Chapter 10 Principal component analysis
From this section, the machine learning will be discussed by foods data, the data from Mid-infrared spectroscopy will be used for Geographical origin of Extra Virgin Olive Oils (Quadram open dataset,2003).
First we use R for data clean and wrangling. Python will also be used for PCA plots.
10.1 Load dataset
## Load date
setwd("C:/local_R/Dataset")
oil <- read.csv('FTIR_Spectra_olive_oils.csv',header = F,
stringsAsFactors = T)
print(oil[1:10,1:5])## V1 V2 V3 V4 V5
## 1 Sample Number: 1 1 2 2
## 2 Group Code: 1 1 1 1
## 3 Wavenumbers Greece Greece Greece Greece
## 4 798.892 0.127523009 0.126498181 0.130411785 0.130022227
## 5 800.8215 0.127949615 0.127130974 0.130675401 0.130406662
## 6 802.751 0.129282219 0.128510777 0.13201661 0.132018029
## 7 804.6805 0.131174169 0.13033991 0.133824061 0.134007275
## 8 806.61 0.133590328 0.132527221 0.136095296 0.136270568
## 9 808.5395 0.136425525 0.135308508 0.138943757 0.13887477
## 10 810.469 0.139357827 0.13835292 0.141722779 0.14148113210.2 Data wrangling
The raw data form are not suitable for R programming, therefore we need to transpose it ti suitable form.
## V1 V2 V3 V4 V5
## V1 Sample Number: Group Code: Wavenumbers 798.892 800.8215
## V2 1 1 Greece 0.127523009 0.127949615
## V3 1 1 Greece 0.126498181 0.127130974
## V4 2 1 Greece 0.130411785 0.130675401
## V5 2 1 Greece 0.130022227 0.130406662
## V6 3 1 Greece 0.128601989 0.128789565
## V7 3 1 Greece 0.128217254 0.128282253
## V8 4 1 Greece 0.126174933 0.126732773
## V9 4 1 Greece 0.126466053 0.126915413
## V10 5 1 Greece 0.127060105 0.127551128## rename some col names to make data analysis easier
colnames(oil_transpose)=oil_transpose[1,]
oil_transpose <- oil_transpose[-1,]
library(dplyr)
oil_transpose <- rename(oil_transpose,c("Group" ="Group Code:",
"Number" ="Sample Number:",
"Countries" ="Wavenumbers"))
## Get transposed dataset
oil.data <- oil_transpose
print(oil.data[1:10,1:5])## Number Group Countries 798.892 800.8215
## V2 1 1 Greece 0.127523009 0.127949615
## V3 1 1 Greece 0.126498181 0.127130974
## V4 2 1 Greece 0.130411785 0.130675401
## V5 2 1 Greece 0.130022227 0.130406662
## V6 3 1 Greece 0.128601989 0.128789565
## V7 3 1 Greece 0.128217254 0.128282253
## V8 4 1 Greece 0.126174933 0.126732773
## V9 4 1 Greece 0.126466053 0.126915413
## V10 5 1 Greece 0.127060105 0.127551128
## V11 5 1 Greece 0.126812707 0.12746074310.3 Principal Component Analysis using R
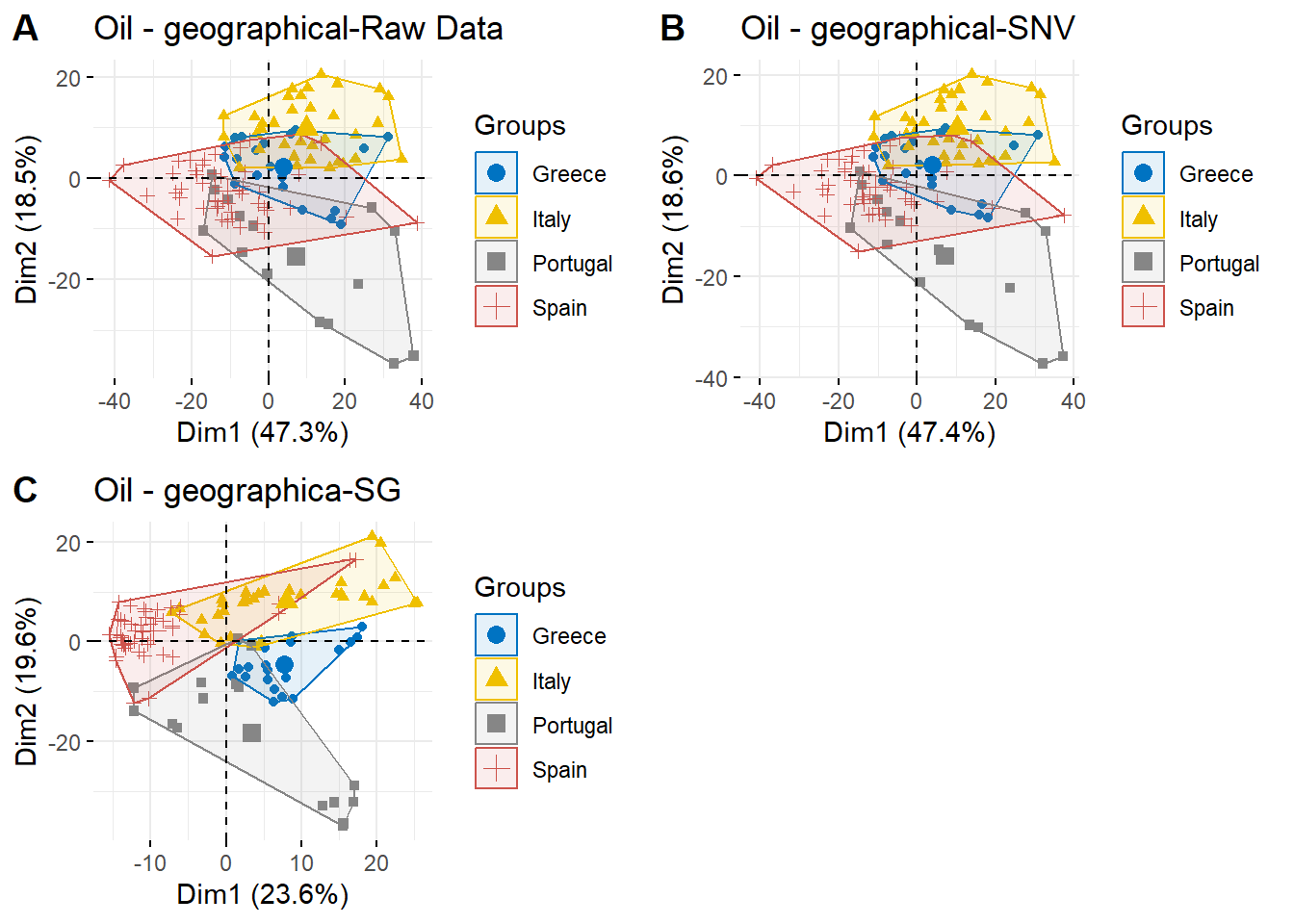
The PCA is used to reduce the dimensionality of the spectral value, and initially explore the distribution of the sample.
The original data and related transformated data (MSC, SNV and Savitzky-Golay filtering) are used to obtain the PCA plots.
library(missMDA)
library(factoextra)
library(FactoMineR)
library(ggrepel)
library(ggplot2)
library(mixOmics)
library(EMSC)
library(pls)
library(prospectr)10.3.1 Raw data PCA
Have a look at spectral of different pre-processing for MIR.
colors = c("#CD0000", "#EE7600", "#FFFF00", "#008B45")
col <- as.factor(oil.data$Countries)
group <- c("Greece","Italy","Portuga", "Spain")RAW spectral
raw_spc <- oil.data[,4:573]
x <- as.numeric(colnames(raw_spc))
y <- t(raw_spc[ , ])
par(mar=(c(5,7,4,2)))
matplot(x, y,col = colors,
type = 'l', lty = 1,
xlab = 'Wavelength (nm)', ylab = 'Absorbance',main = "Raw",
font.lab = 1,
cex.main = 1, cex.axis = 1, cex.lab=2)
legend("topleft",
title = "Country",
xpd = F,ncol = 1,
legend = group,
col = col,
fill = colors,
bty = "n",
cex = 1.5,
text.col = "black", text.font = 1,
horiz = F)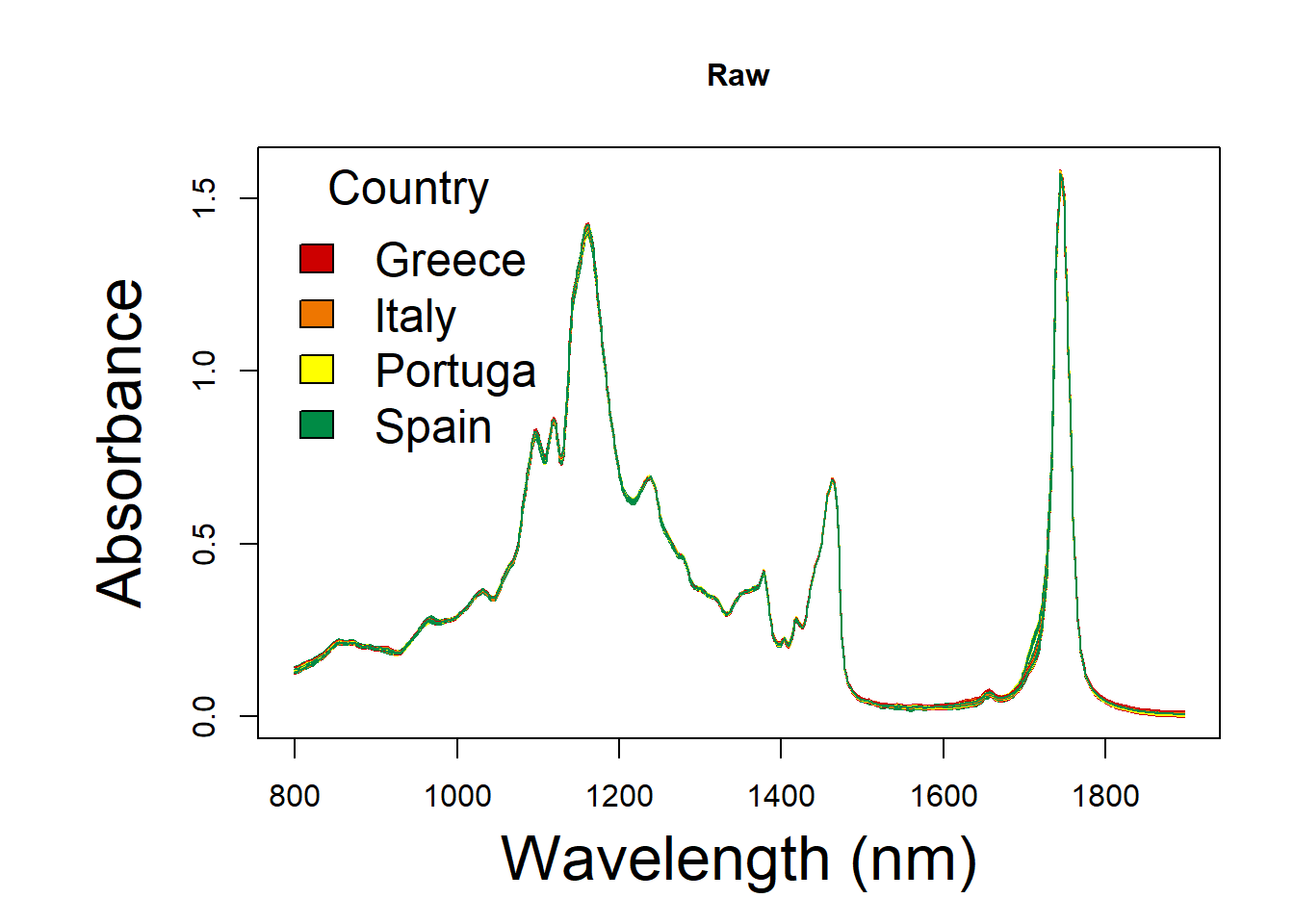
## 798.892 800.8215 802.751 804.6805 806.61
## 1 0.1275230 0.1279496 0.1292822 0.1311742 0.1335903
## 2 0.1264982 0.1271310 0.1285108 0.1303399 0.1325272
## 3 0.1304118 0.1306754 0.1320166 0.1338241 0.1360953
## 4 0.1300222 0.1304067 0.1320180 0.1340073 0.1362706
## 5 0.1286020 0.1287896 0.1300223 0.1320119 0.1344266
## 6 0.1282173 0.1282823 0.1296366 0.1317986 0.1340615
## 7 0.1261749 0.1267328 0.1282438 0.1298927 0.1317546
## 8 0.1264661 0.1269154 0.1282541 0.1299583 0.1320672
## 9 0.1270601 0.1275511 0.1289000 0.1306090 0.1329558
## 10 0.1268127 0.1274607 0.1287653 0.1306390 0.1331313The PCA plot of raw Spectral
g1 <- fviz_pca_ind(oil.pca, geom.ind = "point",
col.ind = oil.data$Countries, # color by groups
palette = 'jco',
addEllipses = TRUE, ellipse.type = "convex",
legend.title = "Groups")+
ggtitle("Oil - geographical-Raw Data")
g1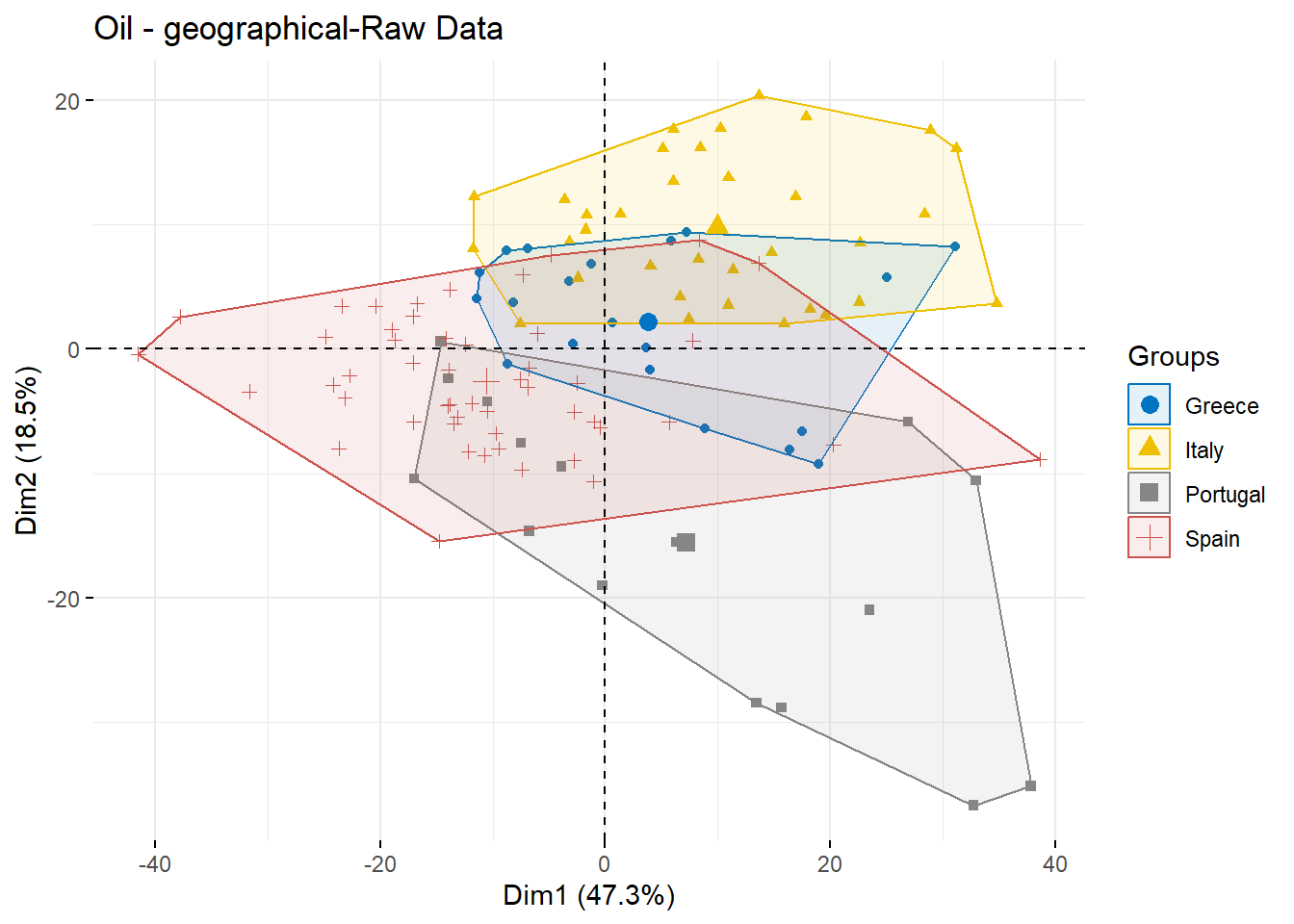
10.3.2 Standard Normal Variate
SNV spectral
x <- as.numeric(colnames(oil.snv))
y <- t(oil.snv[ , ])
par(mar=(c(5,7,4,2)))
matplot(x, y,col = colors, ylim = c(0,2),
type = 'l', lty = 1,
xlab = 'Wavelength (nm)', ylab = 'Absorbance',main = "SNV",
font.lab = 1,
cex.main = 1, cex.axis = 1, cex.lab=2)
legend("topleft",
title = "Country",
xpd = F,ncol = 1,
legend = group,
col = col,
fill = colors,
bty = "n",
cex = 1.5,
text.col = "black", text.font = 1,
horiz = F)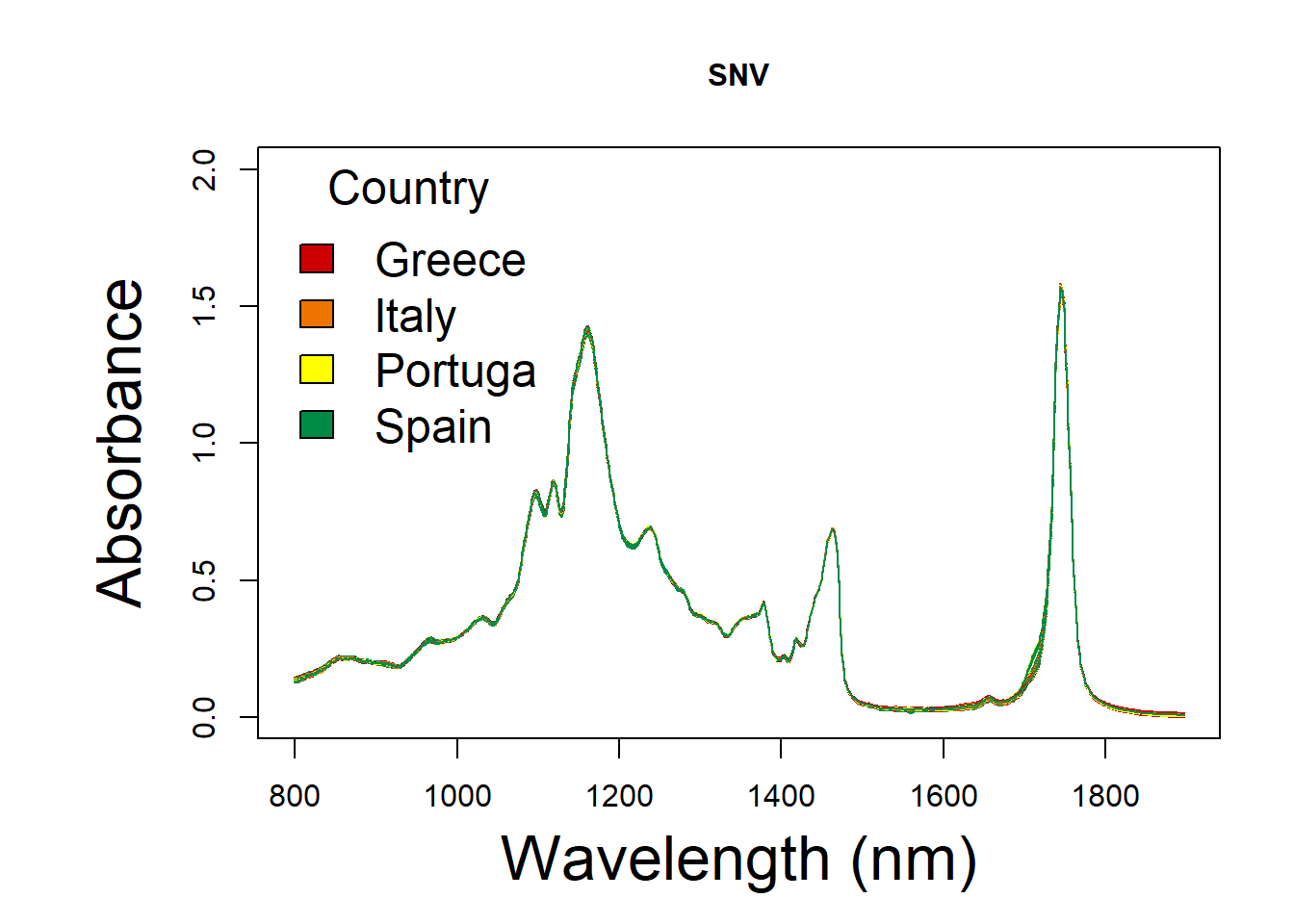
The PCA plot of SNV Spectral
g3 <- fviz_pca_ind(oil.snv.pca, geom.ind = "point",
col.ind = oil.snv$Countries, # color by groups
palette = 'jco',
addEllipses = TRUE, ellipse.type = "convex",
legend.title = "Groups")+
ggtitle("Oil - geographical-SNV")g3
10.3.3 Savitzky-Golay filtering
df_sg <- savitzkyGolay(
X = df_snv,
m = 1,
p = 3,
w = 11,
delta.wav = 2
)
oil.sg <- cbind(oil.data[,1:3], df_sg)Savitzky-Golay spectral
x <- as.numeric(colnames(oil.sg))
y <- t(oil.sg[ , ])
par(mar=(c(5,7,4,2)))
matplot(x, y,col = colors, ylim = c(-0.3,0.3),
type = 'l', lty = 1,
xlab = 'Wavelength (nm)', ylab = 'Absorbance',main = "Savitzky-Golay",
font.lab = 1,
cex.main = 1, cex.axis = 1, cex.lab=2)
legend("topleft",
title = "Country",
xpd = F,ncol = 2,
legend = group,
col = col,
fill = colors,
bty = "n",
cex = 1.5,
text.col = "black", text.font = 1,
horiz = F)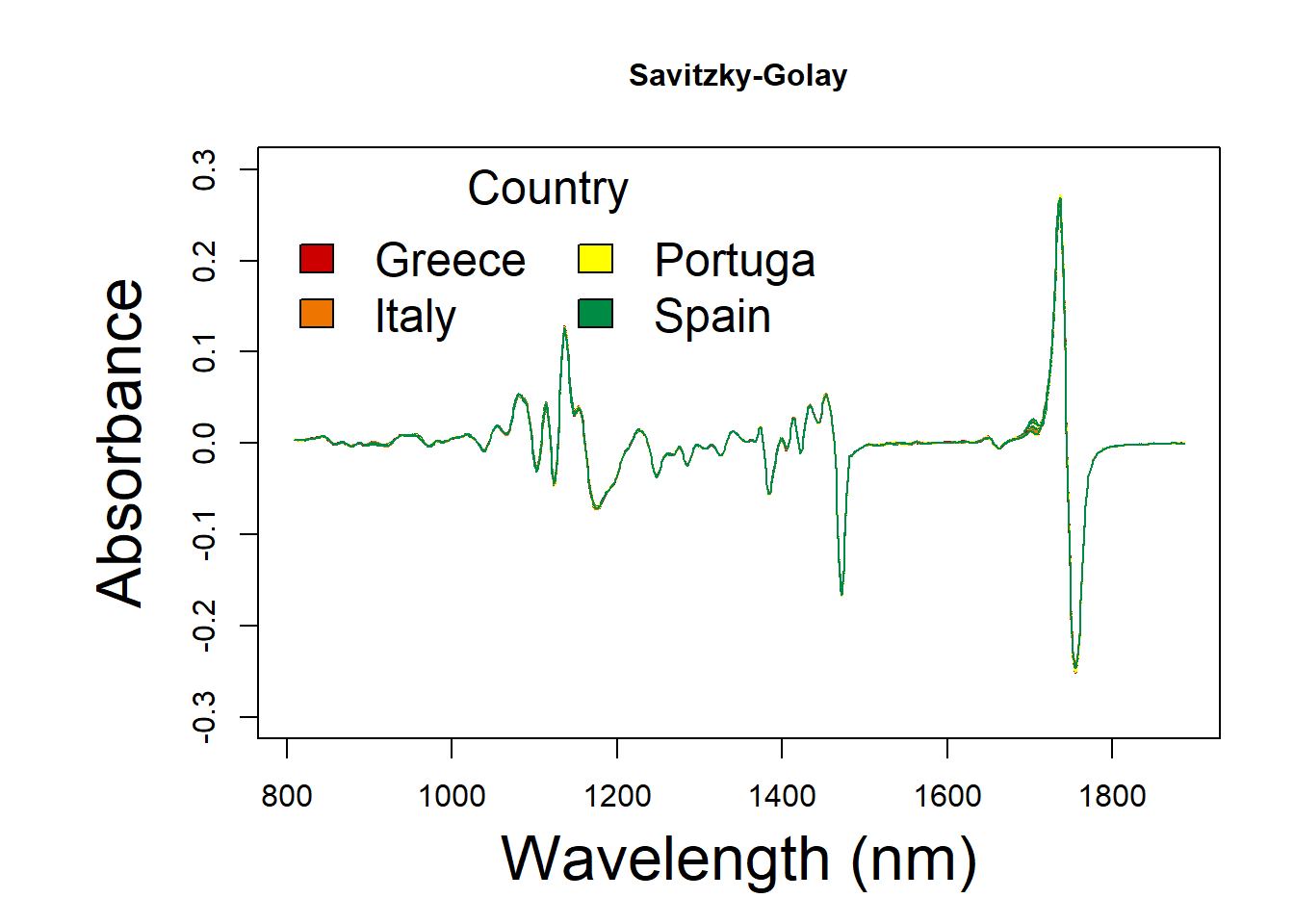
p4
The PCA plot of Savitzky-Golay Spectral:
g4 <- fviz_pca_ind(oil.sg.pca, geom.ind = "point",
col.ind = oil.sg$Countries, # color by groups
palette = 'jco',
addEllipses = TRUE, ellipse.type = "convex",
legend.title = "Groups")+
ggtitle("Oil - geographica-SG")g4