Functional Connectivity
Sustained connectivity using resting-state functional MRI data are analyzed using both network level and finer-grained brain parcellation schemes. Preprocessing involves removing volumes with head motion of greater than 0.2 mm, as well as frames before and after the head motion. Time series are then extracted from volumes for each participant. Multiple volumes can be concatenated if available.
A finer parcellation is computed that consists of 333 regions in the cerebral cortex, 14 subject-specific subcortical regions from FreeSurfer derived segmentation (bilateral thalamus, caudate, putamen, amygdala, hippocampus, pallidum, and nucleus accumbens), and 14 bilateral cerebellar representations of a 7-network parcellation. This combined parcellation scheme incorporates major cortical, subcortical, and cerebellar gray matter ROIs numbering 361 regions in total. Functional connectivity is estimated as the Pearson correlation coefficient between mean time series from each ROI.
Pearson’s correlation measures the linear association between two variables. Because the correlation is bounded between [-1, 1], the sampling distribution for highly correlated variables is highly skewed. In 1921, R. A. Fisher studied the correlation of bivariate normal data and discovered a wonderful transformation that converts the skewed distribution of the sample correlation (r) into a distribution that is approximately normal. Resting state correlation coefficients are Fisher transformed. Values will range between 0 and 2.
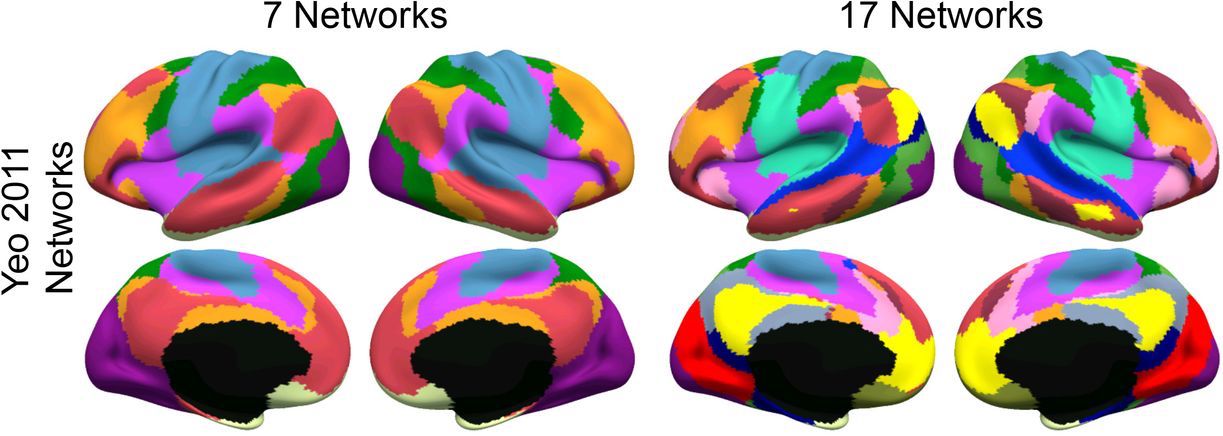
For network analyses, the mean functional connectivity between ROIs for each intrinsic network is calculated and then between-network functional connectivity is calculated as the Pearson correlation coefficient between mean time series from each network. Correlations are Fisher-transformed. Each functional network is treated as a single ROI. Mean functional connectivity outputs are available for 7 and 17 networks. The 7 networks are as follows: (1) Visual; (2) Somatomotor; (3) Dorsal Attention; (4) Ventral Attention; (5) Limbic; (6) Frontoparietal; and (7) Default. The 17 networks are as follows: (1) Central visual; (2) Peripheral visual; (3) Dorsal somatomotor; (4) Ventral somatomotor; (5) Posterior dorsal attention; (6) Somatomotor association; (7) Posterior ventral attention; (8) Anterior ventral attention; (9) Medial temporal–limbic; (10) Orbitofrontal–limbic; (11) Medial superior parietal; (12) Medial frontoparietal; (13) Lateral frontoparietal; (14) Lateral temporal–default mode network (DMN); (15) Ventral–DMN; (16) Dorsal–DMN; and (17) Lateral–DMN.
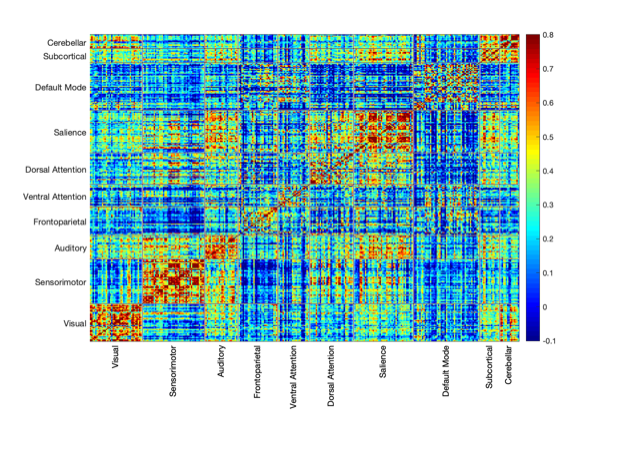
A 385x385 matrix consisting of Fisher transformed correlation coefficients representing functional connectivity is provided for each subject. The matrices can be split into 361x361 ROIs (shown above) and 17x17 networks. The larger 385x385 matrix consists of: 333 ROIs, 14 subcortical regions, 14 bilateral cerebellar representations of a 7-network parcellation, 7 brain networks from Yeo, 17 brain networks from Yeo. Within-network functional connectivity can be estimated from the mean of all ROI time series within a network of interest.
| ParcelID | Hem | Community |
|---|---|---|
| 10, 64, 65, 66, 67, 68, 69, 70, 77, 102, 104 | L | Auditory |
| 171, 224, 227, 230, 232, 233, 239, 244, 268, 269, 329, 330 | R | Auditory |
| 180, 181, 185, 187, 188, 192, 196, 198, 219, 223, 234, 235, 238, 245, 246, 248, 249, 274, 317, 318 | R | CinguloOperc |
| 21, 22, 27, 28, 34, 40, 63, 71, 72, 76, 81, 82, 84, 101, 103, 105, 111, 112, 147, 153 | L | CinguloOperc |
| 12, 89, 93 | L | CinguloParietal |
| 173, 254 | R | CinguloParietal |
| 1, 4, 6, 25, 26, 44, 94, 114, 116, 117, 126, 127, 145, 146, 150, 151, 152, 154, 156, 157 | L | Default |
| 162, 165, 184, 186, 200, 220, 225, 257, 259, 278, 279, 290, 315, 316, 321, 322, 323, 324, 325, 326, 331 | R | Default |
| 189, 199, 203, 208, 211, 236, 250, 252, 253, 262, 266, 271, 275 | R | Dorsal Attention |
| 41, 42, 43, 49, 51, 52, 55, 74, 87, 88, 91, 92, 95, 100, 106, 107, 110, 113, 155 | L | Dorsal Attention |
| 167, 168, 170, 182, 240, 260, 261, 272, 273, 276, 277, 319, 320, 327, 328 | R | FrontoParietal |
| 7, 9, 24, 78, 96, 108, 109, 148, 149 | L | FrontoParietal |
| 11, 18, 19, 73, 115, 118, 119, 120, 121, 122, 123, 124, 125, 128, 129, 133, 134, 135, 142, 144, 159 | L | None |
| 172, 178, 179, 280, 281, 282, 283, 284, 285, 286, 287, 288, 289, 291, 292, 296, 297, 300, 301, 302, 303, 304, 305, 306, 312, 314 | R | None |
| 13, 14, 130, 143 | L | Retrosplenial Temporal |
| 174, 294, 295, 313 | R | Retrosplenial Temporal |
| 183, 247 | R | Salience |
| 29, 83 | L | Salience |
| 163, 190, 191, 193, 194, 195, 201, 202, 204, 205, 206, 207, 209, 210, 213, 214, 215, 216, 217, 270 | R | Somatomotor Hand |
| 2, 30, 31, 32, 33, 35, 36, 37, 38, 45, 46, 47, 48, 50, 54, 56, 57, 58 | L | Somatomotor Hand |
| 164, 197, 212, 218 | R | Somatomotor Mouth |
| 3, 39, 53, 59 | L | Somatomotor Mouth |
| 221, 222, 226, 228, 229, 231, 237, 241, 242, 243, 332, 333 | R | Ventral Attention |
| 23, 60, 61, 62, 75, 79, 80, 85, 86, 158, 161 | L | Ventral Attention |
| 166, 169, 175, 176, 177, 251, 255, 256, 258, 263, 264, 265, 267, 293, 298, 299, 307, 308, 309, 310, 311 | R | Visual |
| 5, 8, 15, 16, 17, 20, 90, 97, 98, 99, 131, 132, 136, 137, 138, 139, 140, 141 | L | Visual |
The follow R code can be adapted to average the correlation coefficients for each of the intrinsic networks:
# Packages
require(ppcor)
# Generate pseudo 100-by-50 correlation matrix
M <- matrix(rnorm(n=5000, mean=0, sd=1), 100, 50)
colnames(M) <- paste0('V',seq(1,50))
# View top of matrix
head(M)
# Generate correlation matrix
CM <- cor(M)
# Or a partial correlation matrix if you prefer
pCM <- ppcor::pcor(M)$estimate
# View CM
head(CM)
#### Function to get average network connectivity ####
# Function requires the correlation matrix, CM, and the IDs of the nodes of interst listed in nodes_of_interest variable
nodes_of_interest <- c('V17', 'V20', 'V33', 'V34', 'V35')
calculate_mean_network_connectivity <- function(CM, nodes){
# subset correlation matrix by nodes of interest
cm_ss <- CM[nodes, nodes]
# get upper triangle values
cm_ss_utri <- cm_ss[upper.tri(cm_ss)]
# return mean of upper triangle
return( mean(cm_ss_utri) )
}
# Apply the function
single_subject_result <- calculate_mean_network_connectivity(CM = CM, nodes = nodes_of_interest)
print(single_subject_result)
#### Extend this to multiple subjects, each with their own correlation matrices ####
nodes_of_interest <- c('V17', 'V20', 'V33', 'V34', 'V35')
# Generate list of 10 connectivity matrices of dimension 100-by-50 and return correlation matrices
CM_List <- lapply(1:10, function(f){
M <- matrix(rnorm(n=5000, mean=0, sd=1), 100, 50)
colnames(M) <- paste0('V',seq(1,50))
CM <- cor(M)
return(CM)
})
# Apply previous function, calculate_mean_network_connectivity, to all 10 matrices while subsetting for nodes of interest
multiple_subject_result <- sapply(CM_List, function(x) calculate_mean_network_connectivity(CM = x, nodes = nodes_of_interest))
print(multiple_subject_result)Subcortical ROIs
| V334 | V335 | V336 | V337 | V338 | V339 | V340 | V341 | V342 | V343 | V344 | V345 | V346 | V347 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 334 | 2.000 | 0.462 | 0.392 | 0.576 | 0.338 | 0.118 | 0.340 | 1.646 | 0.535 | 0.364 | 0.414 | 0.313 | 0.079 | 0.389 |
| 335 | 0.462 | 2.000 | 0.727 | 0.653 | 0.650 | 0.472 | 0.527 | 0.434 | 1.080 | 0.598 | 0.483 | 0.570 | 0.430 | 0.496 |
| 336 | 0.392 | 0.727 | 2.000 | 1.073 | 0.855 | 0.726 | 0.643 | 0.408 | 0.699 | 1.421 | 1.016 | 0.897 | 0.700 | 0.630 |
| 337 | 0.576 | 0.653 | 1.073 | 2.000 | 0.821 | 0.743 | 0.770 | 0.573 | 0.603 | 0.922 | 1.194 | 0.784 | 0.684 | 0.801 |
| 338 | 0.338 | 0.650 | 0.855 | 0.821 | 2.000 | 1.376 | 0.731 | 0.365 | 0.622 | 0.769 | 1.029 | 1.911 | 1.209 | 0.693 |
| 339 | 0.118 | 0.472 | 0.726 | 0.743 | 1.376 | 2.000 | 0.600 | 0.183 | 0.421 | 0.671 | 0.875 | 1.283 | 1.650 | 0.577 |
| 340 | 0.340 | 0.527 | 0.643 | 0.770 | 0.731 | 0.600 | 2.000 | 0.363 | 0.625 | 0.599 | 0.730 | 0.742 | 0.603 | 1.566 |
| 341 | 1.646 | 0.434 | 0.408 | 0.573 | 0.365 | 0.183 | 0.363 | 2.000 | 0.571 | 0.419 | 0.447 | 0.335 | 0.141 | 0.379 |
| 342 | 0.535 | 1.080 | 0.699 | 0.603 | 0.622 | 0.421 | 0.625 | 0.571 | 2.000 | 0.705 | 0.529 | 0.580 | 0.393 | 0.661 |
| 343 | 0.364 | 0.598 | 1.421 | 0.922 | 0.769 | 0.671 | 0.599 | 0.419 | 0.705 | 2.000 | 0.939 | 0.797 | 0.628 | 0.638 |
| 344 | 0.414 | 0.483 | 1.016 | 1.194 | 1.029 | 0.875 | 0.730 | 0.447 | 0.529 | 0.939 | 2.000 | 0.982 | 0.827 | 0.720 |
| 345 | 0.313 | 0.570 | 0.897 | 0.784 | 1.911 | 1.283 | 0.742 | 0.335 | 0.580 | 0.797 | 0.982 | 2.000 | 1.223 | 0.719 |
| 346 | 0.079 | 0.430 | 0.700 | 0.684 | 1.209 | 1.650 | 0.603 | 0.141 | 0.393 | 0.628 | 0.827 | 1.223 | 2.000 | 0.571 |
| 347 | 0.389 | 0.496 | 0.630 | 0.801 | 0.693 | 0.577 | 1.566 | 0.379 | 0.661 | 0.638 | 0.720 | 0.719 | 0.571 | 2.000 |
7-Network Cerebellar Parcellation
| V348 | V349 | V350 | V351 | V352 | V353 | V354 | V355 | V356 | V357 | V358 | V359 | V360 | V361 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 348 | 2.000 | 0.509 | 0.492 | 0.794 | 0.616 | 0.381 | -0.117 | 1.174 | 0.556 | 0.449 | 0.679 | 0.594 | 0.351 | -0.021 |
| 349 | 0.509 | 2.000 | 0.925 | 1.108 | 0.738 | 0.596 | 0.338 | 0.508 | 1.558 | 0.607 | 1.037 | 0.731 | 0.735 | 0.497 |
| 350 | 0.492 | 0.925 | 2.000 | 1.243 | 0.762 | 0.717 | 0.401 | 0.520 | 0.835 | 1.108 | 1.066 | 0.665 | 0.689 | 0.366 |
| 351 | 0.794 | 1.108 | 1.243 | 2.000 | 0.942 | 0.964 | 0.439 | 0.791 | 1.002 | 0.932 | 1.256 | 0.824 | 0.843 | 0.416 |
| 352 | 0.616 | 0.738 | 0.762 | 0.942 | 2.000 | 0.804 | 0.322 | 0.612 | 0.713 | 0.778 | 0.773 | 1.395 | 0.711 | 0.358 |
| 353 | 0.381 | 0.596 | 0.717 | 0.964 | 0.804 | 2.000 | 0.913 | 0.534 | 0.639 | 0.973 | 0.815 | 0.754 | 1.153 | 0.647 |
| 354 | -0.117 | 0.338 | 0.401 | 0.439 | 0.322 | 0.913 | 2.000 | 0.135 | 0.319 | 0.550 | 0.421 | 0.335 | 0.833 | 1.140 |
| 355 | 1.174 | 0.508 | 0.520 | 0.791 | 0.612 | 0.534 | 0.135 | 2.000 | 0.571 | 0.584 | 0.790 | 0.694 | 0.542 | 0.204 |
| 356 | 0.556 | 1.558 | 0.835 | 1.002 | 0.713 | 0.639 | 0.319 | 0.571 | 2.000 | 0.642 | 1.258 | 0.789 | 0.725 | 0.427 |
| 357 | 0.449 | 0.607 | 1.108 | 0.932 | 0.778 | 0.973 | 0.550 | 0.584 | 0.642 | 2.000 | 0.979 | 0.744 | 0.960 | 0.459 |
| 358 | 0.679 | 1.037 | 1.066 | 1.256 | 0.773 | 0.815 | 0.421 | 0.790 | 1.258 | 0.979 | 2.000 | 0.842 | 0.871 | 0.471 |
| 359 | 0.594 | 0.731 | 0.665 | 0.824 | 1.395 | 0.754 | 0.335 | 0.694 | 0.789 | 0.744 | 0.842 | 2.000 | 0.774 | 0.428 |
| 360 | 0.351 | 0.735 | 0.689 | 0.843 | 0.711 | 1.153 | 0.833 | 0.542 | 0.725 | 0.960 | 0.871 | 0.774 | 2.000 | 0.967 |
| 361 | -0.021 | 0.497 | 0.366 | 0.416 | 0.358 | 0.647 | 1.140 | 0.204 | 0.427 | 0.459 | 0.471 | 0.428 | 0.967 | 2.000 |
7 Yeo Networks
| V362 | V363 | V364 | V365 | V366 | V367 | V368 | |
|---|---|---|---|---|---|---|---|
| 362 | 2.000 | 1.173 | 1.478 | 1.016 | 1.155 | 1.065 | 0.975 |
| 363 | 1.173 | 2.000 | 1.356 | 1.270 | 1.069 | 1.126 | 0.999 |
| 364 | 1.478 | 1.356 | 2.000 | 1.118 | 1.195 | 1.444 | 1.139 |
| 365 | 1.016 | 1.270 | 1.118 | 2.000 | 0.787 | 1.080 | 0.858 |
| 366 | 1.155 | 1.069 | 1.195 | 0.787 | 2.000 | 1.066 | 1.168 |
| 367 | 1.065 | 1.126 | 1.444 | 1.080 | 1.066 | 2.000 | 1.282 |
| 368 | 0.975 | 0.999 | 1.139 | 0.858 | 1.168 | 1.282 | 2.000 |
17 Yeo Networks
| V369 | V370 | V371 | V372 | V373 | V374 | V375 | V376 | V377 | V378 | V379 | V380 | V381 | V382 | V383 | V384 | V385 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 369 | 2.000 | 1.000 | 0.961 | 1.017 | 1.462 | 1.143 | 0.846 | 0.680 | 0.957 | 1.107 | 0.547 | 1.028 | 0.853 | 0.732 | 0.993 | 0.802 | 0.640 |
| 370 | 1.000 | 2.000 | 0.937 | 0.929 | 1.055 | 1.020 | 0.817 | 0.736 | 0.825 | 0.672 | 0.685 | 0.738 | 0.585 | 0.899 | 1.121 | 0.863 | 0.635 |
| 371 | 0.961 | 0.937 | 2.000 | 1.568 | 0.997 | 1.361 | 1.126 | 0.698 | 0.898 | 0.873 | 0.539 | 1.038 | 0.904 | 0.868 | 0.919 | 0.799 | 0.786 |
| 372 | 1.017 | 0.929 | 1.568 | 2.000 | 1.049 | 1.343 | 1.485 | 0.835 | 0.952 | 0.854 | 0.510 | 1.140 | 0.872 | 1.012 | 1.008 | 0.845 | 0.818 |
| 373 | 1.462 | 1.055 | 0.997 | 1.049 | 2.000 | 1.362 | 0.931 | 0.808 | 1.056 | 0.927 | 0.728 | 1.361 | 1.057 | 1.061 | 1.339 | 0.979 | 0.891 |
| 374 | 1.143 | 1.020 | 1.361 | 1.343 | 1.362 | 2.000 | 1.253 | 0.867 | 1.048 | 0.874 | 0.524 | 1.280 | 1.049 | 1.056 | 1.061 | 0.931 | 0.960 |
| 375 | 0.846 | 0.817 | 1.126 | 1.485 | 0.931 | 1.253 | 2.000 | 1.138 | 0.767 | 0.614 | 0.450 | 0.977 | 0.758 | 0.827 | 0.902 | 0.735 | 0.666 |
| 376 | 0.680 | 0.736 | 0.698 | 0.835 | 0.808 | 0.867 | 1.138 | 2.000 | 0.633 | 0.464 | 0.518 | 0.698 | 0.712 | 0.650 | 0.748 | 0.737 | 0.676 |
| 377 | 0.957 | 0.825 | 0.898 | 0.952 | 1.056 | 1.048 | 0.767 | 0.633 | 2.000 | 0.954 | 0.559 | 0.933 | 0.922 | 1.020 | 1.075 | 1.003 | 1.075 |
| 378 | 1.107 | 0.672 | 0.873 | 0.854 | 0.927 | 0.874 | 0.614 | 0.464 | 0.954 | 2.000 | 0.563 | 0.833 | 0.913 | 0.638 | 0.805 | 0.807 | 0.726 |
| 379 | 0.547 | 0.685 | 0.539 | 0.510 | 0.728 | 0.524 | 0.450 | 0.518 | 0.559 | 0.563 | 2.000 | 0.482 | 0.568 | 0.688 | 0.825 | 0.981 | 0.593 |
| 380 | 1.028 | 0.738 | 1.038 | 1.140 | 1.361 | 1.280 | 0.977 | 0.698 | 0.933 | 0.833 | 0.482 | 2.000 | 1.216 | 0.975 | 0.998 | 0.804 | 0.952 |
| 381 | 0.853 | 0.585 | 0.904 | 0.872 | 1.057 | 1.049 | 0.758 | 0.712 | 0.922 | 0.913 | 0.568 | 1.216 | 2.000 | 0.879 | 0.871 | 0.999 | 1.220 |
| 382 | 0.732 | 0.899 | 0.868 | 1.012 | 1.061 | 1.056 | 0.827 | 0.650 | 1.020 | 0.638 | 0.688 | 0.975 | 0.879 | 2.000 | 1.182 | 1.049 | 1.199 |
| 383 | 0.993 | 1.121 | 0.919 | 1.008 | 1.339 | 1.061 | 0.902 | 0.748 | 1.075 | 0.805 | 0.825 | 0.998 | 0.871 | 1.182 | 2.000 | 1.246 | 0.976 |
| 384 | 0.802 | 0.863 | 0.799 | 0.845 | 0.979 | 0.931 | 0.735 | 0.737 | 1.003 | 0.807 | 0.981 | 0.804 | 0.999 | 1.049 | 1.246 | 2.000 | 1.159 |
| 385 | 0.640 | 0.635 | 0.786 | 0.818 | 0.891 | 0.960 | 0.666 | 0.676 | 1.075 | 0.726 | 0.593 | 0.952 | 1.220 | 1.199 | 0.976 | 1.159 | 2.000 |
Files
List of files will include a comma-separated values (.csv) file for each participant in the form of a 385 matrix.
- sub-001/cor385.csv
- sub-002/cor385.csv
- […]