes(size = count), alpha = 1/3) +
geom_smooth(se = FALSE)## `geom_smooth()` using method = 'loess' and formula 'y ~ x'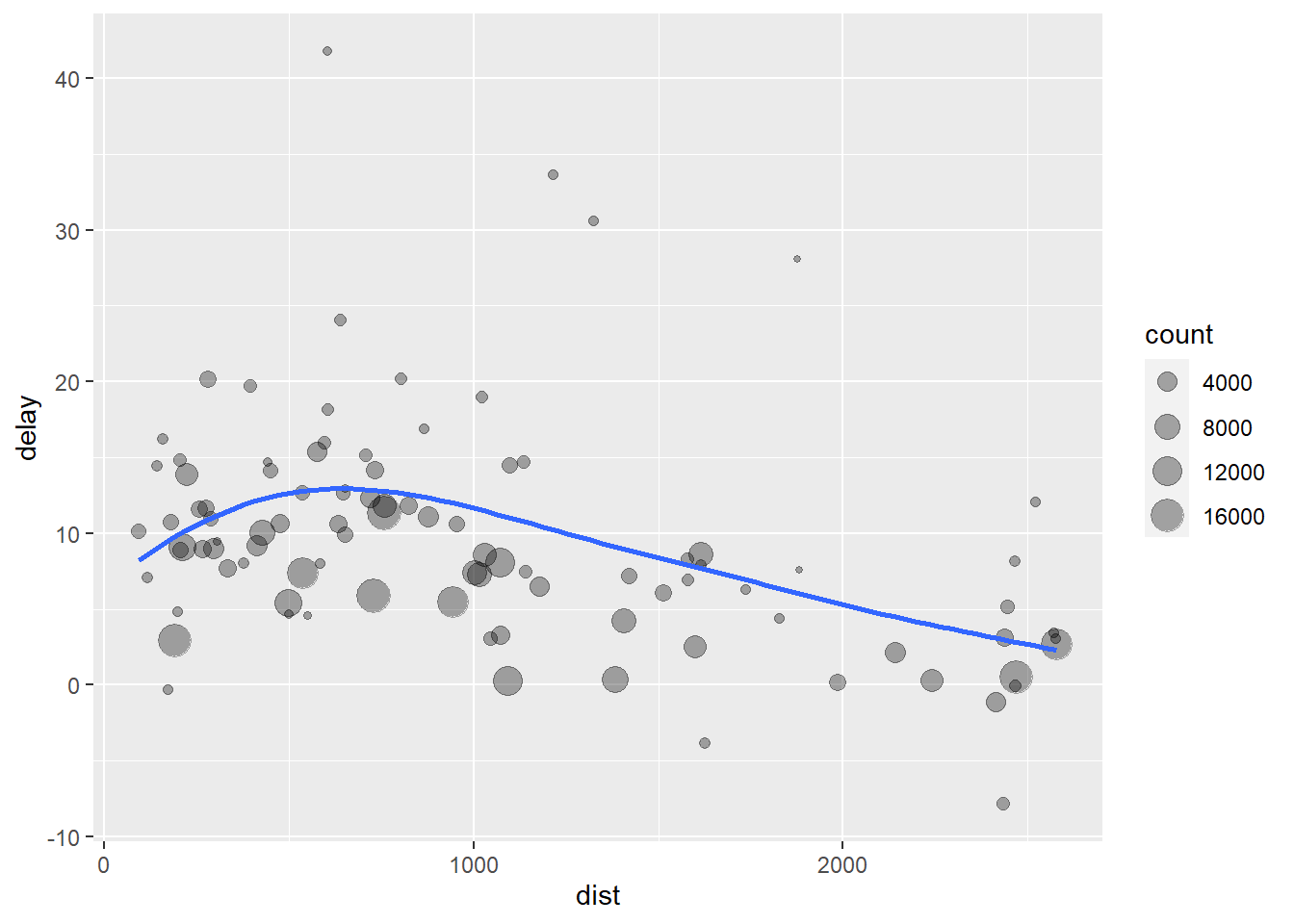
8.8 Mutating joins
A join is a way of matching each row in x to zero, one, or more rows in y
The variables used to match two data frames are called key variables(or keys)
The mutating joins add columns from one data frame y to another data frame x by matching rows based the key variable
inner_join() keeps observations that appear in both data frames
left_join() keeps all observations in x
right_join() keeps all observations in y
full_join() keeps all observations in x and y
tribble() creates a tibble
## # A tibble: 3 x 2
## key val_x
## <dbl> <chr>
## 1 1 x1
## 2 2 x2
## 3 3 x3## # A tibble: 3 x 2
## key val_y
## <dbl> <chr>
## 1 1 y1
## 2 2 y2
## 3 4 y3inner_join() keeps observations that appear in both data frames
## # A tibble: 2 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2## # A tibble: 2 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2left_join() keeps all observations in x
## # A tibble: 3 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 3 x3 <NA>## # A tibble: 3 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 3 x3 <NA>right_join() keeps all observations in y
## # A tibble: 3 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 4 <NA> y3## # A tibble: 3 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 4 <NA> y3full_join() keeps all observations in x and y
## # A tibble: 4 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 3 x3 <NA>
## 4 4 <NA> y3## # A tibble: 4 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 3 x3 <NA>
## 4 4 <NA> y38.9 dyplyr practice
8.9.1 starwars data
## # A tibble: 87 x 14
## name height mass hair_color skin_color eye_color birth_year sex gender homeworld species films vehicles starships
## <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr> <chr> <chr> <list> <list> <list>
## 1 Luke Skyw~ 172 77 blond fair blue 19 male mascul~ Tatooine Human <chr ~ <chr [2~ <chr [2]>
## 2 C-3PO 167 75 <NA> gold yellow 112 none mascul~ Tatooine Droid <chr ~ <chr [0~ <chr [0]>
## 3 R2-D2 96 32 <NA> white, blue red 33 none mascul~ Naboo Droid <chr ~ <chr [0~ <chr [0]>
## 4 Darth Vad~ 202 136 none white yellow 41.9 male mascul~ Tatooine Human <chr ~ <chr [0~ <chr [1]>
## 5 Leia Orga~ 150 49 brown light brown 19 fema~ femini~ Alderaan Human <chr ~ <chr [1~ <chr [0]>
## 6 Owen Lars 178 120 brown, grey light blue 52 male mascul~ Tatooine Human <chr ~ <chr [0~ <chr [0]>
## 7 Beru Whit~ 165 75 brown light blue 47 fema~ femini~ Tatooine Human <chr ~ <chr [0~ <chr [0]>
## 8 R5-D4 97 32 <NA> white, red red NA none mascul~ Tatooine Droid <chr ~ <chr [0~ <chr [0]>
## 9 Biggs Dar~ 183 84 black light brown 24 male mascul~ Tatooine Human <chr ~ <chr [0~ <chr [1]>
## 10 Obi-Wan K~ 182 77 auburn, whi~ fair blue-gray 57 male mascul~ Stewjon Human <chr ~ <chr [1~ <chr [5]>
## # ... with 77 more rowssimilar to str() in Base R
## Rows: 87
## Columns: 14
## $ name <chr> "Luke Skywalker", "C-3PO", "R2-D2", "Darth Vader", "Leia Organa", "Owen Lars", "Beru Whitesun lars", "R5-D...
## $ height <int> 172, 167, 96, 202, 150, 178, 165, 97, 183, 182, 188, 180, 228, 180, 173, 175, 170, 180, 66, 170, 183, 200,...
## $ mass <dbl> 77.0, 75.0, 32.0, 136.0, 49.0, 120.0, 75.0, 32.0, 84.0, 77.0, 84.0, NA, 112.0, 80.0, 74.0, 1358.0, 77.0, 1...
## $ hair_color <chr> "blond", NA, NA, "none", "brown", "brown, grey", "brown", NA, "black", "auburn, white", "blond", "auburn, ...
## $ skin_color <chr> "fair", "gold", "white, blue", "white", "light", "light", "light", "white, red", "light", "fair", "fair", ...
## $ eye_color <chr> "blue", "yellow", "red", "yellow", "brown", "blue", "blue", "red", "brown", "blue-gray", "blue", "blue", "...
## $ birth_year <dbl> 19.0, 112.0, 33.0, 41.9, 19.0, 52.0, 47.0, NA, 24.0, 57.0, 41.9, 64.0, 200.0, 29.0, 44.0, 600.0, 21.0, NA,...
## $ sex <chr> "male", "none", "none", "male", "female", "male", "female", "none", "male", "male", "male", "male", "male"...
## $ gender <chr> "masculine", "masculine", "masculine", "masculine", "feminine", "masculine", "feminine", "masculine", "mas...
## $ homeworld <chr> "Tatooine", "Tatooine", "Naboo", "Tatooine", "Alderaan", "Tatooine", "Tatooine", "Tatooine", "Tatooine", "...
## $ species <chr> "Human", "Droid", "Droid", "Human", "Human", "Human", "Human", "Droid", "Human", "Human", "Human", "Human"...
## $ films <list> [<"The Empire Strikes Back", "Revenge of the Sith", "Return of the Jedi", "A New Hope", "The Force Awaken...
## $ vehicles <list> [<"Snowspeeder", "Imperial Speeder Bike">, <>, <>, <>, "Imperial Speeder Bike", <>, <>, <>, <>, "Tribubbl...
## $ starships <list> [<"X-wing", "Imperial shuttle">, <>, <>, "TIE Advanced x1", <>, <>, <>, <>, "X-wing", <"Jedi starfighter"...Exercise 1. How many humans are contained in starwars overall? (Hint. use count())
## # A tibble: 1 x 1
## n
## <int>
## 1 35## # A tibble: 38 x 2
## species n
## <chr> <int>
## 1 Aleena 1
## 2 Besalisk 1
## 3 Cerean 1
## 4 Chagrian 1
## 5 Clawdite 1
## 6 Droid 6
## 7 Dug 1
## 8 Ewok 1
## 9 Geonosian 1
## 10 Gungan 3
## # ... with 28 more rowsExercise 2. How many humans are contained in starwars by gender?
## # A tibble: 42 x 3
## # Groups: gender, species [42]
## gender species n
## <chr> <chr> <int>
## 1 feminine Clawdite 1
## 2 feminine Droid 1
## 3 feminine Human 9
## 4 feminine Kaminoan 1
## 5 feminine Mirialan 2
## 6 feminine Tholothian 1
## 7 feminine Togruta 1
## 8 feminine Twi'lek 1
## 9 masculine Aleena 1
## 10 masculine Besalisk 1
## # ... with 32 more rowsExercise 3. From which homeworld do the most indidividuals(rows) come from?
## # A tibble: 49 x 2
## # Groups: homeworld [49]
## homeworld n
## <chr> <int>
## 1 Naboo 11
## 2 Tatooine 10
## 3 <NA> 10
## 4 Alderaan 3
## 5 Coruscant 3
## 6 Kamino 3
## 7 Corellia 2
## 8 Kashyyyk 2
## 9 Mirial 2
## 10 Ryloth 2
## # ... with 39 more rowsExercise 4. What is the mean height of all individuals with orange eyes from the most popular homeworld?
## Warning in mean.default(., "height", na.rm = TRUE): argument is not numeric or logical: returning NA## [1] NAExercise 5. Compute the median, mean, and standard deviation of height for all droids
## Warning in mean.default(.): argument is not numeric or logical: returning NA## [1] NA8.9.2 data sets in the ds4psy package
## # A tibble: 295 x 294
## id intervention sex age educ income occasion.0 elapsed.days.0 ahi01.0 ahi02.0 ahi03.0 ahi04.0 ahi05.0 ahi06.0 ahi07.0
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 4 2 35 5 3 0 0 2 3 2 3 3 2 3
## 2 2 1 1 59 1 1 0 0 3 4 3 4 2 3 4
## 3 3 4 1 51 4 3 0 0 3 3 2 4 2 3 4
## 4 4 3 1 50 5 2 0 0 2 2 3 2 2 3 3
## 5 5 2 2 58 5 2 0 0 1 2 3 2 2 2 1
## 6 6 1 1 31 5 1 0 0 2 2 3 3 2 3 3
## 7 7 3 1 44 5 2 0 0 3 2 2 3 3 3 3
## 8 8 2 1 57 4 2 0 0 3 2 2 2 2 1 4
## 9 9 1 1 36 4 3 0 0 2 2 1 2 2 1 3
## 10 10 2 1 45 4 3 0 0 2 2 3 2 3 1 4
## # ... with 285 more rows, and 279 more variables: ahi08.0 <dbl>, ahi09.0 <dbl>, ahi10.0 <dbl>, ahi11.0 <dbl>, ahi12.0 <dbl>,
## # ahi13.0 <dbl>, ahi14.0 <dbl>, ahi15.0 <dbl>, ahi16.0 <dbl>, ahi17.0 <dbl>, ahi18.0 <dbl>, ahi19.0 <dbl>, ahi20.0 <dbl>,
## # ahi21.0 <dbl>, ahi22.0 <dbl>, ahi23.0 <dbl>, ahi24.0 <dbl>, cesd01.0 <dbl>, cesd02.0 <dbl>, cesd03.0 <dbl>, cesd04.0 <dbl>,
## # cesd05.0 <dbl>, cesd06.0 <dbl>, cesd07.0 <dbl>, cesd08.0 <dbl>, cesd09.0 <dbl>, cesd10.0 <dbl>, cesd11.0 <dbl>,
## # cesd12.0 <dbl>, cesd13.0 <dbl>, cesd14.0 <dbl>, cesd15.0 <dbl>, cesd16.0 <dbl>, cesd17.0 <dbl>, cesd18.0 <dbl>,
## # cesd19.0 <dbl>, cesd20.0 <dbl>, ahiTotal.0 <dbl>, cesdTotal.0 <dbl>, occasion.1 <dbl>, elapsed.days.1 <dbl>, ahi01.1 <dbl>,
## # ahi02.1 <dbl>, ahi03.1 <dbl>, ahi04.1 <dbl>, ahi05.1 <dbl>, ahi06.1 <dbl>, ahi07.1 <dbl>, ahi08.1 <dbl>, ahi09.1 <dbl>,
## # ahi10.1 <dbl>, ahi11.1 <dbl>, ahi12.1 <dbl>, ahi13.1 <dbl>, ahi14.1 <dbl>, ahi15.1 <dbl>, ahi16.1 <dbl>, ahi17.1 <dbl>,
## # ahi18.1 <dbl>, ahi19.1 <dbl>, ahi20.1 <dbl>, ahi21.1 <dbl>, ahi22.1 <dbl>, ahi23.1 <dbl>, ahi24.1 <dbl>, cesd01.1 <dbl>,
## # cesd02.1 <dbl>, cesd03.1 <dbl>, cesd04.1 <dbl>, cesd05.1 <dbl>, cesd06.1 <dbl>, cesd07.1 <dbl>, cesd08.1 <dbl>,
## # cesd09.1 <dbl>, cesd10.1 <dbl>, cesd11.1 <dbl>, cesd12.1 <dbl>, cesd13.1 <dbl>, cesd14.1 <dbl>, cesd15.1 <dbl>,
## # cesd16.1 <dbl>, cesd17.1 <dbl>, cesd18.1 <dbl>, cesd19.1 <dbl>, cesd20.1 <dbl>, ahiTotal.1 <dbl>, cesdTotal.1 <dbl>,
## # occasion.2 <dbl>, elapsed.days.2 <dbl>, ahi01.2 <dbl>, ahi02.2 <dbl>, ahi03.2 <dbl>, ahi04.2 <dbl>, ahi05.2 <dbl>,
## # ahi06.2 <dbl>, ahi07.2 <dbl>, ahi08.2 <dbl>, ahi09.2 <dbl>, ahi10.2 <dbl>, ahi11.2 <dbl>, ...Exercise 6. What are the data’s dimensions, variables, and types of variables?
## [1] "list"Exercise 7. From posPsy_wide, select id, intervention, sex, age, educ, income, and 6 ahi01 items across six waves, and assign the name posPsy_wide_subset
\d{2}is a regular expression that representsany two-digits`
## # A tibble: 295 x 12
## id intervention sex age educ income ahi01.0 ahi01.1 ahi01.2 ahi01.3 ahi01.4 ahi01.5
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 4 2 35 5 3 2 3 NA NA NA NA
## 2 2 1 1 59 1 1 3 3 3 3 3 3
## 3 3 4 1 51 4 3 3 NA 3 NA NA NA
## 4 4 3 1 50 5 2 2 3 NA 1 3 NA
## 5 5 2 2 58 5 2 1 1 2 2 2 NA
## 6 6 1 1 31 5 1 2 NA 3 3 NA NA
## 7 7 3 1 44 5 2 3 NA NA NA NA NA
## 8 8 2 1 57 4 2 3 2 2 3 2 NA
## 9 9 1 1 36 4 3 2 NA NA NA NA NA
## 10 10 2 1 45 4 3 2 NA NA NA NA NA
## # ... with 285 more rowsExercise 8. Using the pivot_longer() in tidyr, make posPsy_wide_subset longer
posPsy_wide_subset %>%
pivot_longer(
cols = starts_with("ahi"),
names_to = "ahi",
values_to = "mesaurement"
)## # A tibble: 1,770 x 8
## id intervention sex age educ income ahi mesaurement
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <dbl>
## 1 1 4 2 35 5 3 ahi01.0 2
## 2 1 4 2 35 5 3 ahi01.1 3
## 3 1 4 2 35 5 3 ahi01.2 NA
## 4 1 4 2 35 5 3 ahi01.3 NA
## 5 1 4 2 35 5 3 ahi01.4 NA
## 6 1 4 2 35 5 3 ahi01.5 NA
## 7 2 1 1 59 1 1 ahi01.0 3
## 8 2 1 1 59 1 1 ahi01.1 3
## 9 2 1 1 59 1 1 ahi01.2 3
## 10 2 1 1 59 1 1 ahi01.3 3
## # ... with 1,760 more rowsExercise 9. Using separate() in tidyr, create a variable Wave that indicates the measurement index (i.e., 0, 1, 2, 3, 4, 5)