Chapter 5 Functions
Every operation on a value, vector, object, and other structures in R is done using functions. You’ve already used several: c() is a function, as are <-, +, -, *, etc. A function is a special kind of object that takes some arguments and uses them to run the code contained inside of it. Every function has the form function.name() and the arguments are given inside the brackets. Arguments must be separated with a comma. Different functions take different number of various arguments. The help file brought up by the ?... gives a list of arguments used by the given function along with the description of what the arguments do and what legal values they take.
Let’s look at the most basic functions in R: the assignment operator <- and the concatenate function c(). Yes, <- is a function, even though the way it’s used differs from most other functions. It takes exactly two arguments – a name of an object to the left of it and an object or a value to the right of it. the output of this function is an object with the provided name containing the values assigned:
# arg 1: name function arg 2: value
x <- 3
# print x
x## [1] 3# arg 1: name function arg 2: object + value
y <- x + 7
y## [1] 10The c() function, on the other hand takes an arbitrary number of arguments, including none at all. The arguments can either be single values (numeric, character, logical, NA), or objects. The function then outputs a single vector consisting of all the arguments:
c(1, 5, x, 9, y)## [1] 1 5 3 9 105.1 Packages
All functions in R, except the ones you write yourself or copy from online forums, come in packages. These are essentially folders that contain all the code that gets run whenever you use a function along with help files and some other data. Basic R installation comes with several packages and every time you open R or RStudio, some of these will get loaded, making the functions these packages contain available for you to use. That’s why you don’t have to worry about packages when using functions such as c(), mean(), or plot().
Other functions however come in packages that either don’t get loaded automatically upon startup or need to be installed. For instance, a very handy package for data visualisation that is not installed automatically is ggplot2. If we want to make use of the many functions in this package, we need to first install it using the install.packages() command:
install.packages("ggplot2")Notice that the name of the package is in quotes. This is important. Without the quotes, the command will not work!
You only ever need to install a package once. R will go online, download the package from the package repository and install it on your computer.
Once we’ve installed a package, we need to load it so that R can access the functions provided in the package. This is done using the library() command:
library(ggplot2) # no quotes needed this timePackages have to be loaded every session. If you load a package, you can use its functions until you close R. When you re-open it, you will have to load the package again. For that reason, it is very useful to load all the packages you will need for your data processing, visualisation, and analysis at the very beginning of your script.
Now that we have made ggplot2 available for our current R session, we can use any of its functions. For a quick descriptive plot of the a variable, we can use the qplot() function:
qplot(rnorm(1000, 100, 15)) # random normal variable of 1000 values with mean = 100 and sd = 15## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
If you don’t install or load the package that contains the function you want to use, R will tell you that it cannot find the function. Let’s illustrate this on the example of a function describe() from the package psych which is pre-installed but not loaded at startup:
describe(df$ID)## Error in describe(df$ID): could not find function "describe"Once, we load the psych package, R will be able to find the function:
library(psych)##
## Attaching package: 'psych'## The following objects are masked from 'package:ggplot2':
##
## %+%, alphadescribe(df$ID)## vars n mean sd median trimmed mad min max range skew kurtosis
## X1 1 325 163 93.96 163 163 120.09 1 325 324 0 -1.21
## se
## X1 5.215.2 Using functions
5.2.1 The ()s
As a general rule of thumb, the way to tell R that the thing we are calling is a function is to put brackets – () – after the name, e.g., data.frame(), rnorm(), or factor(). The only exception to this rule are operators – functions that have a convenient infix form – such as the assignment operators (<-, =), mathematical operators (+, ^, %%, …), logical operators (==, >, %in%, …), and a handful of others. Even the subsetting square brackets [] are a function! However, the infix form is just a shorthand and all of these can be used in the standard way functions are used:
2 + 3 # infix form## [1] 5`+`(2, 3) # prefix form, notice the backticks## [1] 5The above is to explain the logic behind a simple principle in R programming: If it is a function (used in its standard prefix form) then it must have ()s. If it is not a function, then it must NOT have them. If you understand this, you will never attempt to run commands like as.factor[x] or my_data(...)!
5.2.2 Specifying arguments
The vast majority of functions require you to give it at least one argument. Arguments of a function are often named. From the point of view of a user of a function, these names are only placeholders for some values we want to pass to the function. In RStudio, you can type the name of a function, open the bracket and then press the Tab key to see a list of arguments the function takes. You can try it now, type, for instance, sd( and press the Tab key. You should see a pop-up list of two arguments – x = and na.rm = – appear.
This means, that to use the sd() function explicitly, we much give it two arguments: a numeric vector and a single logical value (technically, a logical vector of length 1):
sd(x = c(-4, 1, 100, 52, -32, 0.5433, NA), na.rm = FALSE)## [1] NAThe output of the function is NA because our vector contained an NA value and the na.rm = argument that removes the NAs was set to FALSE. Try setting it to TRUE (or T if you’re a lazy typist) and see what R gives you.
Look what happens when we run the following command:
sd(x = c(-4, 1, 100, 52, -32, 0.5433)) # no NAs this time## [1] 47.83839We didn’t specify the value of the na.rm = argument but the code worked anyway. Why might that be…?
5.2.2.1 Default values of arguments
The reason for this behaviour is that functions can have arguments set to some value by default to facilitate the use of the functions in the most common situations by reducing the amount of typing. Look at the documentation for the sd() function (by running ?sd in the console).
You should see that under “Usage” it reads sd(x, na.rm = FALSE). This means that, by default, the na.rm = argument is set to FALSE and if you don’t specify its value manually, the function will run with this setting. Re-visiting our example with the NA value, you can see that the output is as it was before:
sd(c(-4, 1, 100, 52, -32, 0.5433, NA))## [1] NA5.2.2.2 Argument matching
I hope you noticed a tiny change in the way the first argument was specified in the line above (coding is a lot about attention to detail!) – there is no x = in the code.
The reason why R is still able to understand what we meant is argument matching. If no names are given to the arguments, R assumes they are entered in the order in which they were specified when the function was created. This is the same order you can look up in the “Usage” section of the function documentation (using ?)
To give you another example, take rnorm() for instance. If you pull up the documentation (AKA help) of the file with ?rnorm, you’ll see that it takes 3 arguments: n =, mean =, and sd =. The latter two have default values but n = doesn’t so we must provide its value.
Setting the first argument to 10 and omitting the other 2 will generate a vector of 10 numbers drawn from a normal distribution with \(\mu = 0\) and \(\sigma=1\):
rnorm(10)## [1] -1.18835097 1.53782361 -0.31321827 0.14730795 -0.14572524
## [6] -0.53147535 -2.35880152 -0.07096727 1.17249619 0.98135234Lets say we want to change the mean to -5 but keep standard deviation the same. Relying on argument matching, we can do:
rnorm(10, -5)## [1] -5.038285 -5.225887 -4.714281 -5.117829 -3.540936 -3.888444 -4.082252
## [8] -5.097556 -4.353539 -5.719502However, if we want to change the sd = argument to 3 but leave mean = set to 0, we need to let R know this more explicitly. There are several ways to do the same thing but they all rely on the principle that unnamed values will be interpreted in order of arguments:
rnorm(10, 0, 3) # keep mean = 0## [1] 2.65949880 -4.24562258 2.43576330 -0.01174611 -2.82576647
## [6] 0.62116644 1.27132609 3.74690482 -2.59212148 -2.48575096rnorm(10, , 3) # skip mean = (DON'T DO THIS! it's illegible)## [1] -6.2477789 3.2320593 -0.5055326 -0.4662970 1.1038795 -2.1389158
## [7] 2.7513674 -3.5566095 5.7317257 2.6879213If the arguments are named, they can be entered in any order:
rnorm(sd = 2, n = 10, mean = -100)## [1] -100.05658 -94.66810 -99.98974 -99.57405 -100.92782 -97.76991
## [7] -97.75127 -100.71410 -96.86328 -97.06812The important point here is that if you give a function 4 (for example) unnamed values separated with commas, R will try to match them to the first 4 arguments of the function. If the function takes fewer than arguments or if the values are not valid for the respective arguments, R will throw an error:
rnorm(100, 5, -3, 8) # more values than arguments## Error in rnorm(100, 5, -3, 8): unused argument (8)That is, it will throw an error if you’re lucky. If you’re not, you might get all sorts of unexpected behaviours:
rnorm(10, T, -7) # illegal values passed to arguments## Warning in rnorm(10, T, -7): NAs produced## [1] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN5.2.2.3 Passing vectors as arguments
The majority of functions – and you’ve already seen quite a few of these – have at least one argument that can take multiple values. The x = (first) argument of sample() or the labels = argument of factor() are examples of these.
Imagine we want to sample 10 draws with replacement from the words “elf”, “orc”, “hobbit”, and “dwarf”. Your intuition might be to write something like sample("elf", "orc", "hobbit", "dwarf", 10). That will however not work:
sample("elf", "orc", "hobbit", "dwarf", 10, replace = T)## Error in sample("elf", "orc", "hobbit", "dwarf", 10, replace = T): unused arguments ("dwarf", 10)
Take a moment to ponder why this produces an error…
Yes, you’re right, it has to do with argument matching! R interprets the above command as you passing five arguments to the sample() function, which only takes three arguments. Moreover, the second argument size = must be a positive number, replace = must be a single logical value, and prob = is a vector of numbers between 0 and 1 that must add up to 1 and the vector must be of the same length as the vector passed to the first x = argument. As you can see, our command fails on most of these criteria.
So, how do we tell R that we want to pass the four races of Middle-earth to the first argument of sample()? Well, we need to bind them into a single vector using the c() function:
sample(c("elf", "orc", "hobbit", "dwarf"), 10, T)## [1] "elf" "dwarf" "hobbit" "dwarf" "dwarf" "dwarf" "orc"
## [8] "elf" "dwarf" "orc"Remember: If you want to pass a vector of values into a single argument of a function, you need to use an object in your environment containing the vector or a function that outputs a vector. The basic one is c() but others work too, e.g., sample(5:50, 10) (the : operator returns a vector containing a complete sequence of integers between the specified values).
5.2.2.4 Passing objects as arguments
Everything in R is an object and thus the values passed as arguments to functions are also objects. It is completely up to you whether you want to create the object ad hoc for the purpose of only passing it to a function or whether you want to pass to a function an object you already have in your environment. For example, if our four races are of particular interest to us and we want to keep them for future use, we can assign them to the environment under some name:
ME_races <- c("elf", "orc", "hobbit", "dwarf")
ME_races # here they are## [1] "elf" "orc" "hobbit" "dwarf"Then, we can just use them as arguments to functions:
factor(sample(1:4, 20, T), labels = ME_races)## [1] hobbit orc hobbit hobbit elf hobbit orc orc hobbit hobbit
## [11] dwarf elf dwarf dwarf hobbit elf orc dwarf elf elf
## Levels: elf orc hobbit dwarf5.3 Function output
5.3.1 Command is a representation of its output
Any complete command in R, such as the one above is merely a symbolic representation of the output it returns. Understanding this is crucial! Just like in a natural language, there are many ways to say the same thing, there are multiple ways of producing the same output in R. It’s not called a programming language for nothing!
5.3.1.1 One-way street
Another important thing to realise is that, given that there are many ways to do the same thing in R, there is a sort of directionality to the relationship between a command and its output. If you know what a function does, you can unambiguously tell what the output will be given specified arguments. However, once the output is produced, there is no way R can tell what command was used.
Imagine you have three bakers making the same kind of bread: one uses the traditional kneading method, one uses the slap-and-fold technique, and one uses a mixer. If you know the recipe and the procedure they are using, you will be able to tell what they’re making. However, once you have your three loaves in front of you, you won’t be able to say which came from which baker. It’s the same thing with commands in R!
This is the reason why some commands look like they’re repeating things beyond necessity. Take, for instance, this line:
mat[lower.tri(mat)] <- "L"The lower.tri() function takes a matrix as its first argument and returns a matrix of logicals with the same dimensions as the matrix provided. Once it returns its output, R has no way of knowing what matrix was used to produce it and so it has no clue that it has anything to do with our matrix mat. That’s why, if we want to modify the lower triangle of mat, we do it this way.
Obviously, nothing is stopping you from creating the logical matrix by means of some other approach and then use it to subset mat but the above solution is both more elegant and more intelligible.
5.3.2 Knowe thine output as thou knowest thyself
Because more often than not you will be using function to create some object only so that you can feed it into another function, it is essential that you understand what you are asking R to do and know what result you are expecting. There should be no surprises!
A good way to practice is to say to yourself what the output of a command will be before you run it. For instance, the command factor(sample(1:4, 20, T), labels = ME_races) returns a vector of class factor and length 20 containing randomly sampled values labelled according to the four races of Middle-earth we worked with.
5.4 Output is an object
Notice that in the code above we passed the sample(1:4, 20, T) command as the first argument of factor(). This works because – as we mentioned earlier – a command is merely a symbolic representation of its output and because everything in R is an object. This means that function output is also an object. Depending on the particular function, the output can be anything from, e.g., a logical vector of length 1 through long vectors and matrices to huge data frames and complex lists of lists of lists…
For instance, the t.test() function returns a list that contains all the information about the test you might ever need:
t_output <- t.test(rnorm(100, 5, 2), mu = 0)
str(t_output) # see the structure of the object## List of 9
## $ statistic : Named num 27.8
## ..- attr(*, "names")= chr "t"
## $ parameter : Named num 99
## ..- attr(*, "names")= chr "df"
## $ p.value : num 1.44e-48
## $ conf.int : num [1:2] 4.78 5.52
## ..- attr(*, "conf.level")= num 0.95
## $ estimate : Named num 5.15
## ..- attr(*, "names")= chr "mean of x"
## $ null.value : Named num 0
## ..- attr(*, "names")= chr "mean"
## $ alternative: chr "two.sided"
## $ method : chr "One Sample t-test"
## $ data.name : chr "rnorm(100, 5, 2)"
## - attr(*, "class")= chr "htest"If we want to know the p-value of the above test, we can simply query the list accordingly:
t_output$p.value## [1] 1.442878e-48Because the command only represents the output object, it can be accessed in the same way. Say you are running some kind of simulation study and are only interested in the t-statistic of the test. Instead of saving the entire output into some kind of named object in the environment, you can simply save the t:
t_stat <- t.test(rnorm(100, 5, 2), mu = 0)$statistic
t_stat## t
## 27.837335.4.1 Where does the output go?
Let’s go back to discussing factor(). There’s another important issue that sometime causes a level of consternation among novice R users. Imagine we have a data set and we want to designate one of its columns as a factor so that R knows that the column contains a categorical variable.
df <- data.frame(id = 1:10, x = rnorm(10))
df## id x
## 1 1 0.6052863
## 2 2 -0.5805976
## 3 3 0.5701260
## 4 4 -0.7590538
## 5 5 -1.8907664
## 6 6 0.7089412
## 7 7 -1.1660943
## 8 8 2.1680322
## 9 9 1.3509332
## 10 10 0.7968681An intuitive way of turning id into a factor might be:
factor(df$id)## [1] 1 2 3 4 5 6 7 8 9 10
## Levels: 1 2 3 4 5 6 7 8 9 10This, however, does not work:
class(df$id)## [1] "integer"The reason for this has to do with the fact that the argument-output relationship is directional. Once the object inside df$id is passed to factor(), R forgets about the fact that it had anything to do with df or one of its columns. It has therefore no way of knowing that you want to be modifying a column of a data frame. Because of that, the only place factor() can return the output to is the one it uses by default.
5.4.1.1 Console
The vast majority of functions return their output into the console. factor() is one of these functions. That’s why when you type in the command above, you will see the printout of the output in the console. Once it’s been returned, the output is forgotten about – R can’t see the console or read from it!
This is why factor(df$id) does not turn the id column of id into a factor.
5.4.1.2 Environment
A small number of functions return a named object to the global R environment, where you can see, access, and work with it. The only one you will need to use for a long time to come (possibly ever) is the already familiar assignment operator <-.
You can use <- to create new objects in the environment or re-assign values to already existing names. So, if you want to turn the id column of df into a factor you need to reassign some new object to df$id. What object? Well, the one returned by the factor(...) command above:
df$id <- factor(df$id)As you can see, there is no printout now because the output of factor() has been passed into the assignment function which directed it into the df$id object. Let’s make sure it really worked:
class(df$id)## [1] "factor"5.4.1.3 Graphical device
Functions that create graphics return their output into something called the graphical device. It is basically the thing responsible for drawing stuff on the screen of your computer. You’ve already encountered some of these functions – plot(), par(), lines(), abline().
5.4.1.4 Files
Finally, there are functions that can write output into all sorts of files. For instance, if you want to save a data frame into a .csv file, you can use the read.csv() function.
Of course, you can redirect where particular output gets sent, just like we did with df$id <- factor(...). For instance, you can save a plot into the global environment using assignment:
my_plot <- hist(rnorm(1000))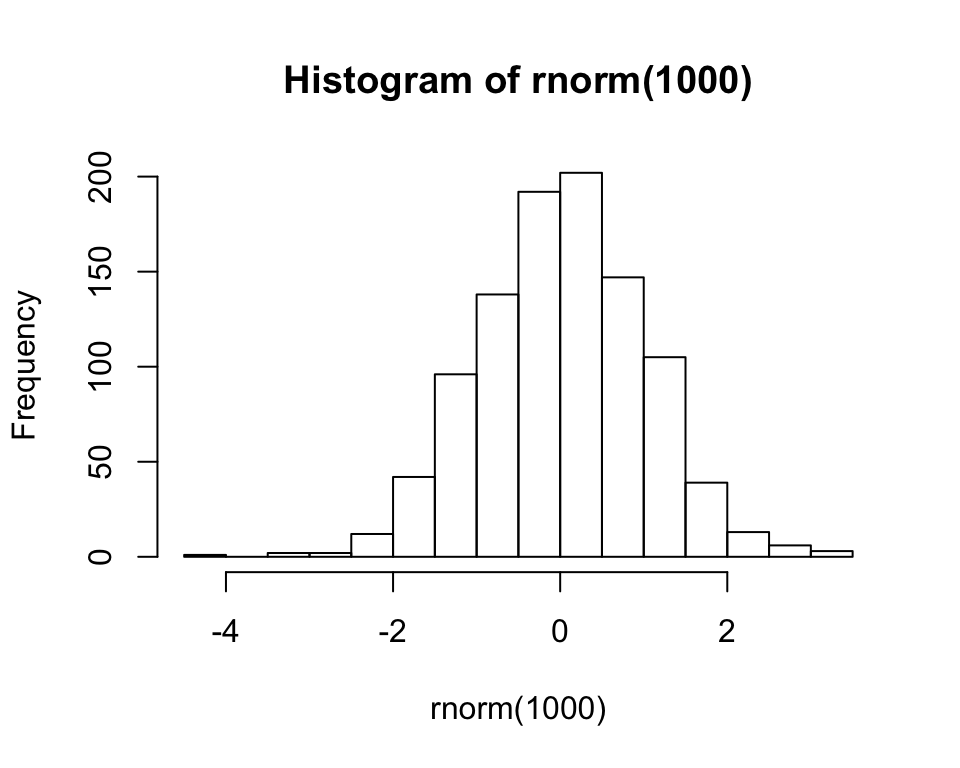
my_plot## $breaks
## [1] -4.5 -4.0 -3.5 -3.0 -2.5 -2.0 -1.5 -1.0 -0.5 0.0 0.5 1.0 1.5 2.0
## [15] 2.5 3.0 3.5
##
## $counts
## [1] 1 0 2 2 12 42 96 138 192 202 147 105 39 13 6 3
##
## $density
## [1] 0.002 0.000 0.004 0.004 0.024 0.084 0.192 0.276 0.384 0.404 0.294
## [12] 0.210 0.078 0.026 0.012 0.006
##
## $mids
## [1] -4.25 -3.75 -3.25 -2.75 -2.25 -1.75 -1.25 -0.75 -0.25 0.25 0.75
## [12] 1.25 1.75 2.25 2.75 3.25
##
## $xname
## [1] "rnorm(1000)"
##
## $equidist
## [1] TRUE
##
## attr(,"class")
## [1] "histogram"Alternatively, you can export it by creating a new graphical device inside of a file:
# create a graphical device in a new my_plot.png
# file in the working directory
png("my_plot.png")
hist(rnorm(100)) # create plot
# close the graphical device
dev.off()