Preprocessing Diffusion Images
WARNING! Information and code contained in this section is meant as a general guide only and not explicit instructions for how to preprocess DWI images. It is important for any pipeline to check that all steps completed as intended.
T1
T2
DWI
DWI vs. DTI
Diffusion weighted imaging
- Measures water movement within a voxel of tissue
- Intensity value represents rate of water diffusion
Diffusion tensor imaging
- Specific type of modeling of diffusion weighted images based on the theory that water molecules diffuse differently within different types of tissue
Convert DICOM to NIFTI
DWI data are acquired across the whole brain by repeating the acquisition while varying the orientation or magnitude of the diffusion gradients. When converting DWI DICOM to NIfTI certain scan parameters must be extracted from the DICOM file: diffusion weighting (b-val) and gradient direction (b-vector).
Diffusion Weighting (bval)
The b-values are the amount of diffusion weighting used for each volume. Increasing the b-value leads to:
- Increased contrast b/w areas of higher and lower diffusivity in principle
- Decreased signal-to-noise ratio => Less reliable estimation of diffusion measures in practice
- Typical diffusion weighting is b ~ 1000 sec/mm2
0 0 0 0 0 0 0 0 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000
Gradient Direction (bvec)
Your b-vectors are the gradient directions that you collect, often predetermined by the scanner depending on how many total directions you choose to collect.
0 0 0 0 0 0 0 0 -0.157293 -0.866884 -0.288312 -0.321709 0.540555 0.131401 0.224512 0.829429 0.0141989 -0.153198 0.836844 0.0424971 0.690601 0.257194 -0.278801 -0.0543019 0.234627 -0.672272 0.57914 0.701167 -0.528531 -0.568571 -0.998727 -0.122108 0.908787 -0.886351 0.911169 -0.607895 0.419914 -0.915661 -0.867206 0.507935 0.65263 -0.482829 0.184596 0.873698 -0.889691 0.451762 0.678609 0.894383 -0.422352 -0.833252 0.290195 0.379917 -0.435847 0.190695 -0.226299 -0.211885 -0.555467 0.309998 0.621338 -0.041801 -0.705353 0.306804 -0.691036 -0.65772 -0.240721 -0.0356014 0.996059 0.616992 -0.561711
0 0 0 0 0 0 0 0 -0.406337 -0.498106 -0.731736 0.413121 0.408009 0.355336 -0.0955482 0.320832 0.987839 0.0502477 -0.152406 0.502994 -0.0757262 0.795206 0.920048 0.746701 -0.745187 0.133129 0.151618 -0.700337 -0.323875 -0.0650361 -0.0459666 -0.980465 -0.264574 0.307366 -0.390704 -0.744293 0.603837 0.379694 -0.286925 -0.381866 -0.639006 -0.863732 -0.70336 0.486465 -0.162551 0.88721 0.628054 0.199374 0.888361 0.15891 0.113213 -0.912387 0.398604 0.929858 -0.800652 0.783225 -0.491331 -0.322375 0.723233 -0.953358 0.699882 -0.901674 -0.576735 0.600742 -0.0237355 -0.463934 0.0513951 -0.543639 0.625415
0 0 0 0 0 0 0 0 -0.900083 0.0200564 0.617607 0.851959 -0.73575 0.925457 0.969776 0.45729 -0.154832 -0.986917 0.5258 -0.863244 -0.719261 -0.549089 0.275285 0.66294 0.624216 0.728236 0.801005 -0.133764 0.784703 -0.82006 -0.0207777 -0.154201 -0.322656 0.346279 0.13085 -0.276569 0.677535 -0.131901 -0.406973 0.772127 0.407119 0.144372 -0.686447 0.00173833 0.426646 -0.0936476 -0.380839 -0.40041 -0.180096 -0.529565 -0.950247 0.152357 -0.80694 0.314643 -0.554748 -0.58452 -0.670858 -0.894414 0.301451 0.298934 0.112435 -0.304722 0.435714 -0.454437 0.970304 0.885154 0.0722801 -0.569015 0.541606
dcm2niix
When you run the latest version of dcm2niix it will automatically generates these files during DICOM conversion.
Depending on the brand of the scanner, e.g., Siemens or GE, these scan parameters may not always be applied to the images correctly. It is important to verify tensors after preprocessing.
Symlink
Preprocessing DWI creates a lot of files and anything that can be done to reduce the number of files is helpful. Create a derivative directory, pDWI, and instead of copying your files from BIDS, create a symlink.
cd ${DERIV_DIR}/${SUBJID}/
ln -s ${BIDS_DIR}/${SUBJID}/dwi/${SUBJID}_dwi.nii.gz dwi.nii.gz
ln -s ${BIDS_DIR}/${SUBJID}/dwi/${SUBJID}_dwi.bval dwi.bval
ln -s ${BIDS_DIR}/${SUBJID}/dwi/${SUBJID}_dwi.bvec dwi.bvecVerify Volumes
It is very easy to have missing volumes in the DWI. It is critical that the numbers match between the NIFTI image and the bvec and bval files.
$ c4d dwi.nii.gz -info
Image #1: dim = [104, 104, 69, 63]; bb = {[-103.445 91.427 -36.6199 0], [104.555 299.427 101.38 819]}; vox = [2, 2, 2, 13]; range = [0, 2910]; orient = Oblique, closest to RPIThis NIFTI image has 63 volumes with a FOV of 104 x 104 x 69. Make sure there’s 63 entries in the bval file:
$ cat dwi.bval | wc -w
63Finally, make sure you have the same number of volumes in bvec file:
$ head dwi.bvec | wc -w
63Pad Image
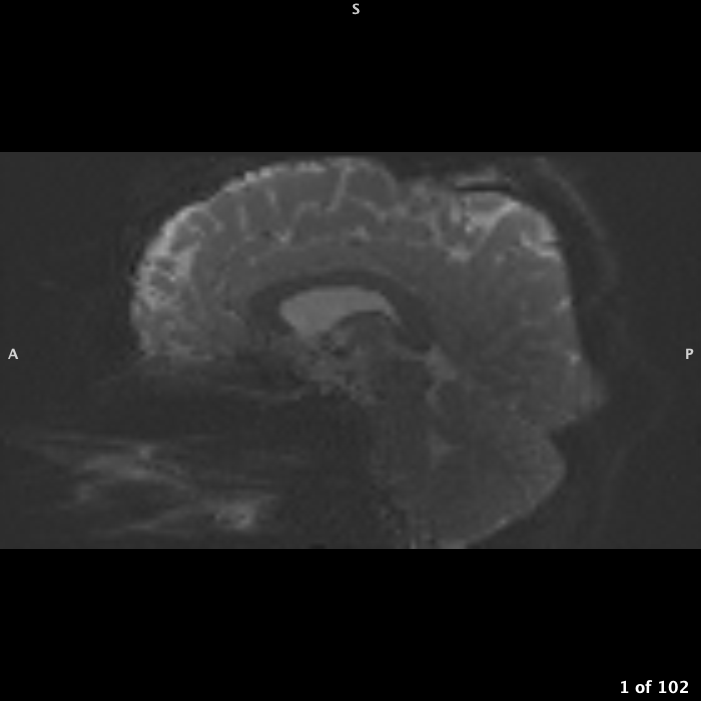
If your FOV box is right up against skull or even cutting off part of skull but not brain, you will need to pad the image to give background space:
$ c4d \
dwi.nii.gz \
-pad 10% 10% 0 \
-o dwi_padded.nii.gzExtract B0
You want to create a mask based on just the b0 and not the whole 4D image.
FSL
Using FSL:
fslroi dwi.nii.gz dwi_bse.nii.gz 0 1 # if padded use that image insteadpnlNipype
There’s also an option for creating a b0 image average if multiple were collected during the scan.
~/build/pnlNipype/exec/nifti_bse \
-i dwi.nii.gz \ # if padded use that image instead
--bvals dwi.bval \
-o dwi_bseMask B0
Must be as accurate as possible.
FSL
Every study will be a little different and the f value will have to be changed.
bet dwi_bse.nii.gz dwi_bse -f 0.1 -g 0 -m -n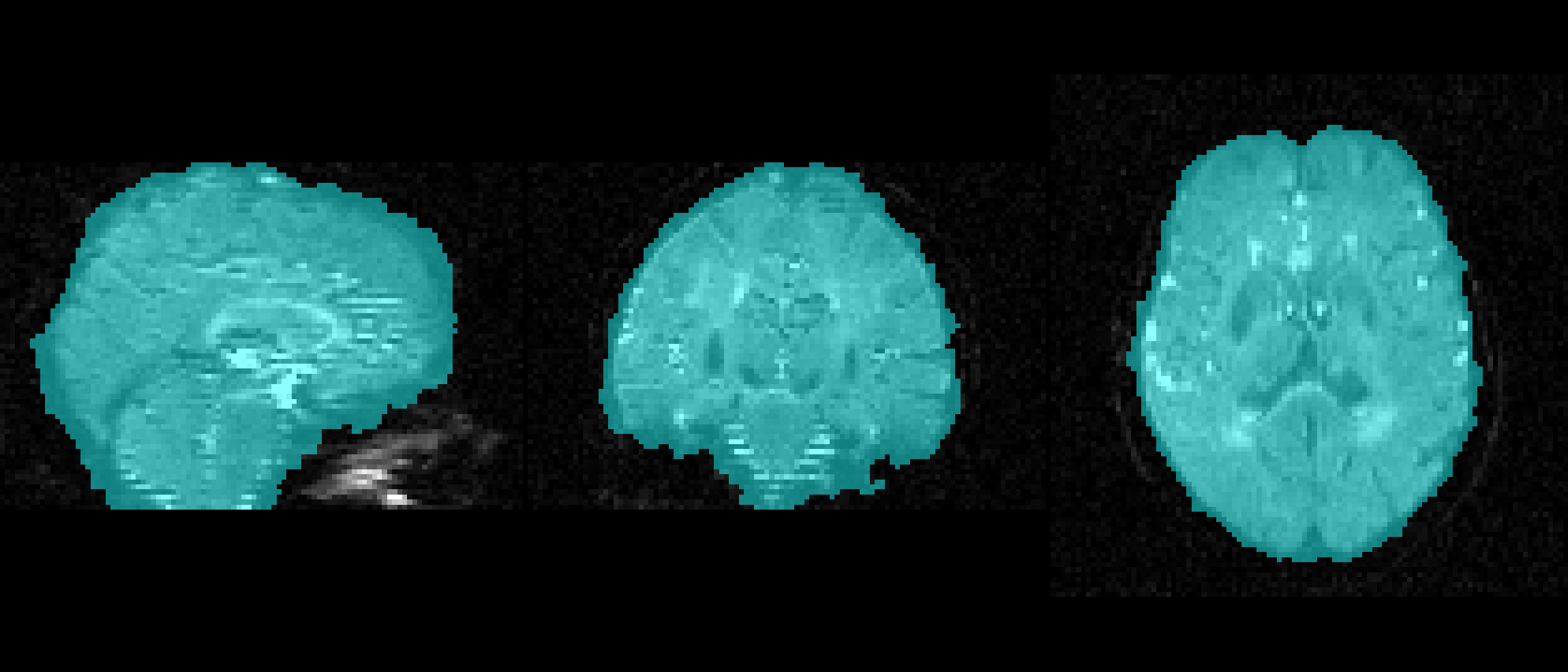 f = 0.1
f = 0.1
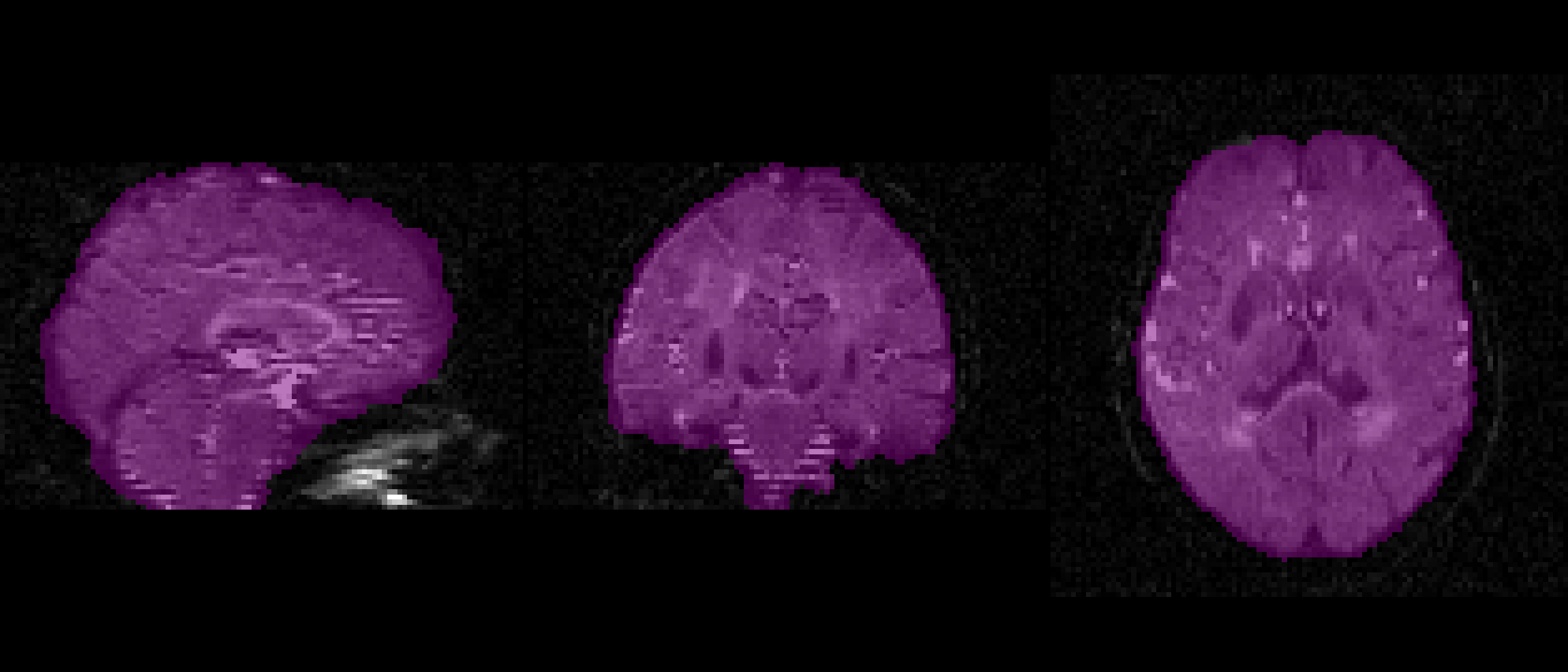 f = 0.2
f = 0.2
 f = 0.3
f = 0.3
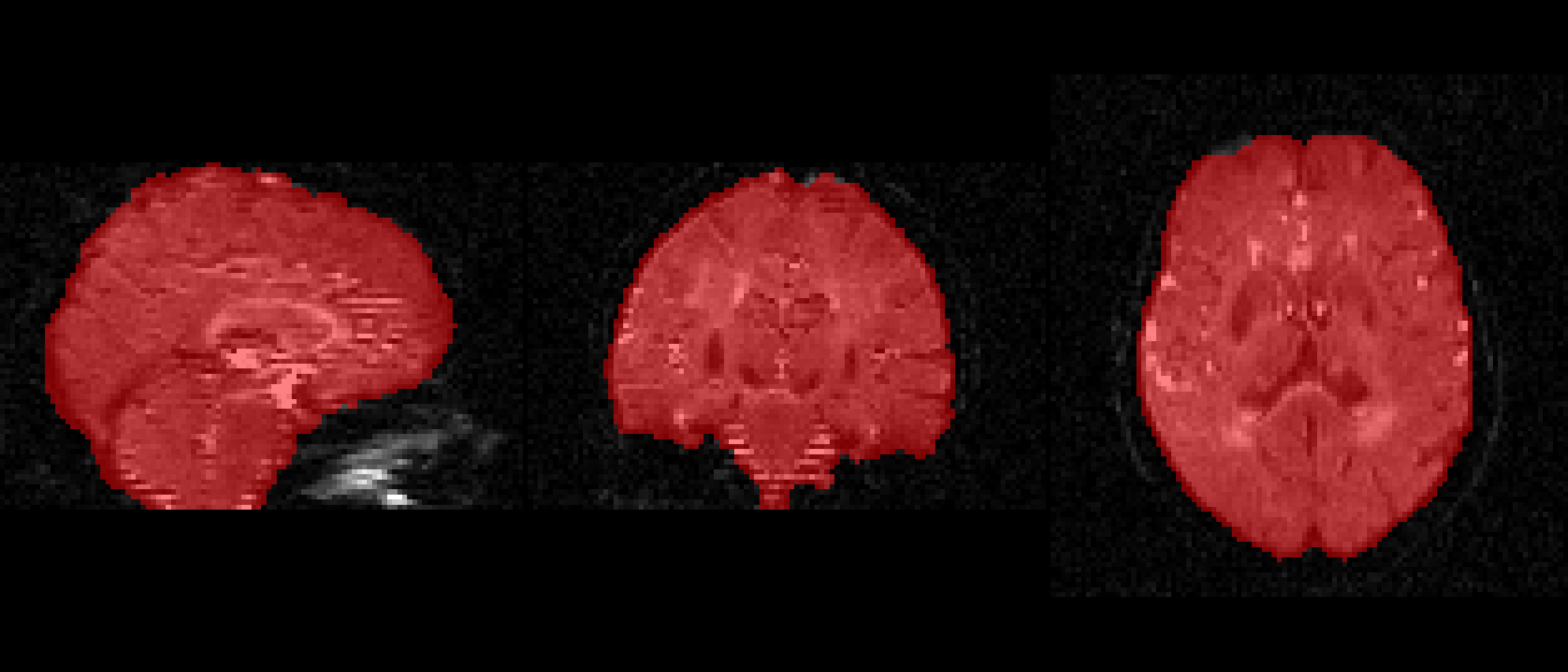 f = 0.4
f = 0.4
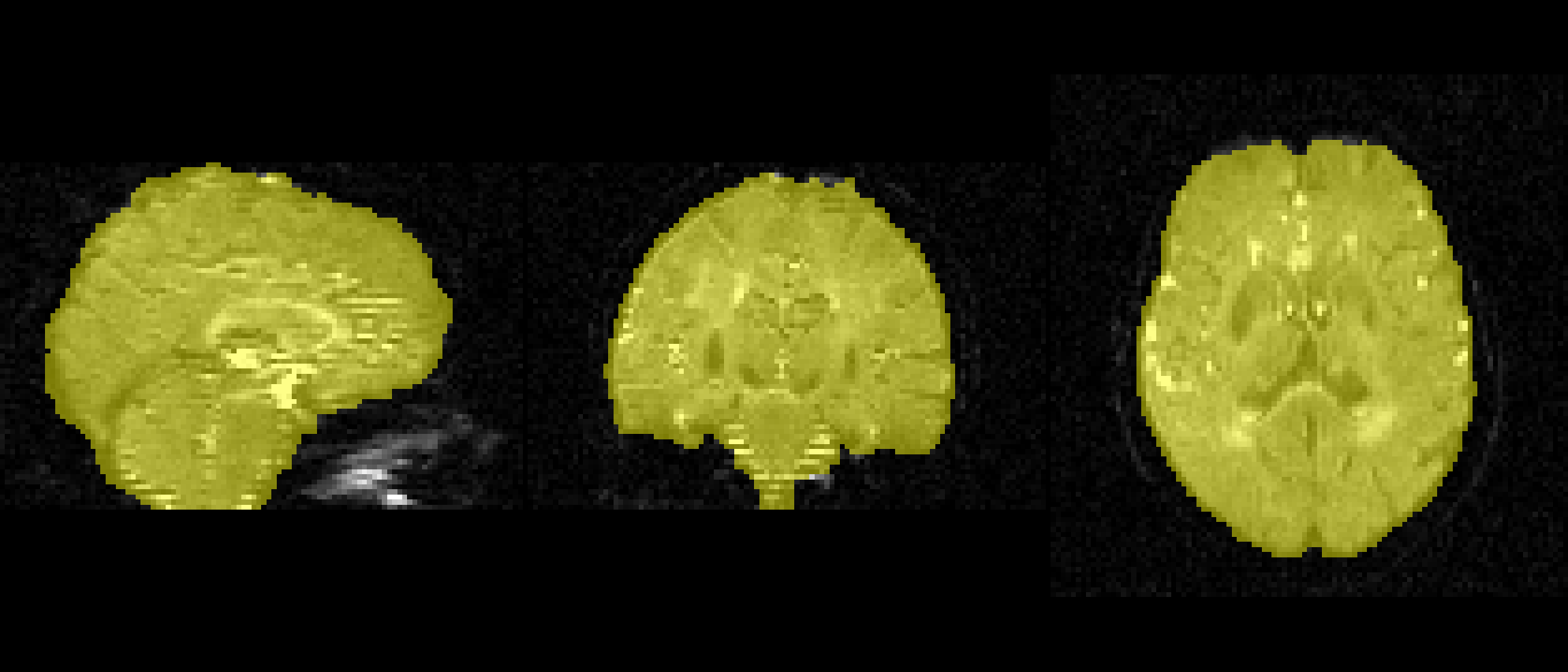 f = 0.5
f = 0.5
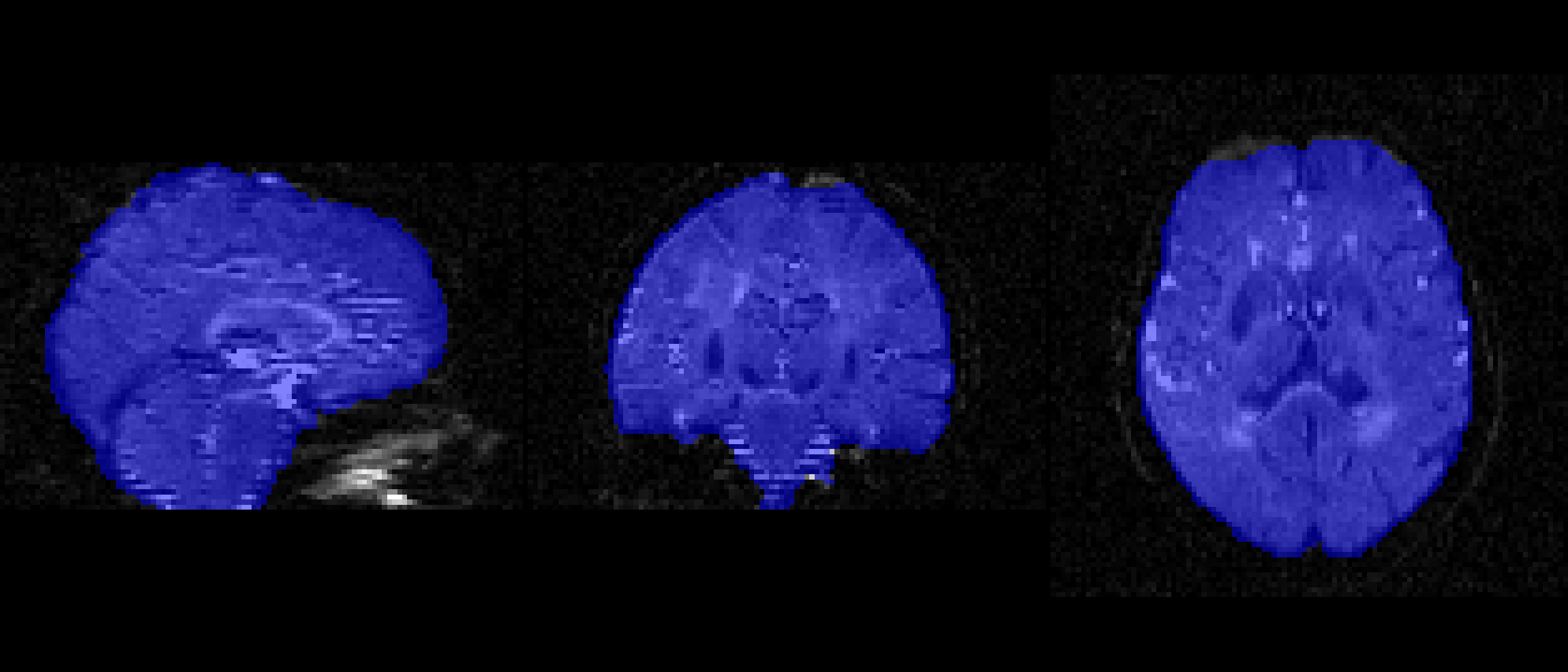 f = 0.6
f = 0.6
pnlNipype
Using nifti_bet_mask that packages FSL:
~/build/pnlNipype/exec/nifti_bet_mask \
-i dwi_bse.nii.gz \
--bvals dwi.bval \
-o dwiUsing nifti_atlas that packages ANTs:
~/build/pnlNipype/exec/nifti_atlas \
-t dwi_bse.nii.gz \
--train ~/apps/pnlbwh/pnlNipype/trainingDataT2Masks-12a14d9/trainingDataT2Masks-hdr.csv \
-o dwiMRtrix3
Currently this program seems the most robust for brain masking: https://mrtrix.readthedocs.io/en/latest/reference/commands/dwi2mask.html#dwi2mask
Clean mask
Sometimes all that’s needed is to erode / shrink the mask a little to make it better
c3d \
dwi_mask.nii.gz \
-erode 1 1x1x1vox \
-o dwi_mask_cleaned.nii.gzEddy Current Correction
This correction will help fix eddy current-induced distortions and subject movements during the scan.
Set acqparams.txt
All information can be derived / found in the DWI JSON file. For your acqparams.txt file, each row consists of a vector (three values) that specify what the phase-encode (PE) axis is. The fourth element in each row is the time (in seconds) between reading the center of the first echo and reading the center of the last echo. The fourth element is also the reciprocal of the PE bandwidth/pixel. Remember that to calculate the reciprocoal you take: (1 BandwidthPerPixelPhaseEncode)
printf "0 1 0 0.095" > acqparams.txt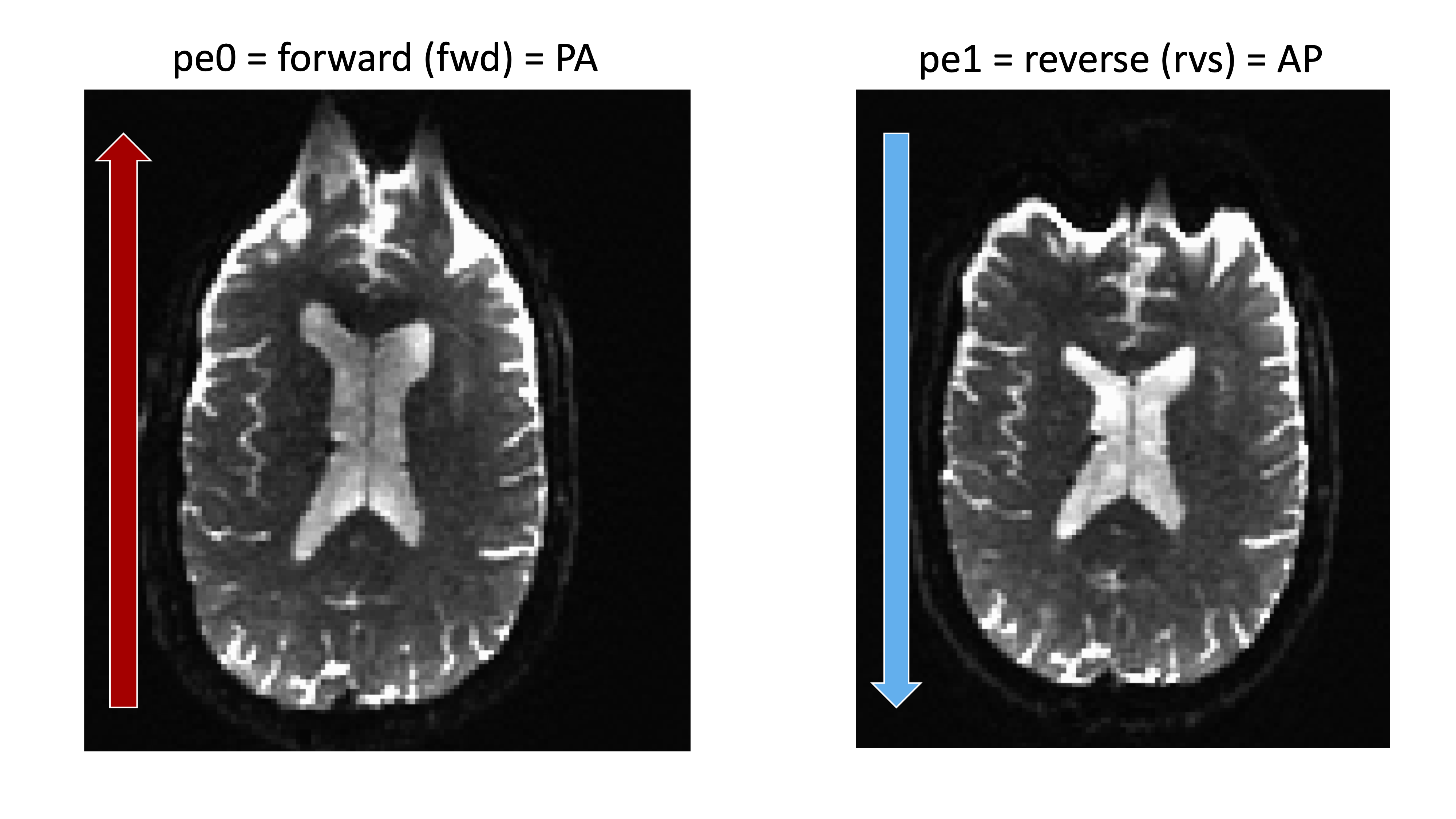
Forward
- Posterior to Anterior
- Phase Encoding is j
- acqparams.txt is 0 1 0
Reverse
- Anterior to Posterior
- Phase Encoding is j-
- acqparams.txt is 0 -1 0
Looking at the JSON file:
- “PhaseEncodingDirection”: “j-,”
- “BandwidthPerPixelPhaseEncode”: 13.935,
- (1 = 0.07176175099)
printf "0 -1 0 0.072" > acqparams.txtSet index.txt
Create an index file that tells eddy which line of the “acqparams.txt” file to use for each volume of data. In our example above, the index file has a single line with 63 1’s separated by spaces:
myVar=($(wc -w dwi.bval))
indx=""
for ((i=1; i<=$myVar; i+=1)); do indx="$indx 1"; done
echo $indx > index.txtEddy
The only command you should use is eddy_openmp or eddy_cuda. Older versions, eddy_correct, cannot handle multishell or high B0 values and does not correct for high motion participants. FSL recommends NEVER using eddy_correct for this reason.
time eddy_cuda8.0 \
--imain=dwi_xc.nii.gz \
--acqp=acqparams.txt \
--index=index.txt \
--mask=dwi_bse_mask.nii.gz \
--bvecs=dwi_xc.bvec \
--bvals=dwi_xc.bval \
--out=dwi_eddy \
--cnr_maps \
--repol \
--mporder=16 \
--estimate_move_by_susceptibility \
--verboseIt is CRITICAL that a new bvec file is generated after eddy correct since whole volume alignment was done. You cannot do tractography with unaligned tensors. Luckily eddy_openmp and eddy_cuda output a rotated bvec file. The older version, eddy_correct, does not output a rotated bvec file. Just don’t use eddy_correct.
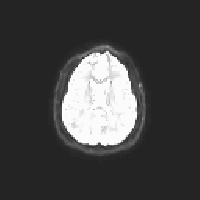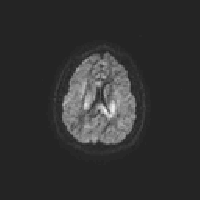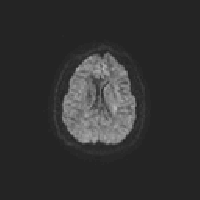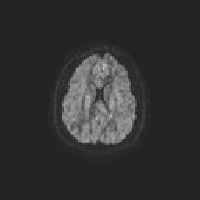
One way to check eddy correction is to create a gif image that scans through each volume along the same slice. I’ve create an R script that can save a gif for each participant in your study:
library(oro.nifti)
library(neurobase)
setwd('/fslhome/intj5/compute/data/BRAINSCOPE/pnlNipype/')
dirs=list.files(path=".")
for (x in 1:length(dirs)){
setwd(paste('/fslhome/intj5/compute/data/BRAINSCOPE/pnlNipype/', dirs[x], sep = ""))
getwd()
img <- readNIfTI("dwi_eddy.nii.gz")
png(file="eddy%02d.png", width=200, height=200)
for (i in 1:img@dim_[5]){
oro.nifti::image(robust_window(img),z = 35,w = i,plot.type = "single")
}
dev.off()
system("convert -delay 8 *.png eddy.gif")
file.remove(list.files(pattern=".png"))
}
Eddy Quality Control
For full information: https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/eddyqc/UsersGuide
Participant
for i in $(ls <path-to-dir>/pDWI/ | grep "sub-"); do
SUBJ_DIR=<path-to-dir>/pDWI/${i}
eddy_quad \
${SUBJ_DIR}/dwi_eddy \
-idx ${SUBJ_DIR}/index.txt \
-par ${SUBJ_DIR}/acqparams.txt \
-m ${SUBJ_DIR}/dwi_b0_mask.nii.gz \
-b ${SUBJ_DIR}/dwi.bval \
-v
doneStudy
eddy_squad \
participants.txt \ # List with full-path name to dwi_eddy.qc directories
-g group.txt \
-u \
-o squad_siteThe right way to format the group.txt file:
VarName
0 [1 for continuous]
v_1
v_2
.
.
.
v_nNote that the variable must always be numeric regardless if the variable is categorical or continuous.
EPI Distortion Correction
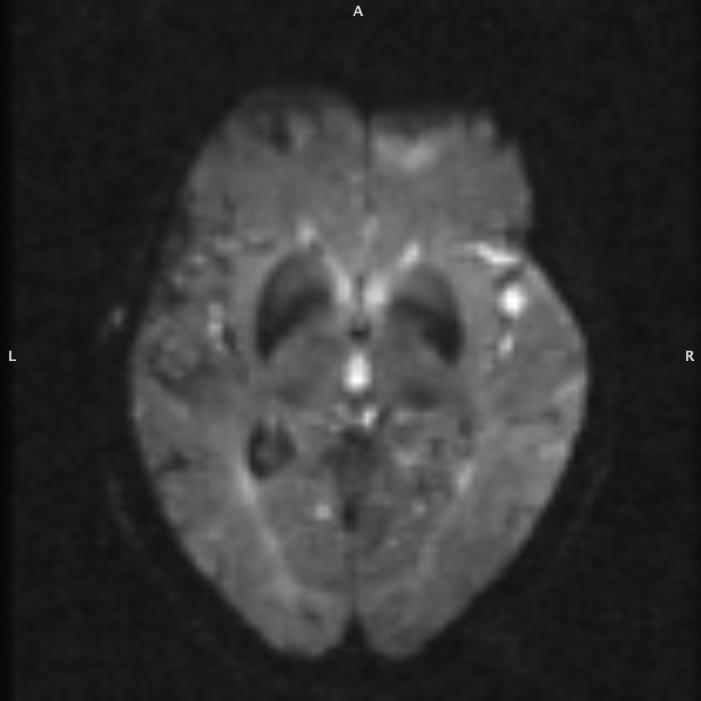
Extract B0 for EPI Distortion Correction
fslroi dwi_eddy.nii.gz dwi_eddy_b0.nii.gz 1 0Create a skull stripped B0 image
It is easier to register the B0 and T1 if they are appropriately skull stripped. The skull will cause registration errors.
Fix Displacement between B0 and T1
Do a rigid alignment between preprocessed T1 and eddy corrected B0 image to eliminate any displacement / motion differences between the two images.
antsRegistrationSyNQuick.sh \
-d 3 \
-f b0.nii.gz \
-m t1.nii.gz \ # skull stripped image from FreeSurfer
-o t1tob0- \
-t aFix Distortions
Non-linearly register B0 to T1 to fix distortions (stretching and squishing). You are not moving the b0 image, only adjusting voxels out of place. You do not need to adjust bvec file afterwards.
antsRegistrationSyNQuick.sh \
-d 3 \
-f t1tob0-Warped.nii.gz \
-m b0.nii.gz \
-o epi- \
-t soApply Transformation
Apply the registration to the entire eddy current corrected image.
antsApplyTransforms \
--dimensionality 3 \
--input-image-type 3 \
--input eddy.nii.gz \
--output epi.nii.gz \
--reference-image t1tob0-Warped.nii.gz \
--interpolation Bspline \
--transform epi-1Warp.nii.gzTopup
Create a Symlink for all your files:
ln -fs ${BIDS_DIR}/dwi/${1}_${2}_acq-AP_dwi.nii.gz AP_scan.nii.gz
ln -fs ${BIDS_DIR}/dwi/${1}_${2}_acq-PA_dwi.nii.gz PA_scan.nii.gz
ln -fs ${BIDS_DIR}/dwi/${1}_${2}_acq-AP_dwi.bval AP_scan.bval
ln -fs ${BIDS_DIR}/dwi/${1}_${2}_acq-AP_dwi.bvec AP_scan.bvec
ln -fs ${BIDS_DIR}/dwi/${1}_${2}_acq-PA_dwi.bval PA_scan.bval
ln -fs ${BIDS_DIR}/dwi/${1}_${2}_acq-PA_dwi.bvec PA_scan.bvecExtract B0 for each scan and combine together:
fslroi AP_scan b0_AP_scan 0 1
fslroi PA_scan b0_PA_scan 0 1
fslmerge -t b0_AP_PA_scan b0_AP_scan b0_PA_scanCreate an acqparams.txt file:
printf "0 -1 0 0.0805839" > acqparams.txt
printf "\n0 1 0 0.0805839" >> acqparams.txtRun topup:
topup \
--imain=b0_AP_PA_scan \
--datain=acqparams.txt \
--out=topup \
--config=b02b0.cnf \
--iout=b0_unwarpedCreate a brain mask:
fslmaths b0_unwarped -Tmean b0_unwarped_mean
bet b0_unwarped_mean b0_unwarped_brain -m -f 0.1Create a single 4D image:
fslmerge -a AP_PA_scan AP_scan PA_scanCombine the bvec and bval files:
paste -d" " AP_scan.bvec PA_scan.bvec >> bvecs
paste -d" " AP_scan.bval PA_scan.bval >> bvalsCreate index.txt:
myVar=($(wc -w AP_scan.bval))
indx=""
for ((i=1; i<=$myVar; i+=1)); do indx="$indx 1"; done
for ((i=1; i<=$myVar; i+=1)); do indx="$indx 2"; done
echo $indx > index.txtRun eddy:
eddy_openmp \
-v \
--imain=AP_PA_scan \
--mask=b0_unwarped_brain_mask \
--index=index.txt \
--acqp=acqparams.txt \
--bvecs=bvecs \
--bvals=bvals \
--topup=topup \
--out=eddy \
--cnr_maps \
--repol \
--mporder=16 \
--estimate_move_by_susceptibility \At this point you do not need to run any epi distortion corrections since topup was included with eddy to fix the distortions.
Generate New Mask
A final mask needs to be generated before tensor fitting.