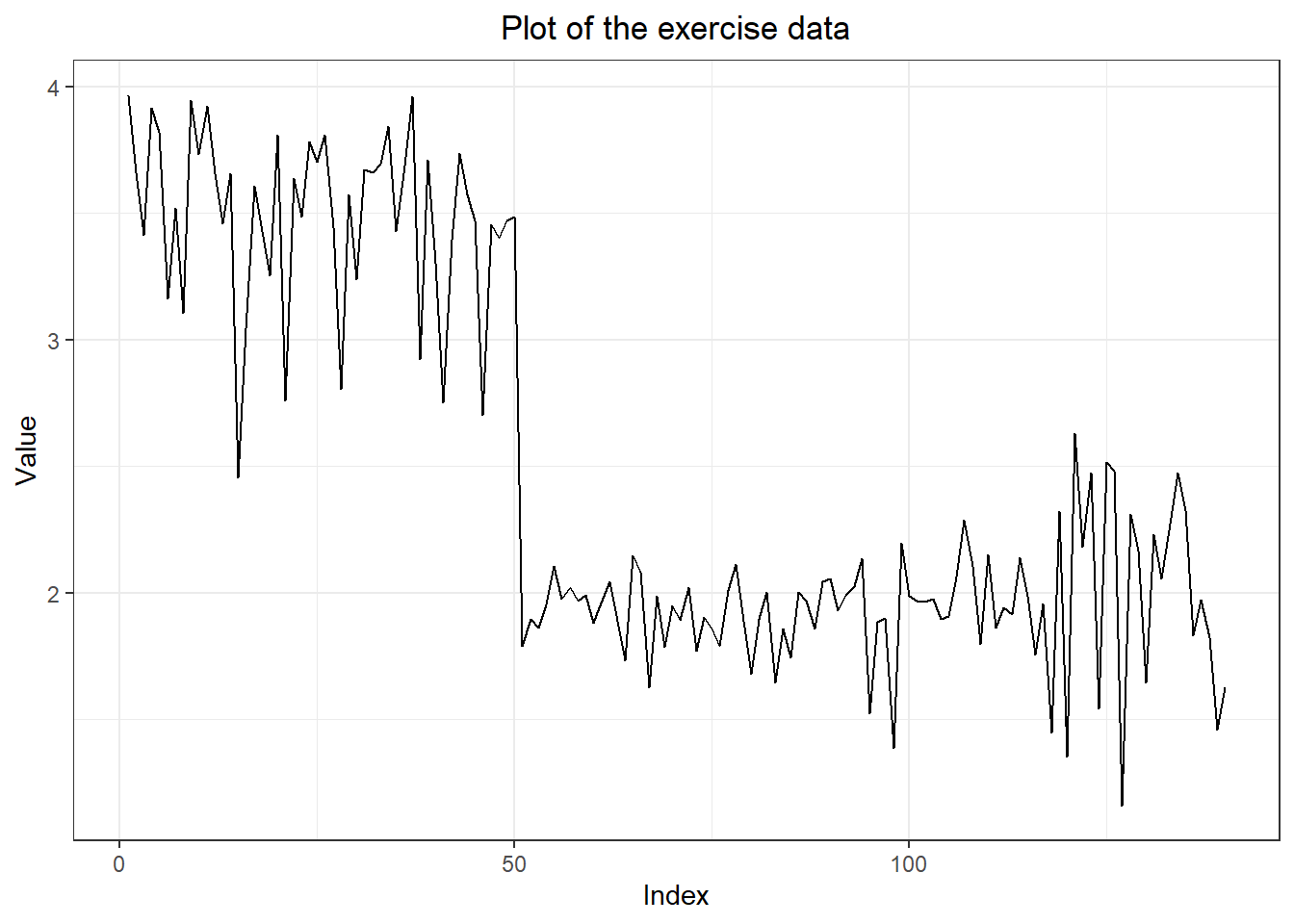
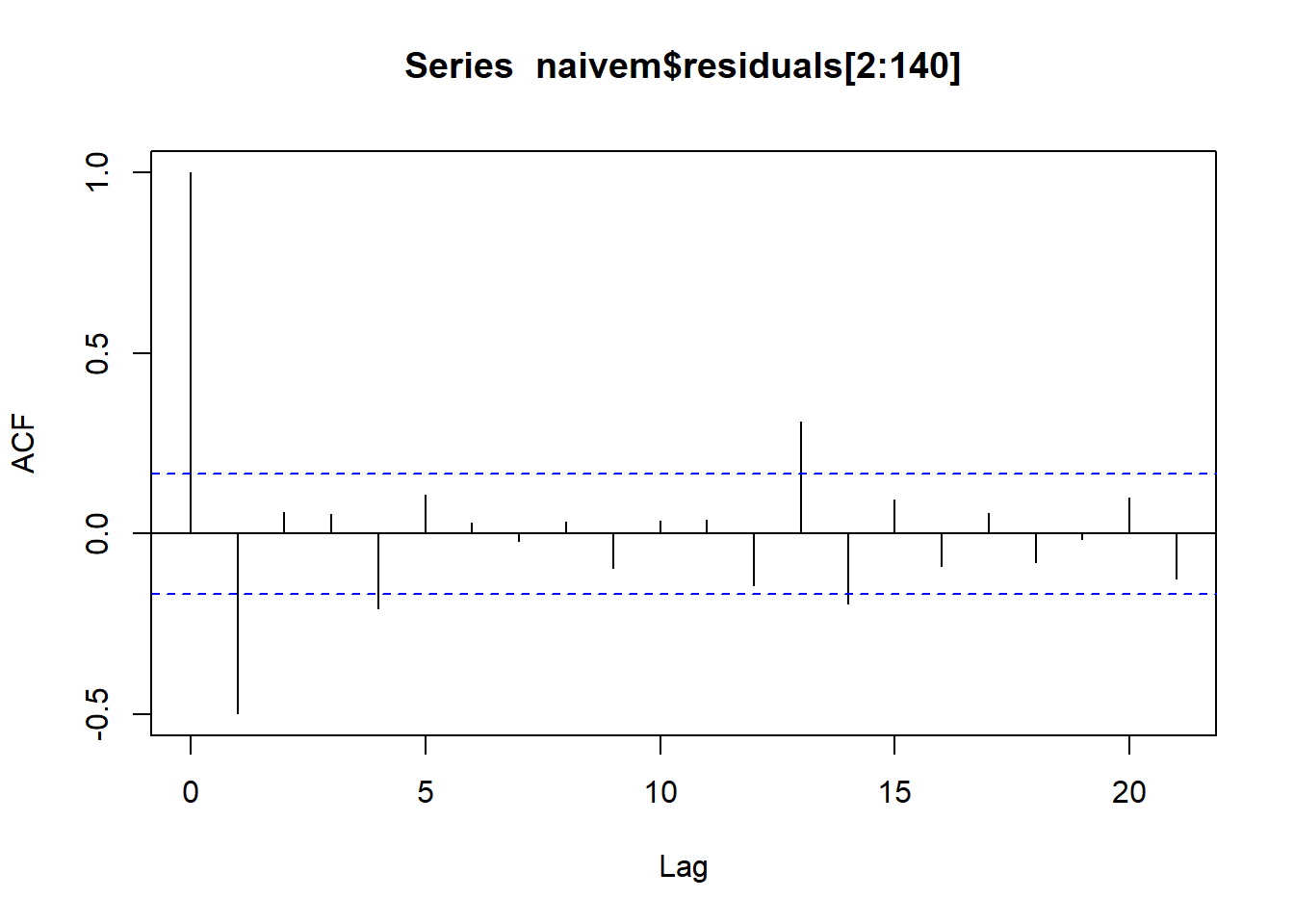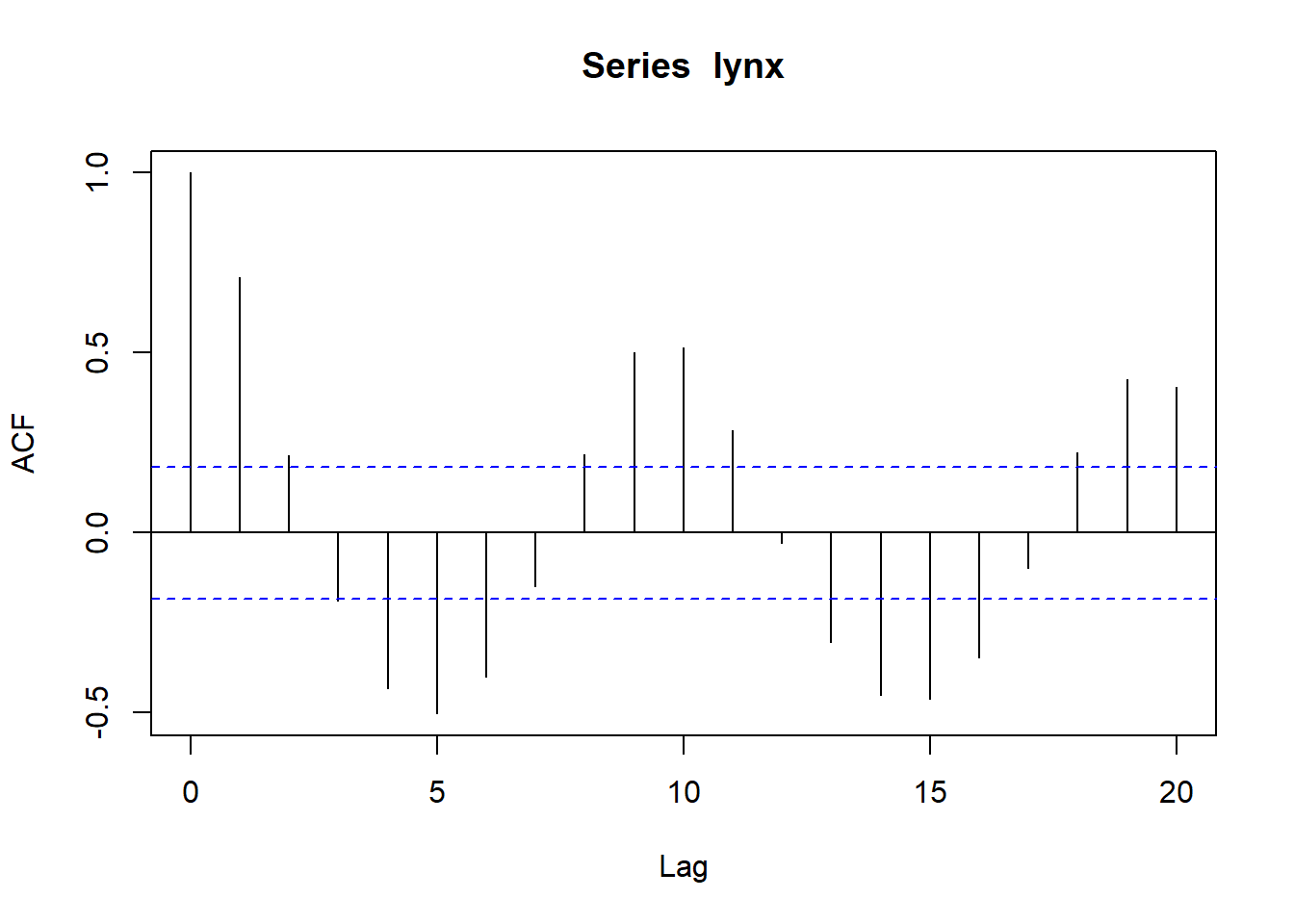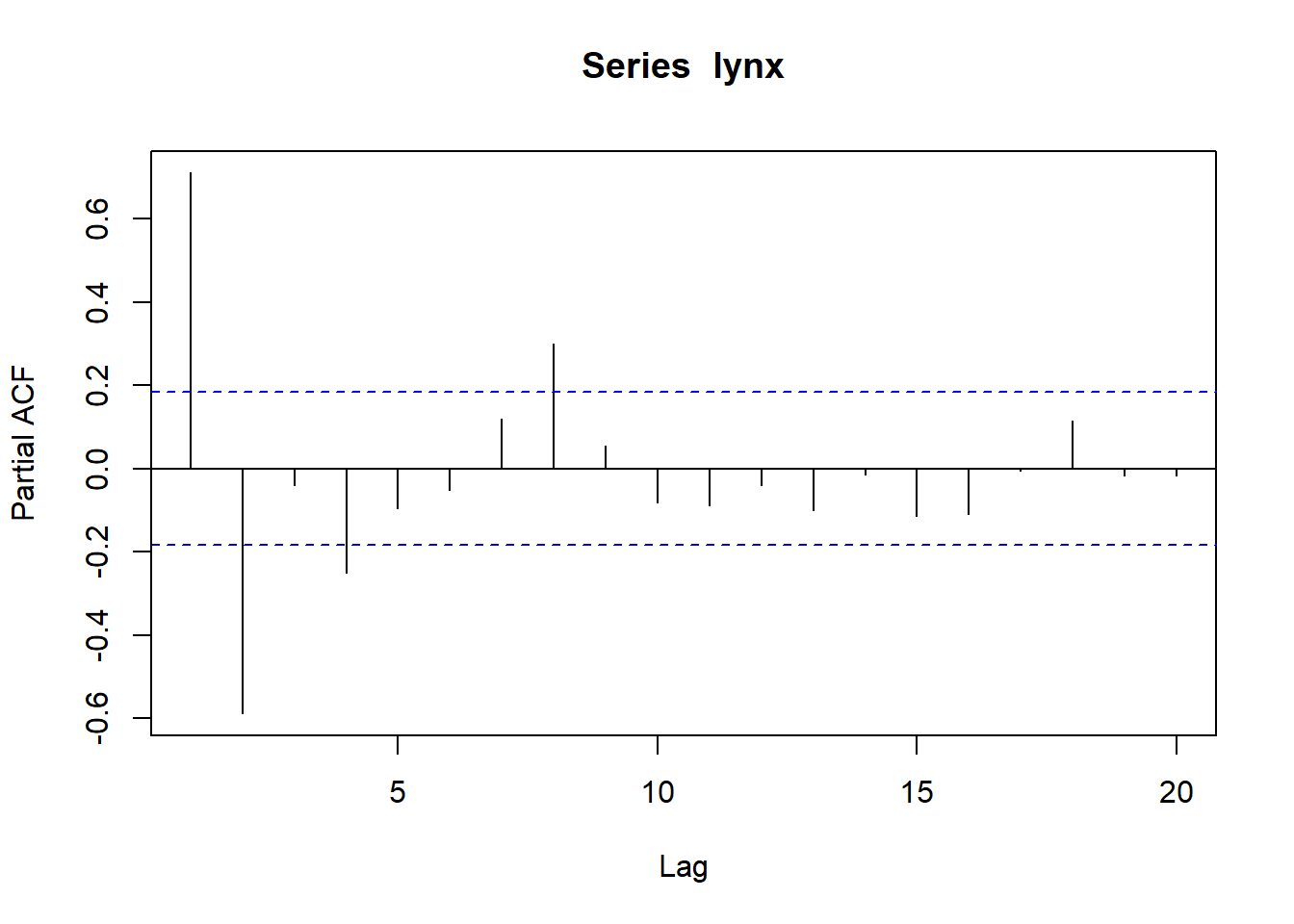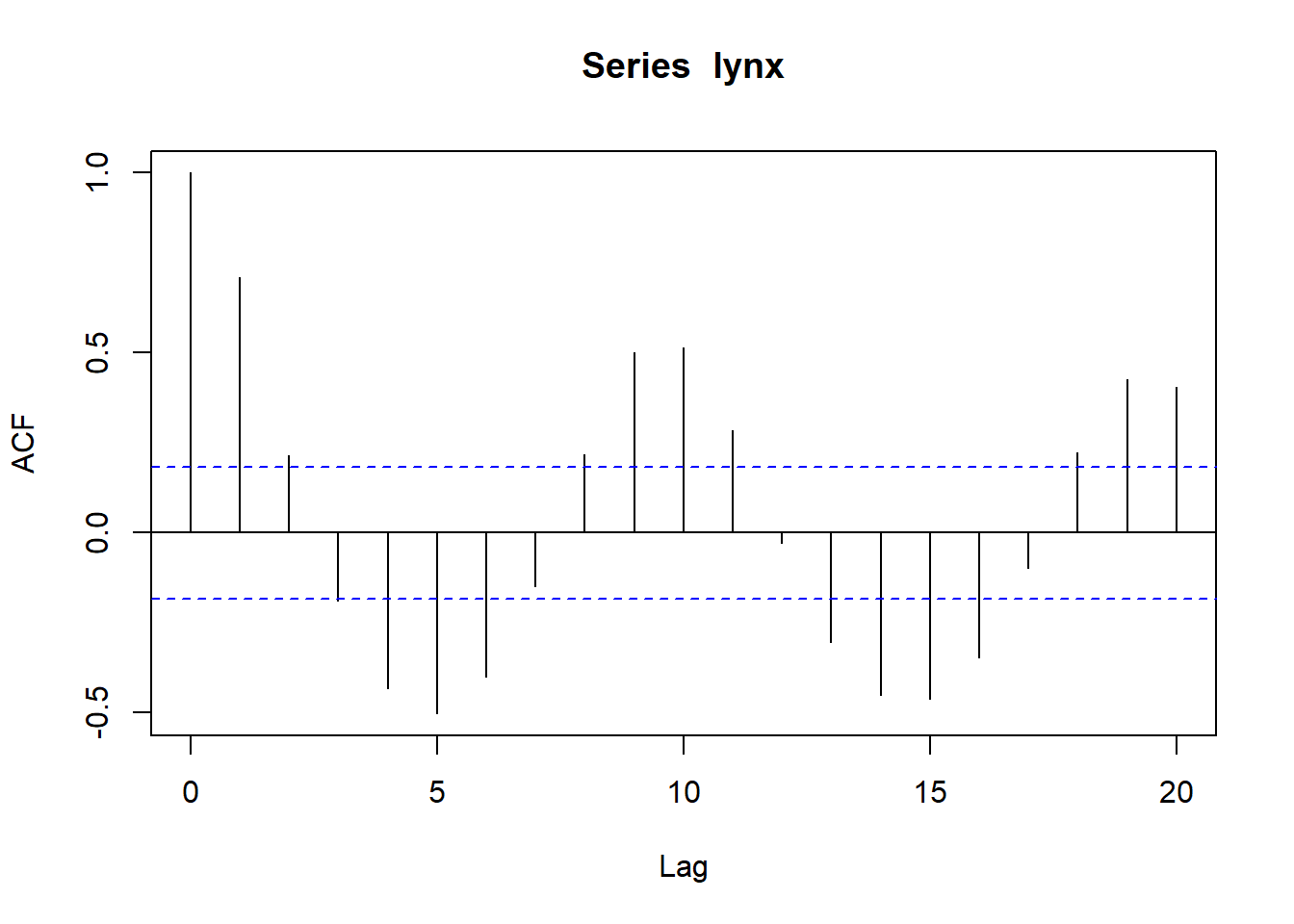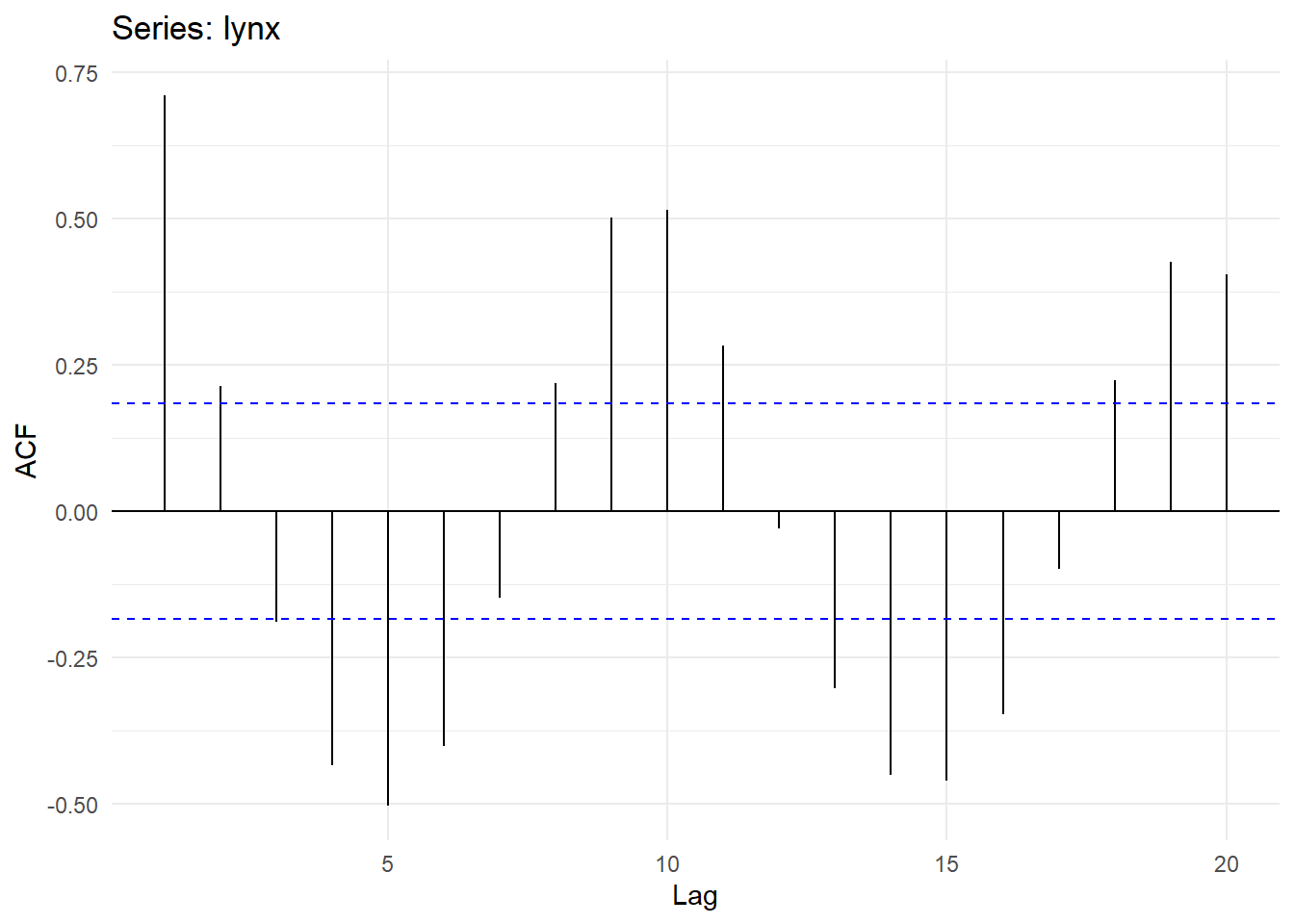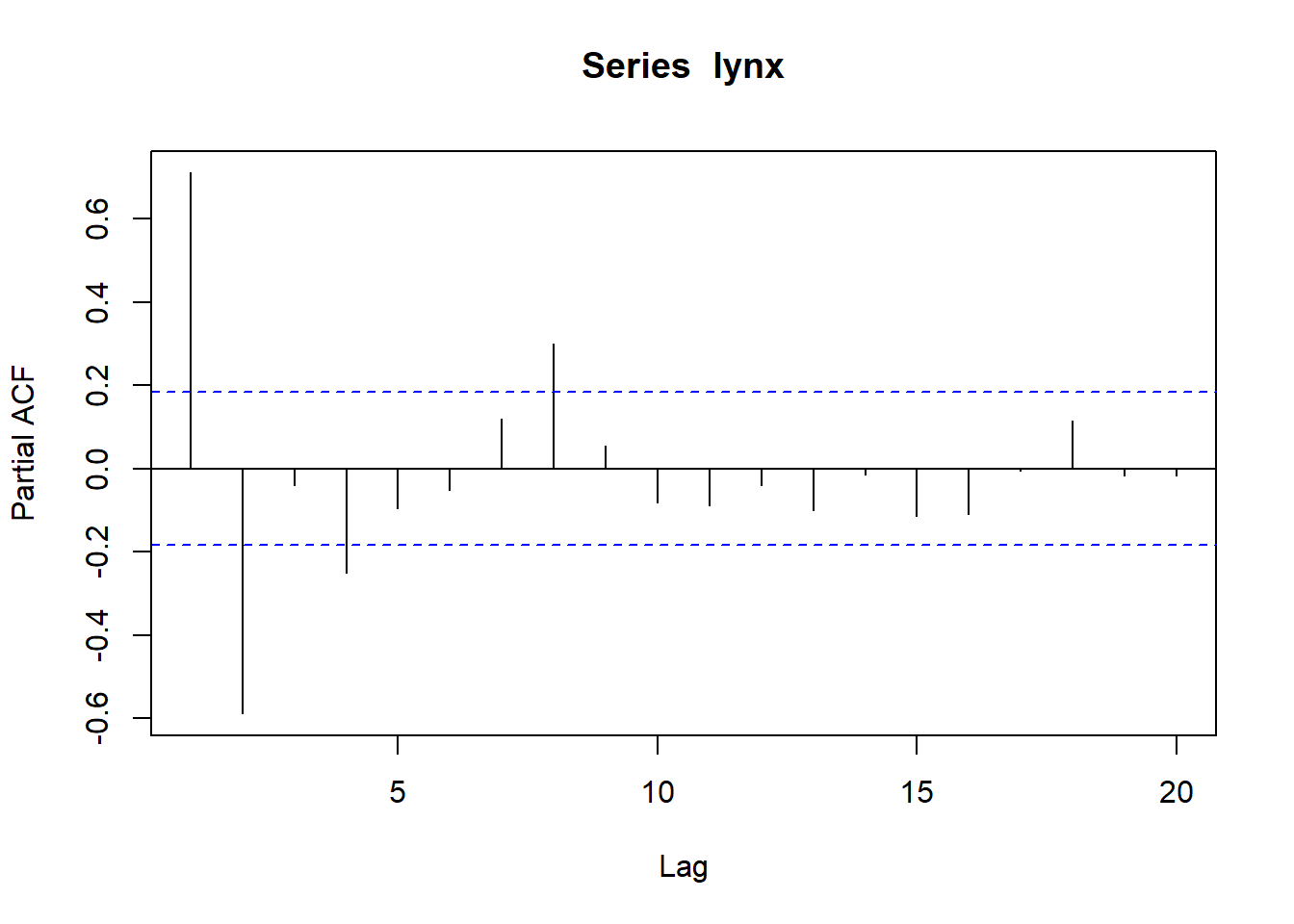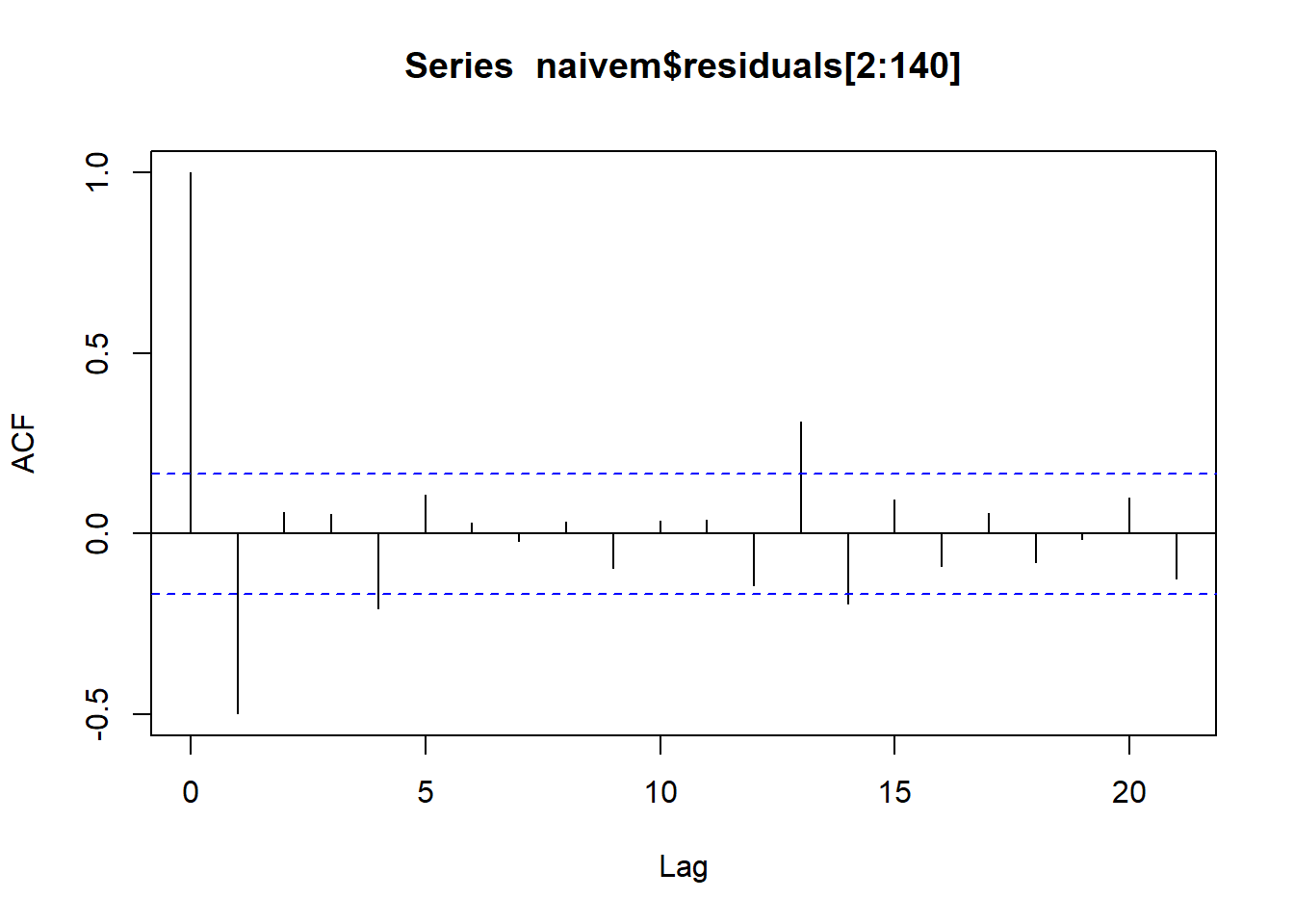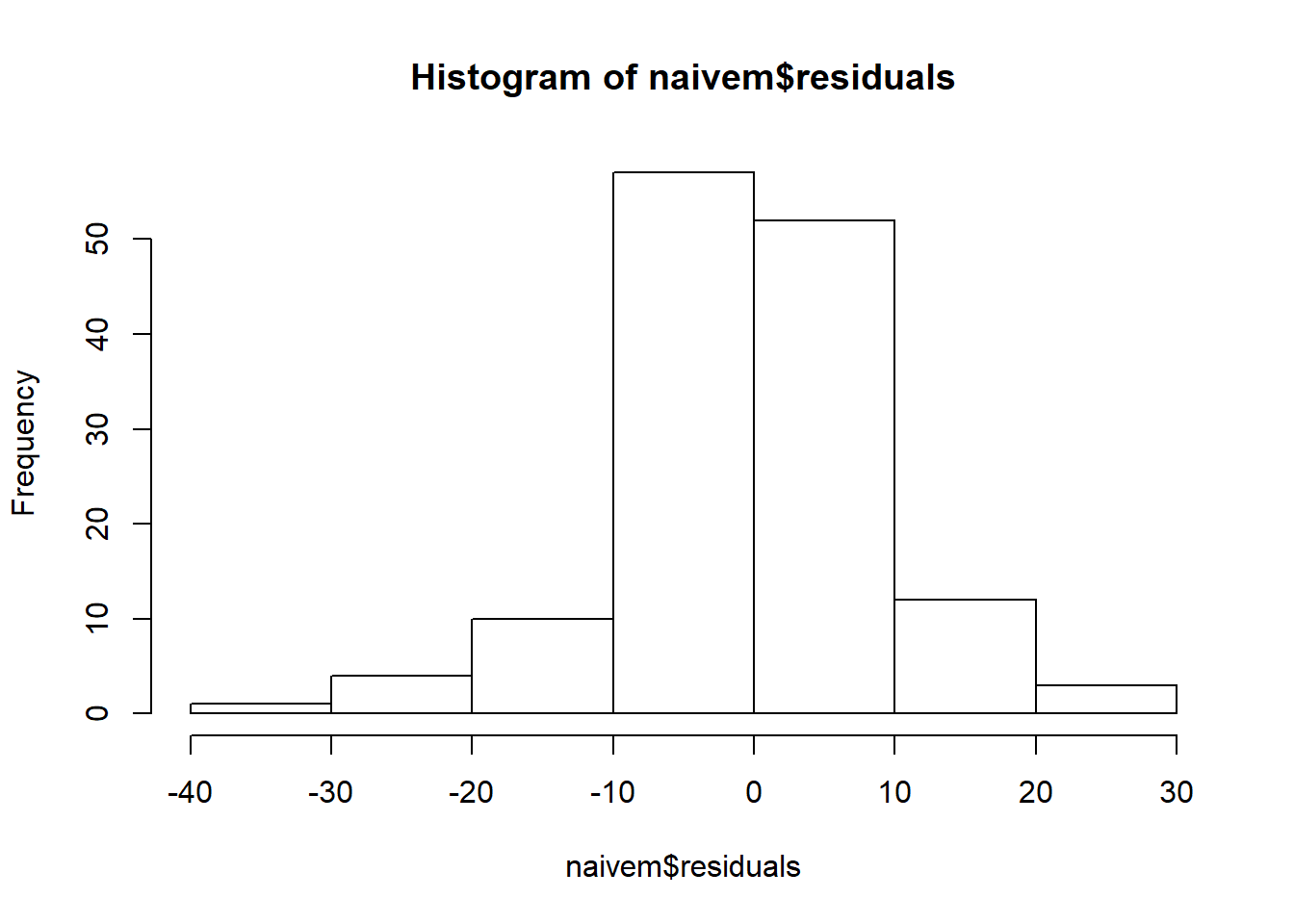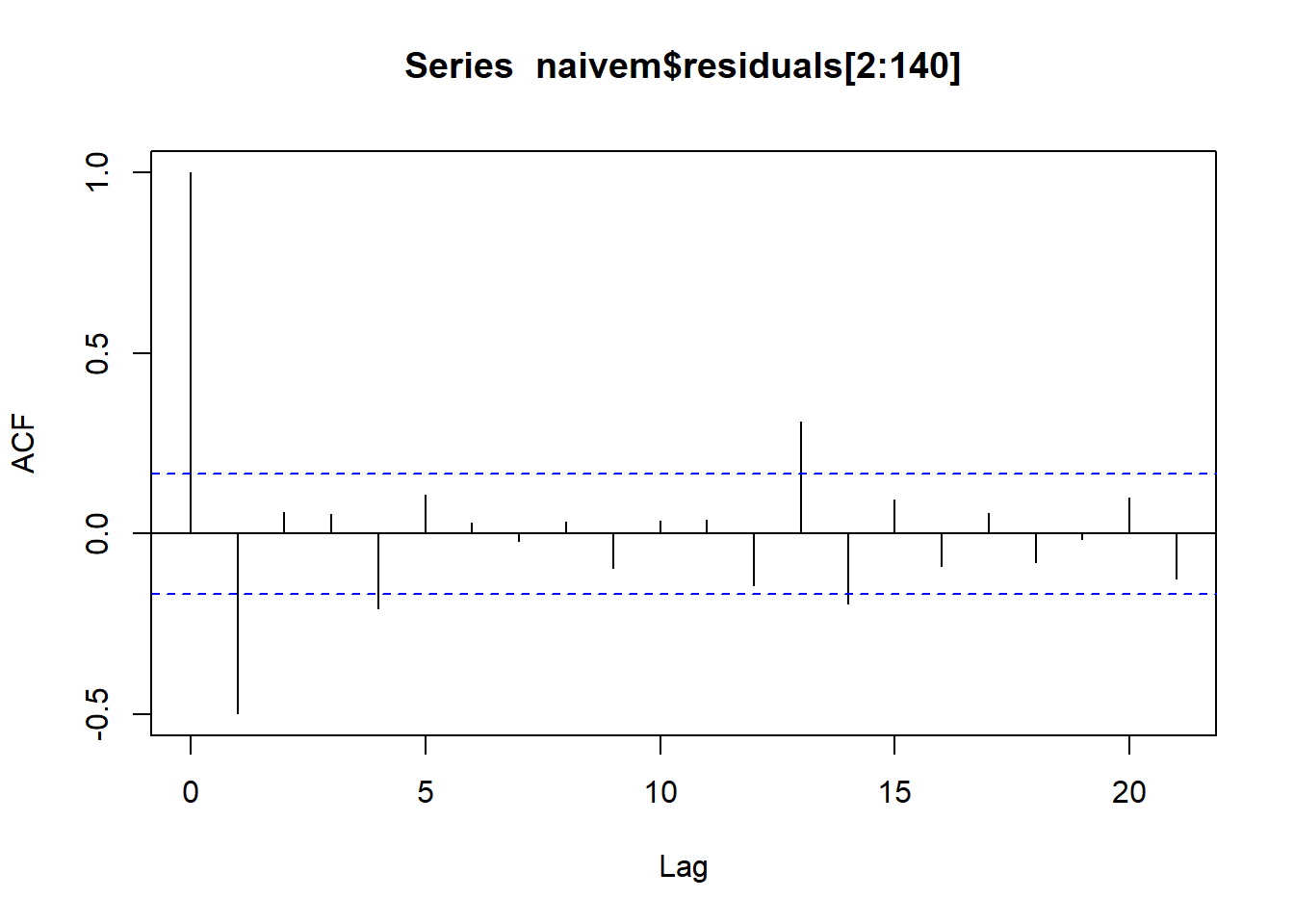Statistical Background For TS Analysis & Forecasting
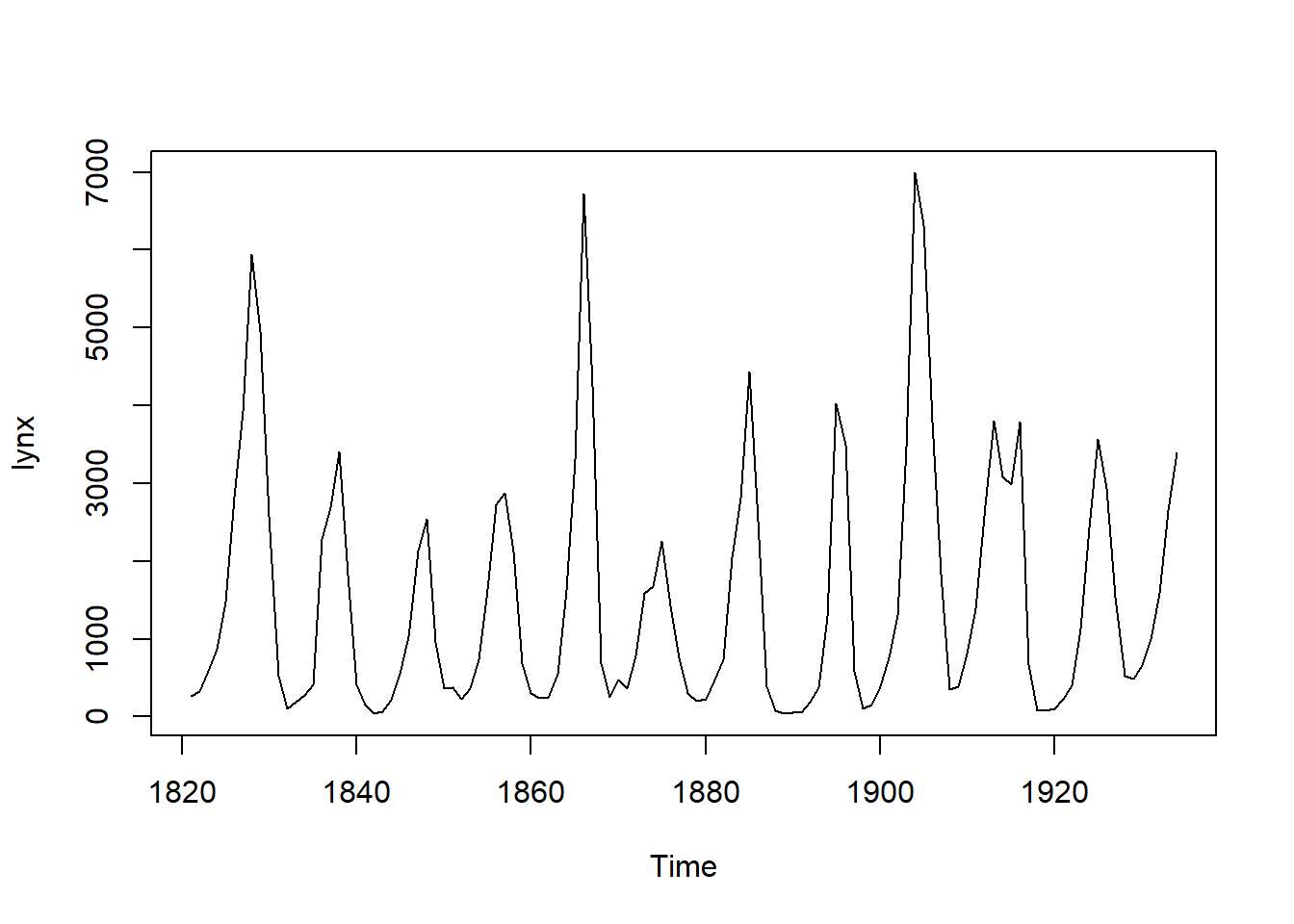
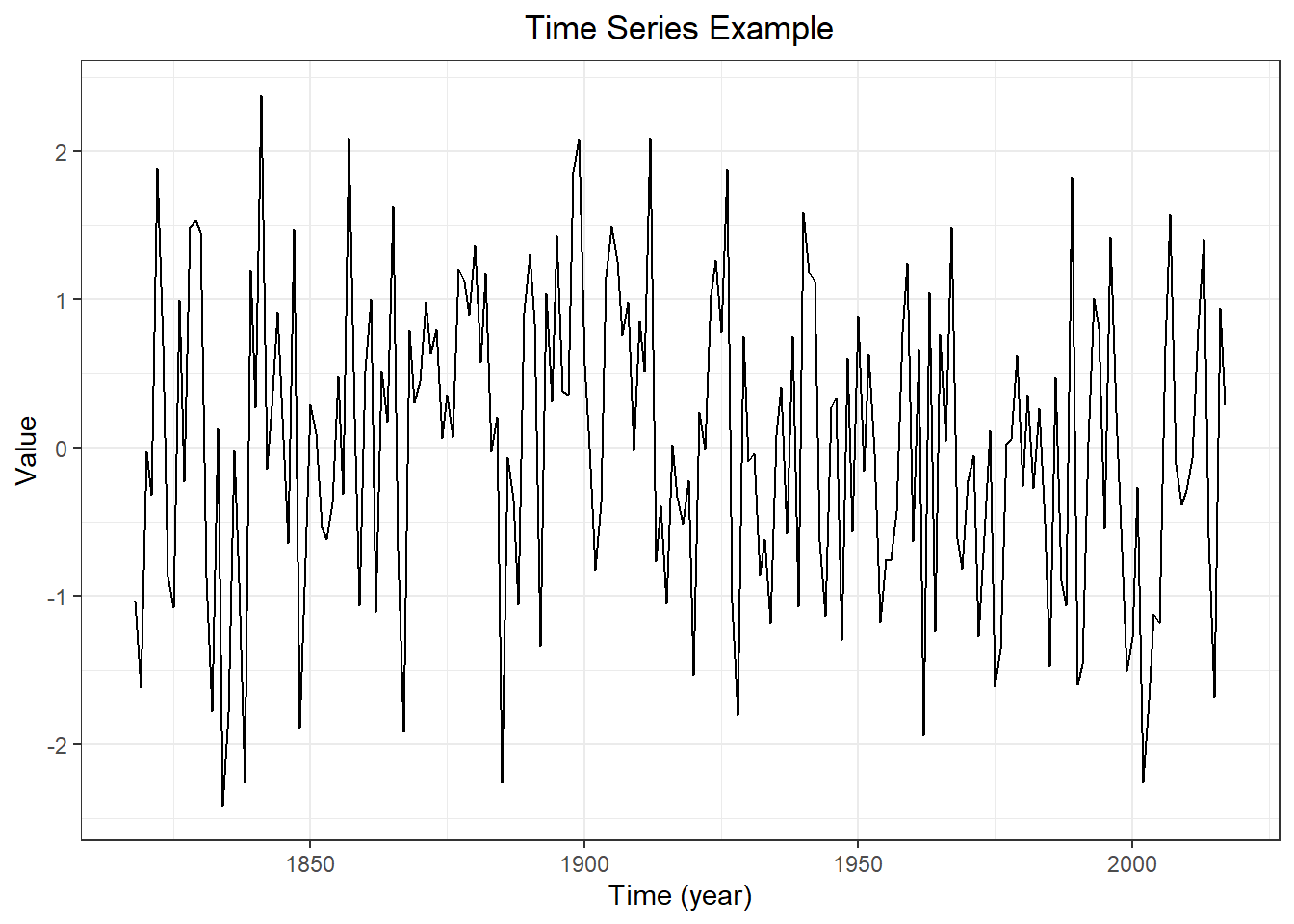
## Time Series:
## Start = 1821
## End = 1934
## Frequency = 1
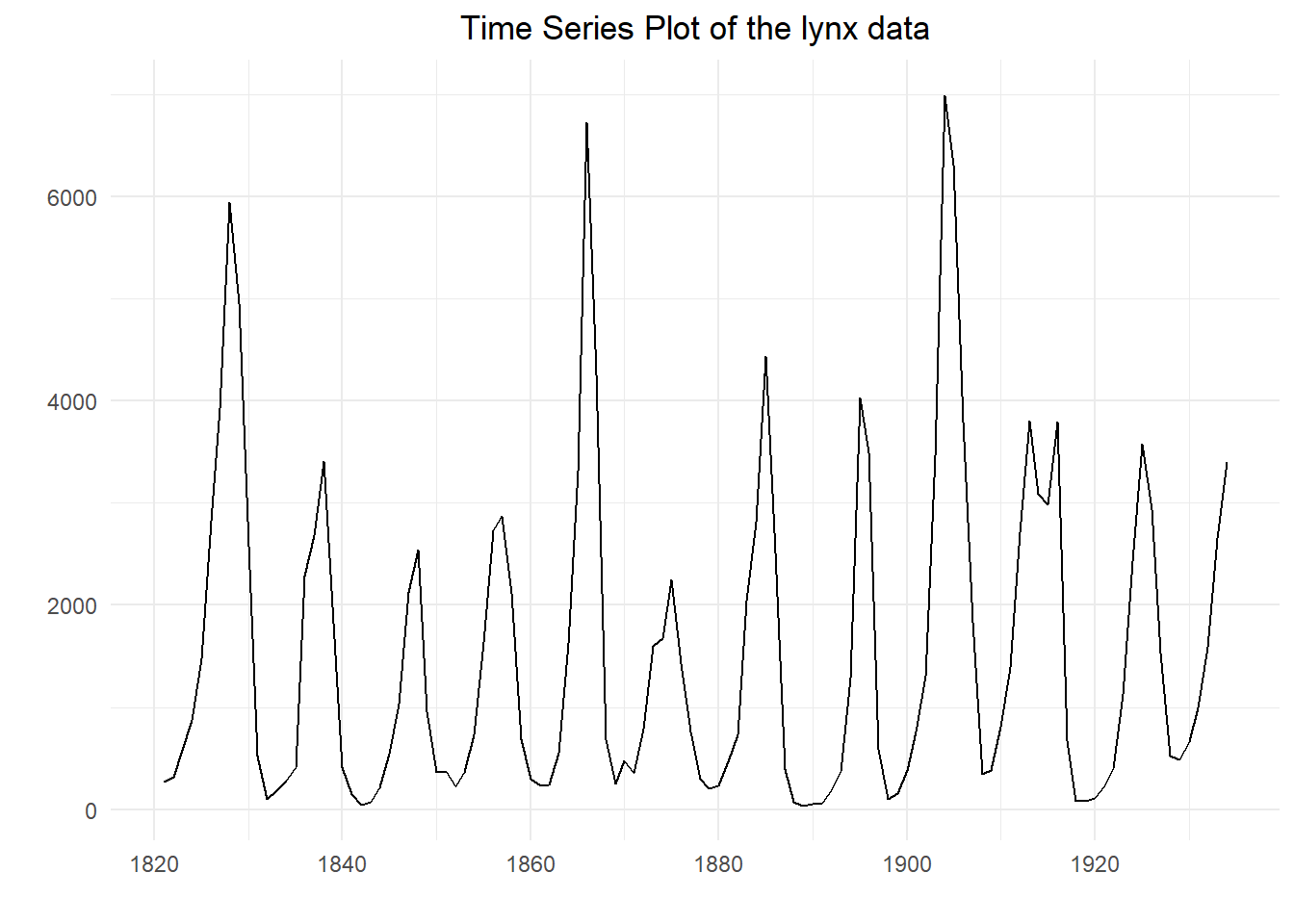
## [1] 269 321 585 871 1475 2821 3928 5943 4950 2577 523 98 184 279
## [15] 409 2285 2685 3409 1824 409 151 45 68 213 546 1033 2129 2536
## [29] 957 361 377 225 360 731 1638 2725 2871 2119 684 299 236 245
## [43] 552 1623 3311 6721 4254 687 255 473 358 784 1594 1676 2251 1426
## [57] 756 299 201 229 469 736 2042 2811 4431 2511 389 73 39 49
## [71] 59 188 377 1292 4031 3495 587 105 153 387 758 1307 3465 6991
## [85] 6313 3794 1836 345 382 808 1388 2713 3800 3091 2985 3790 674 81
## [99] 80 108 229 399 1132 2432 3574 2935 1537 529 485 662 1000 1590
## [113] 2657 3396
## Time Series:
## Start = 1821
## End = 1934
## Frequency = 1
## [1] 1821 1822 1823 1824 1825 1826 1827 1828 1829 1830 1831 1832 1833 1834
## [15] 1835 1836 1837 1838 1839 1840 1841 1842 1843 1844 1845 1846 1847 1848
## [29] 1849 1850 1851 1852 1853 1854 1855 1856 1857 1858 1859 1860 1861 1862
## [43] 1863 1864 1865 1866 1867 1868 1869 1870 1871 1872 1873 1874 1875 1876
## [57] 1877 1878 1879 1880 1881 1882 1883 1884 1885 1886 1887 1888 1889 1890
## [71] 1891 1892 1893 1894 1895 1896 1897 1898 1899 1900 1901 1902 1903 1904
## [85] 1905 1906 1907 1908 1909 1910 1911 1912 1913 1914 1915 1916 1917 1918
## [99] 1919 1920 1921 1922 1923 1924 1925 1926 1927 1928 1929 1930 1931 1932
## [113] 1933 1934
## [1] 114
## Time Series:
## Start = 1929
## End = 1934
## Frequency = 1
## [1] 485 662 1000 1590 2657 3396
## [1] 1538.018
## [1] 771
## [1] 39 45 49 59 68 73 80 81 98 105 108 151 153 184
## [15] 188 201 213 225 229 229 236 245 255 269 279 299 299 321
## [29] 345 358 360 361 377 377 382 387 389 399 409 409 469 473
## [43] 485 523 529 546 552 585 587 662 674 684 687 731 736 756
## [57] 758 784 808 871 957 1000 1033 1132 1292 1307 1388 1426 1475 1537
## [71] 1590 1594 1623 1638 1676 1824 1836 2042 2119 2129 2251 2285 2432 2511
## [85] 2536 2577 2657 2685 2713 2725 2811 2821 2871 2935 2985 3091 3311 3396
## [99] 3409 3465 3495 3574 3790 3794 3800 3928 4031 4254 4431 4950 5943 6313
## [113] 6721 6991
## [1] 758 784
## 0% 25% 50% 75% 100%
## 39.00 348.25 771.00 2566.75 6991.00
## 0% 10% 20% 30% 40% 50% 60% 70% 80% 90%
## 39.0 146.7 259.2 380.5 546.6 771.0 1470.1 2165.6 2818.0 3790.4
## 100%
## 6991.0
## Warning in plot.window(xlim, ylim, log, ...): "plot.conf" is not a
## graphical parameter
## Warning in title(main = main, xlab = xlab, ylab = ylab, ...): "plot.conf"
## is not a graphical parameter
## Warning in axis(1, ...): "plot.conf" is not a graphical parameter
## Warning in axis(2, ...): "plot.conf" is not a graphical parameter
## Warning in box(...): "plot.conf" is not a graphical parameter
lines(naivem$mean, col=123, lwd = 2)
lines(driftm$mean, col=22, lwd = 2)
legend("topleft",lty=1,col=c(4,123,22),
legend=c("Mean method","Naive method","Drift Method"))
## ME RMSE MAE MPE MAPE MASE
## Training set 1.407307e-17 1.003956 0.8164571 77.65393 133.4892 0.7702074
## Test set -2.459828e-01 1.138760 0.9627571 100.70356 102.7884 0.9082199
## ACF1 Theil's U
## Training set 0.1293488 NA
## Test set 0.2415939 0.981051
## ME RMSE MAE MPE MAPE MASE
## Training set -0.0002083116 1.323311 1.060048 -152.73569 730.9655 1.000000
## Test set 0.8731935861 1.413766 1.162537 86.29346 307.9891 1.096683
## ACF1 Theil's U
## Training set -0.4953144 NA
## Test set 0.2415939 2.031079
## ME RMSE MAE MPE MAPE MASE
## Training set -1.957854e-17 1.323311 1.060041 -152.64988 730.8626 0.9999931
## Test set 8.763183e-01 1.415768 1.163981 85.96496 308.7329 1.0980447
## ACF1 Theil's U
## Training set -0.4953144 NA
## Test set 0.2418493 2.03317
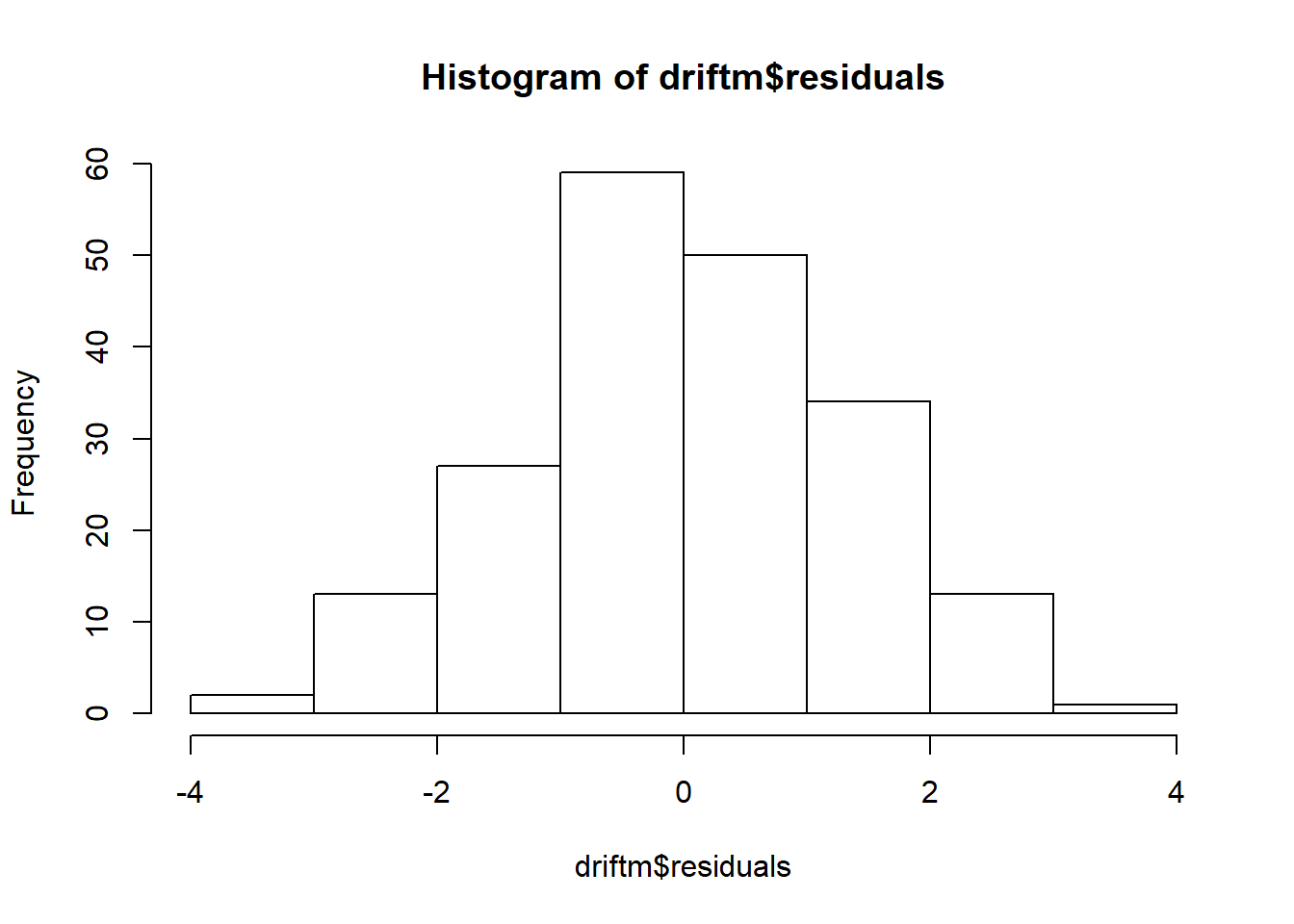
## [1] 1.053807
## [1] -5.95498e-18
## [1] NA
## [1] 1.798592
## [1] 0.006605028
## [1] 1.798592
## [1] -4.502054e-17
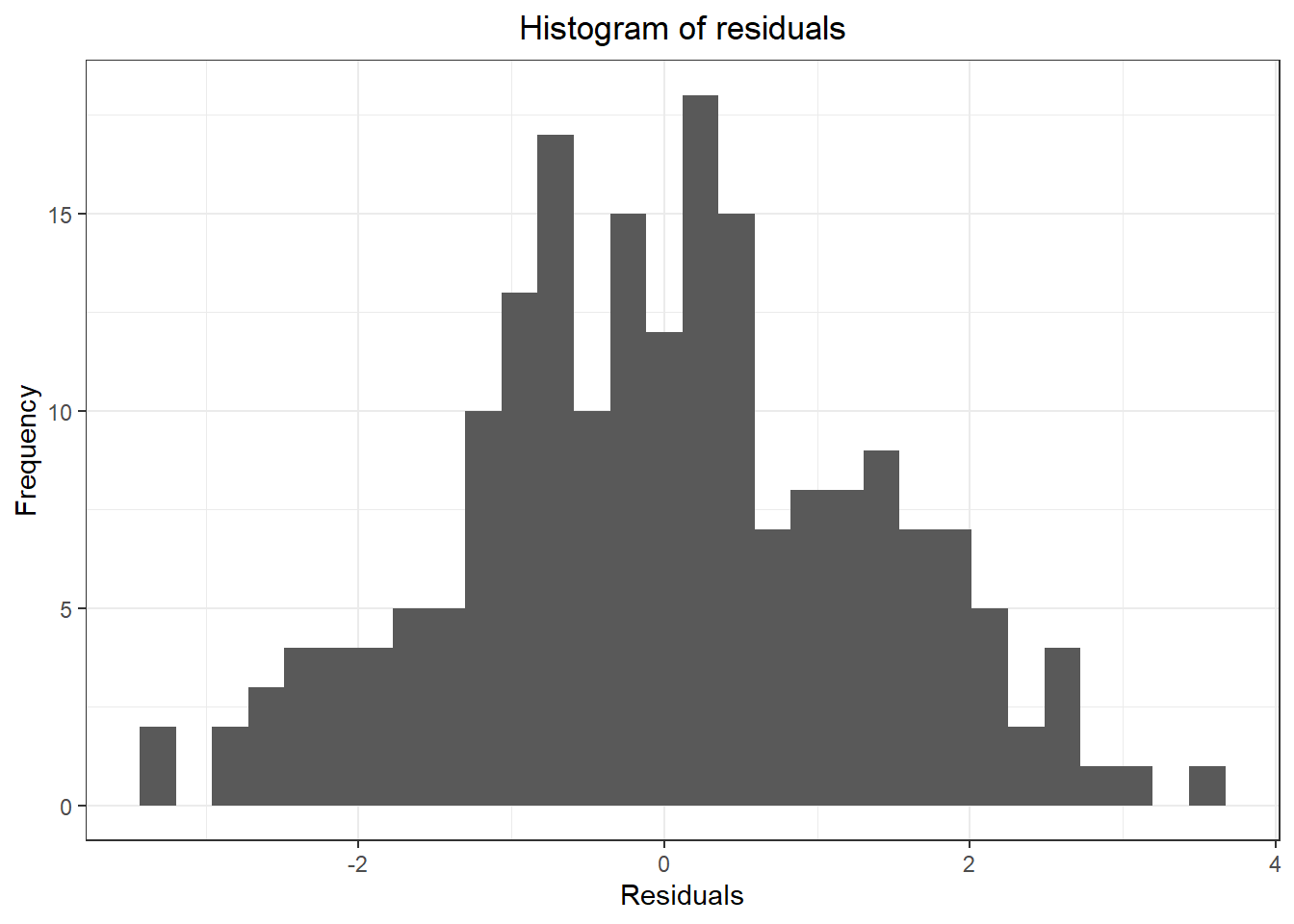
## Don't know how to automatically pick scale for object of type ts. Defaulting to continuous.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## Warning: Removed 1 rows containing non-finite values (stat_bin).
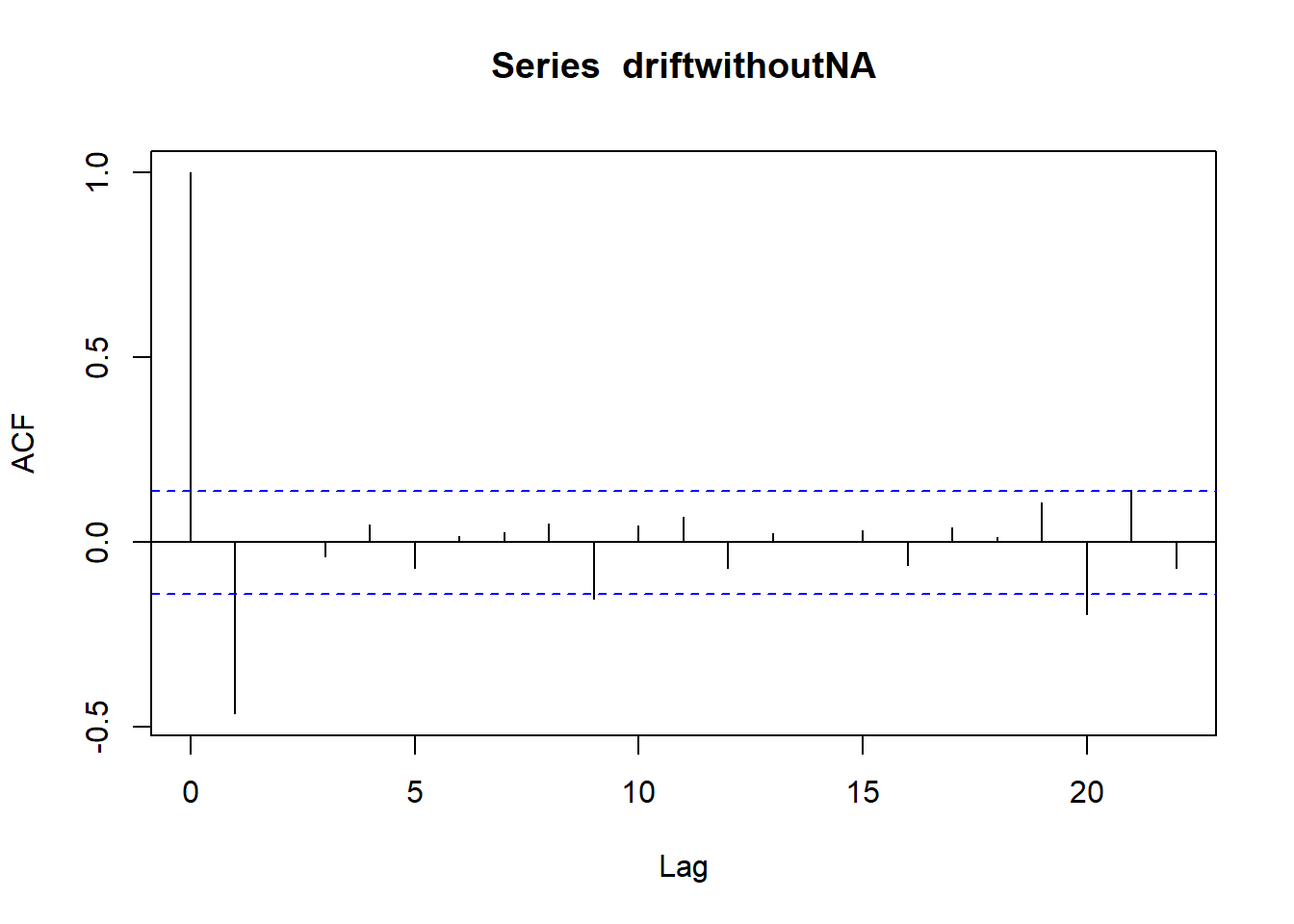
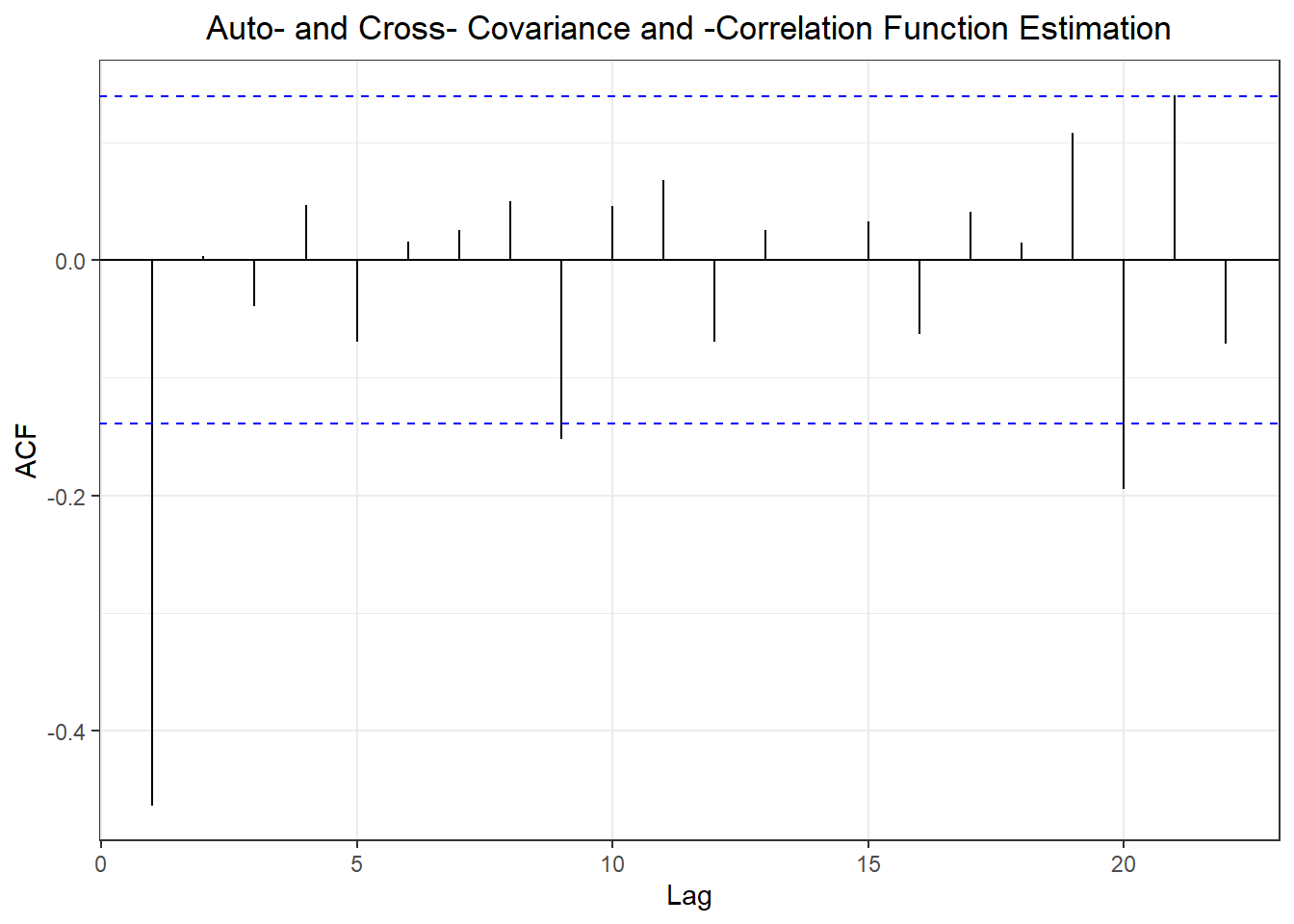
## Warning in adf.test(x): p-value smaller than printed p-value
##
## Augmented Dickey-Fuller Test
##
## data: x
## Dickey-Fuller = -10.647, Lag order = 9, p-value = 0.01
## alternative hypothesis: stationary
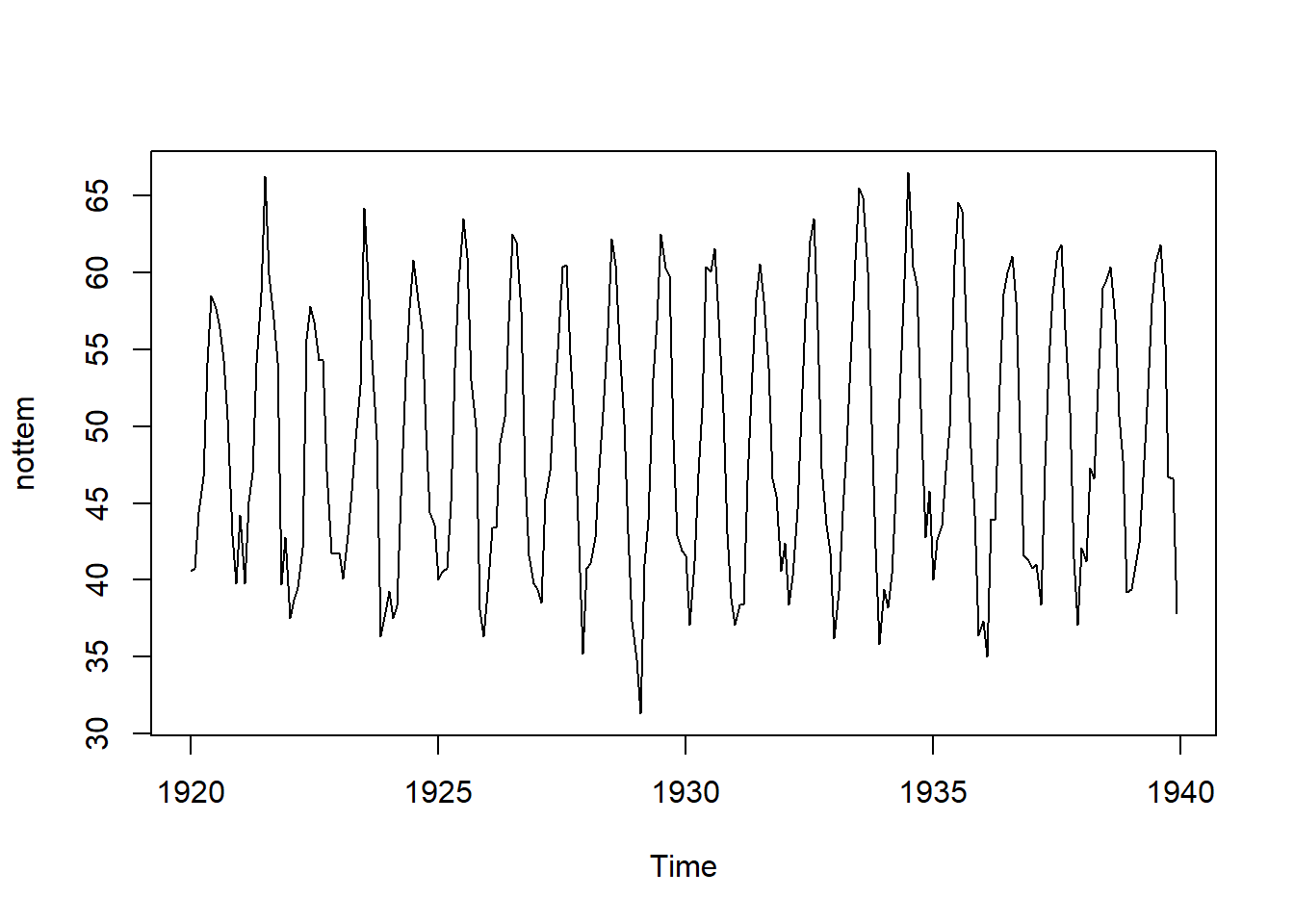
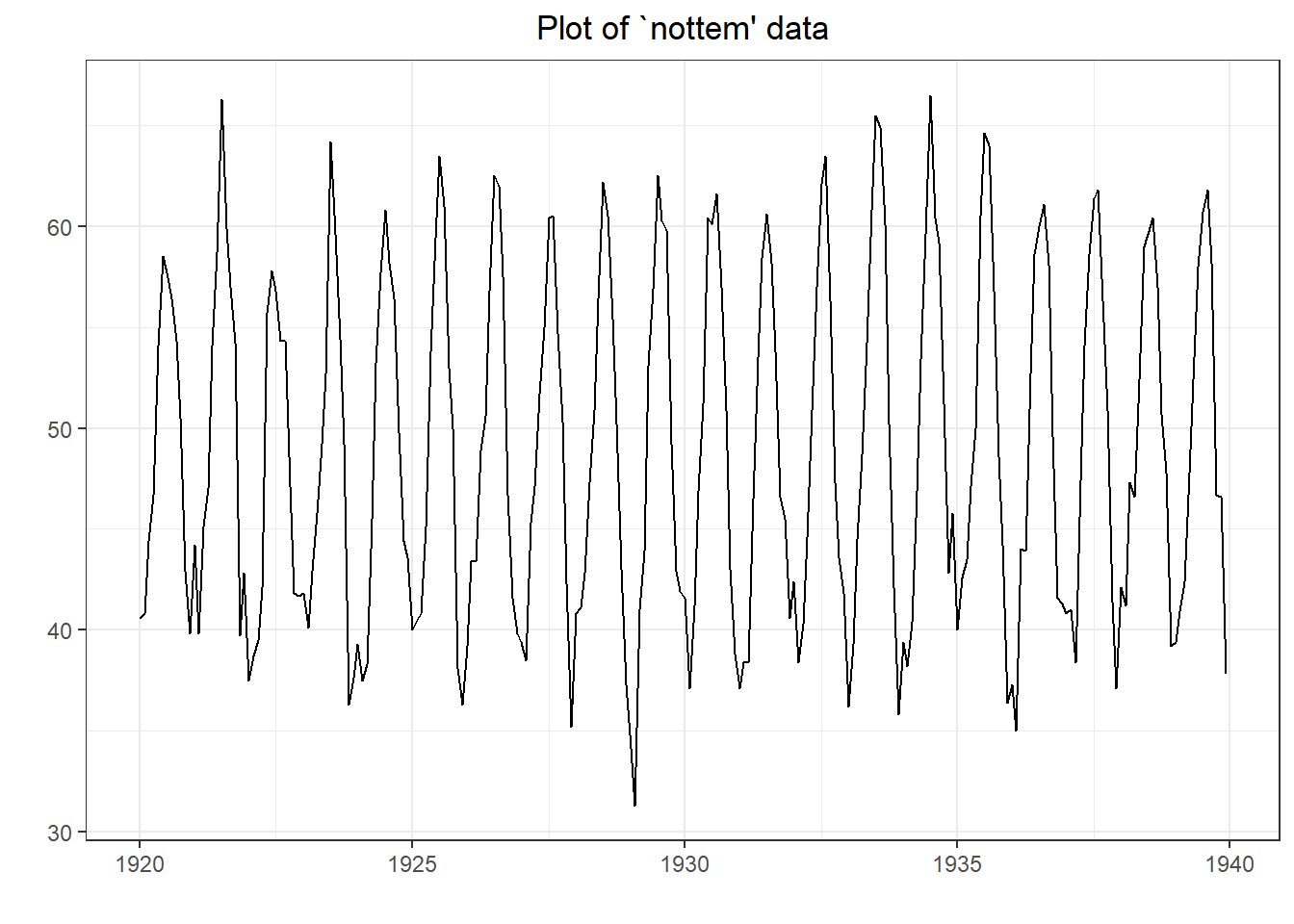
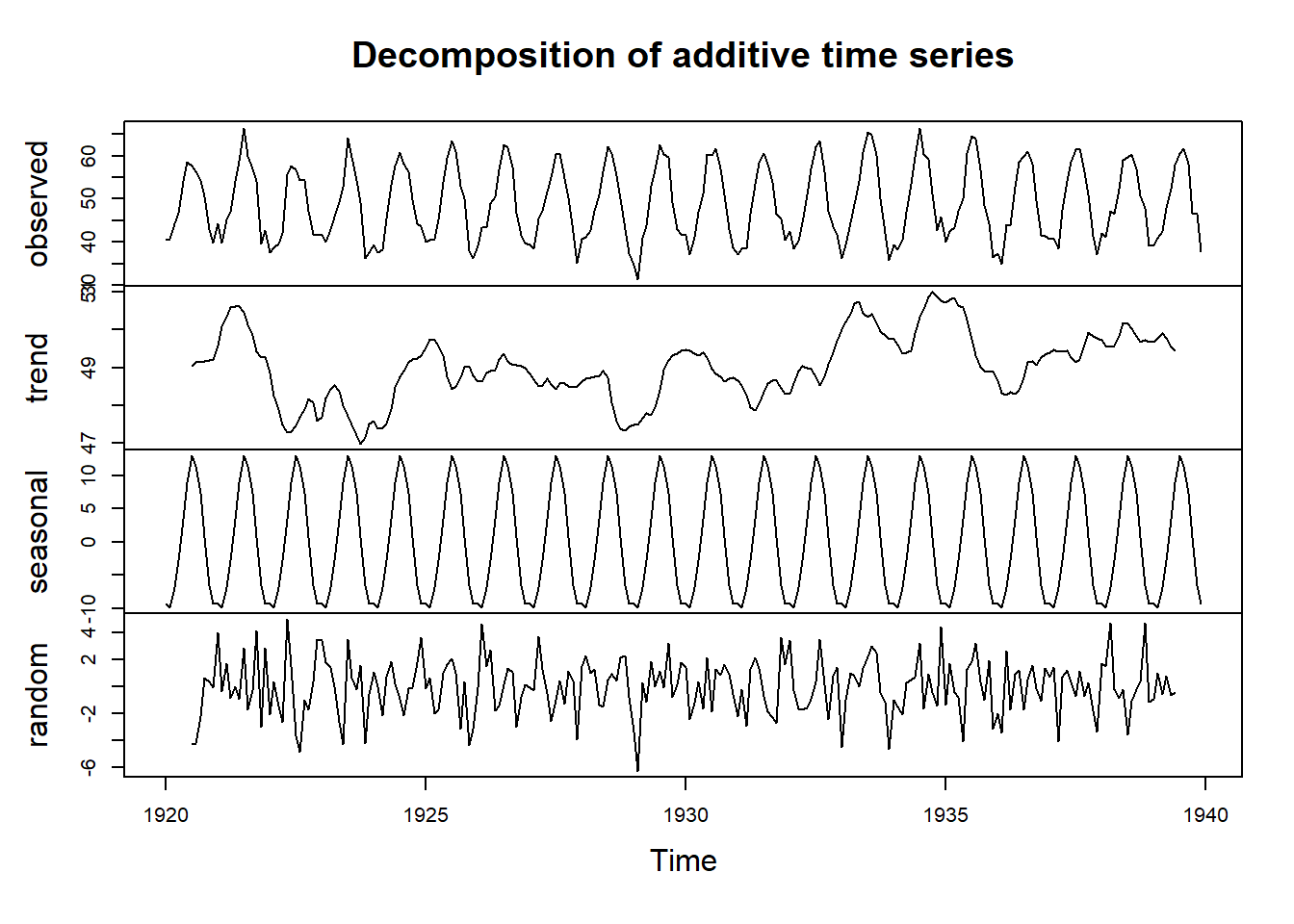
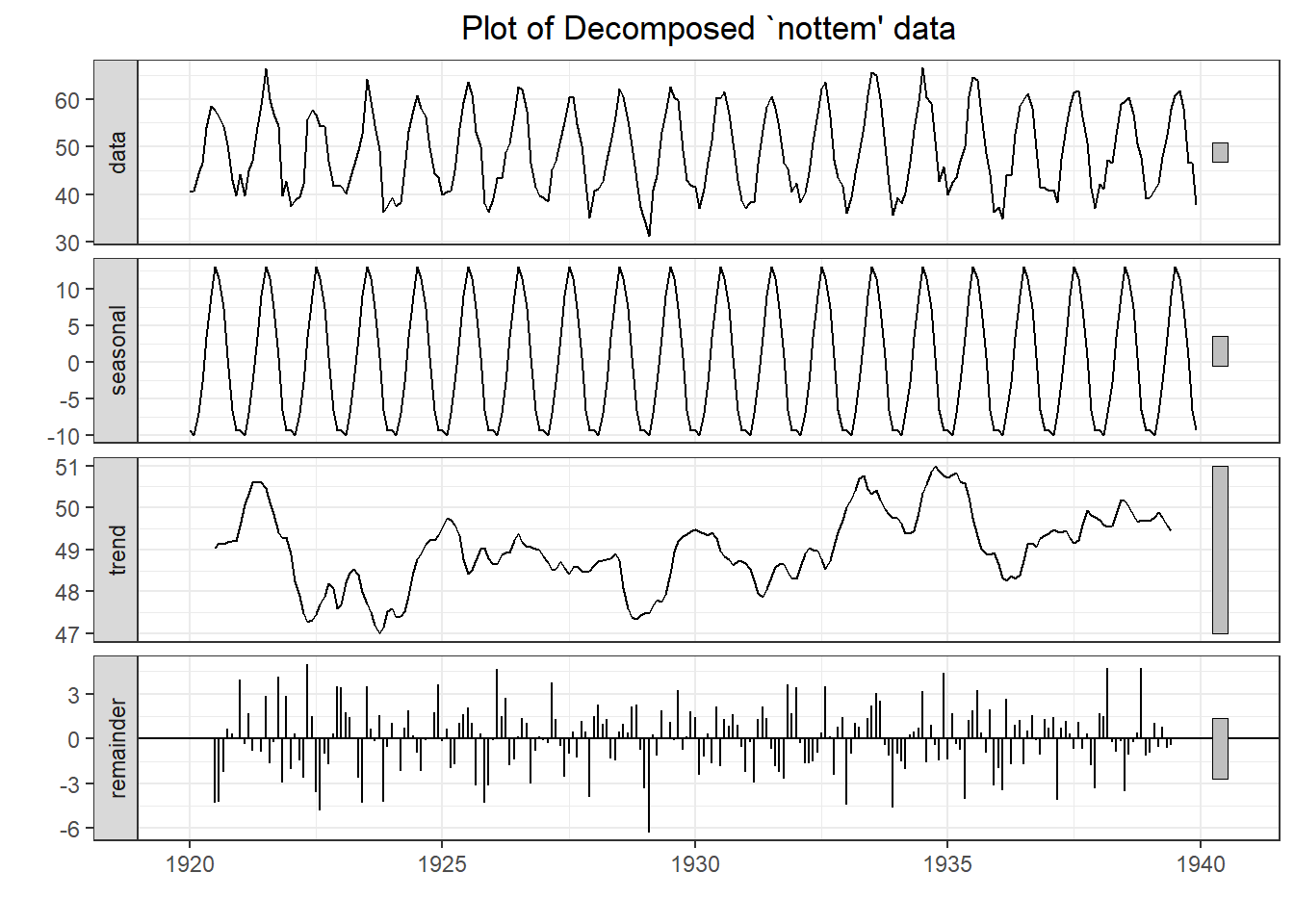
## Warning in adf.test(nottem): p-value smaller than printed p-value
##
## Augmented Dickey-Fuller Test
##
## data: nottem
## Dickey-Fuller = -12.998, Lag order = 6, p-value = 0.01
## alternative hypothesis: stationary
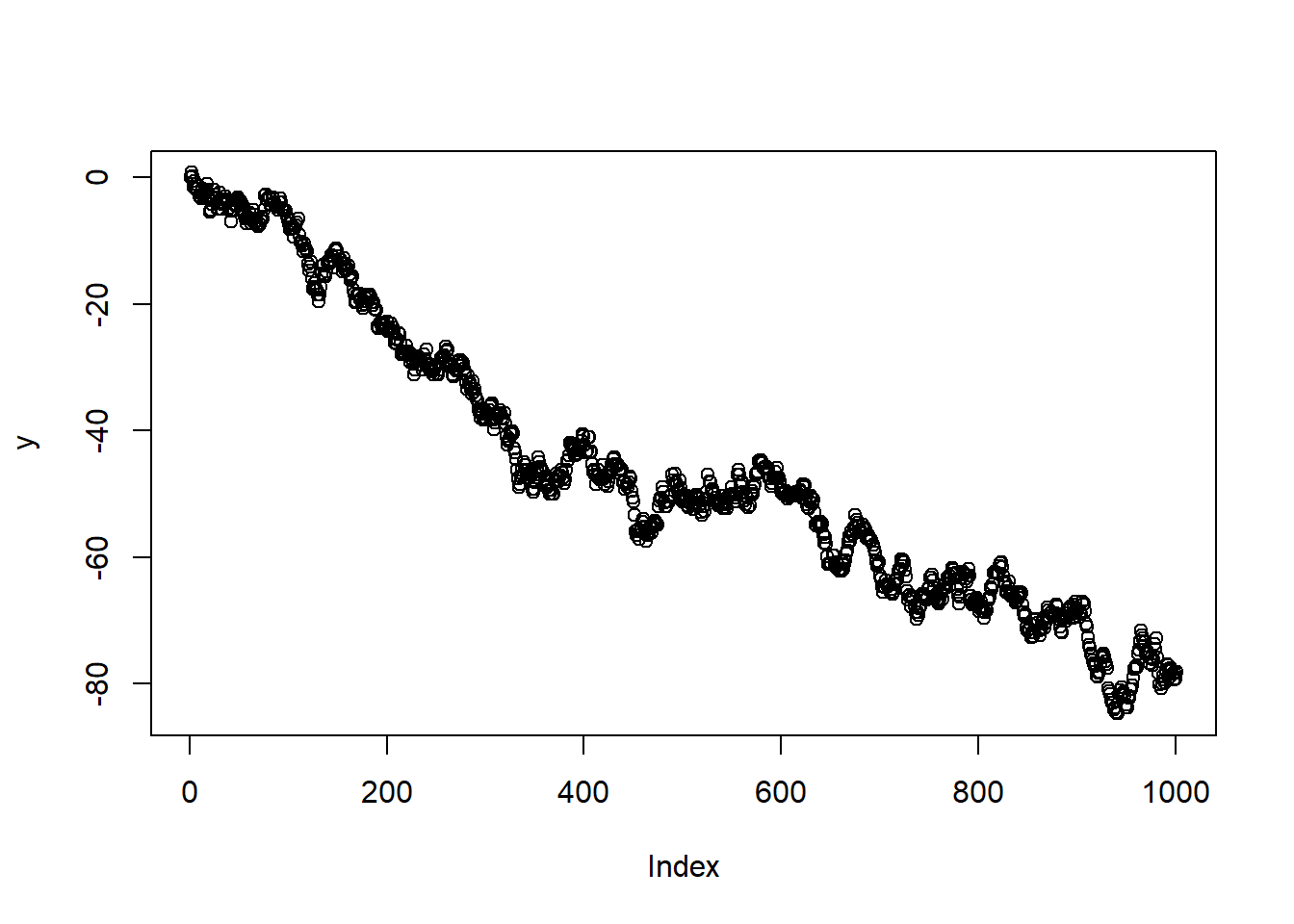
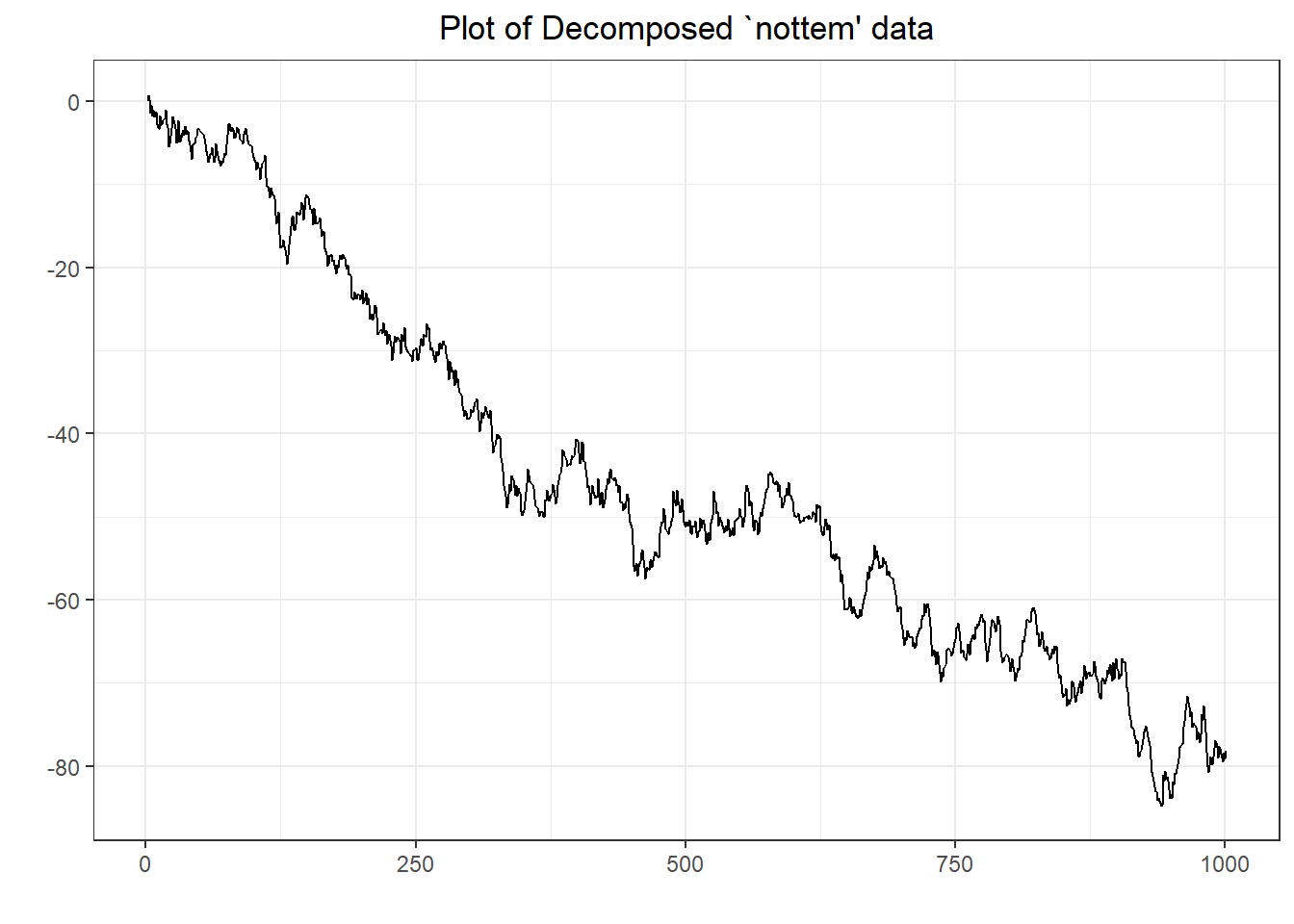
##
## Augmented Dickey-Fuller Test
##
## data: y
## Dickey-Fuller = -2.6432, Lag order = 9, p-value = 0.3061
## alternative hypothesis: stationary
## [1] 114
## Time Series:
## Start = 1821
## End = 1826
## Frequency = 1
## [1] 269 321 585 871 1475 2821
## [1] 321 585 871 1475 2821 3928
## [1] 269 321 585 871 1475 2821
##
## Durbin-Watson test
##
## data: lynx[-114] ~ lynx[-1]
## DW = 1.1296, p-value = 1.148e-06
## alternative hypothesis: true autocorrelation is greater than 0
##
## Durbin-Watson test
##
## data: x[-700] ~ x[-1]
## DW = 1.996, p-value = 0.4789
## alternative hypothesis: true autocorrelation is greater than 0
## [1] 240
##
## Durbin-Watson test
##
## data: nottem[-240] ~ nottem[-1]
## DW = 1.0093, p-value = 5.097e-15
## alternative hypothesis: true autocorrelation is greater than 0
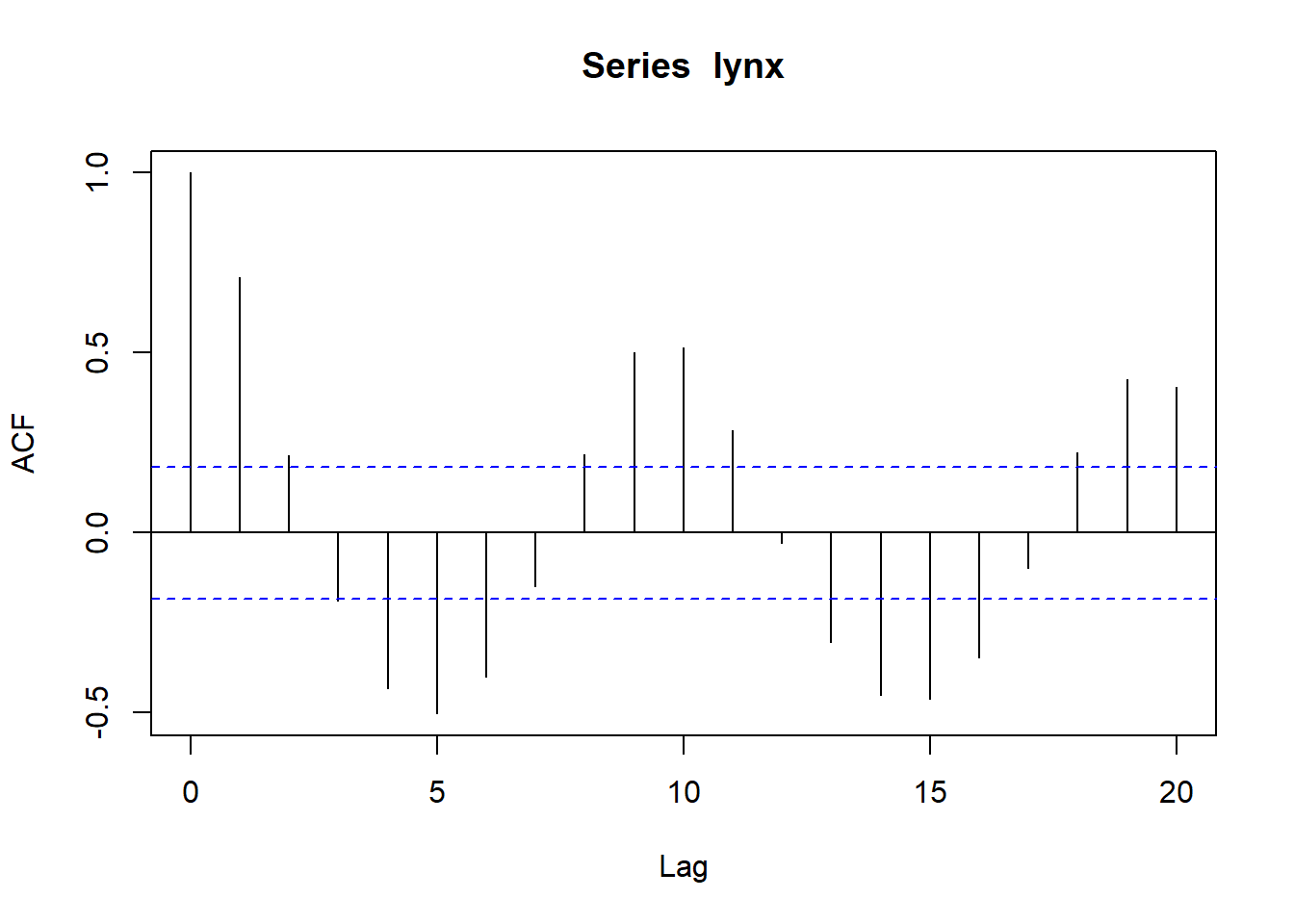
##
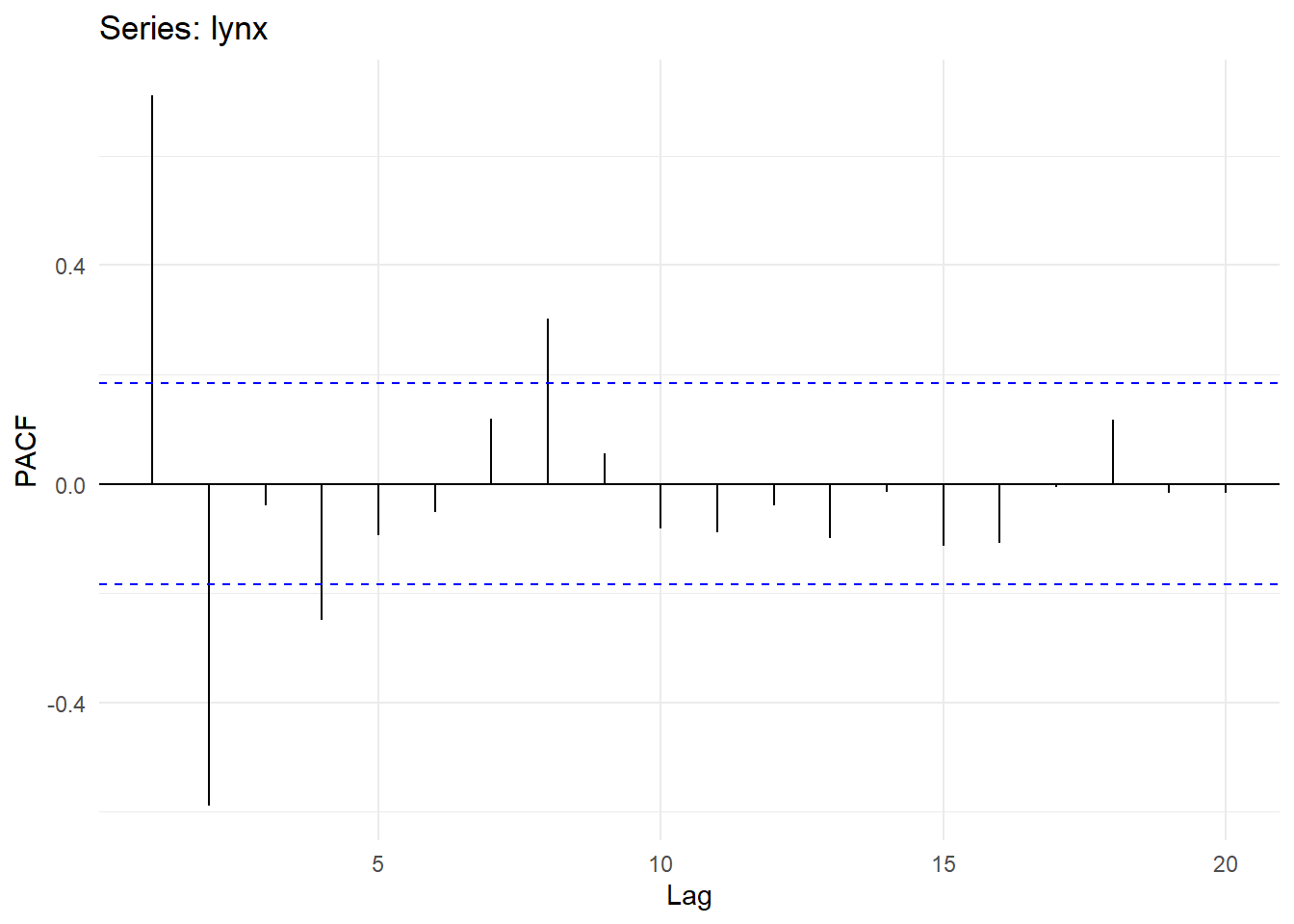
## Partial autocorrelations of series 'lynx', by lag
##
## 1 2 3 4 5 6 7 8 9 10
## 0.711 -0.588 -0.039 -0.250 -0.094 -0.052 0.119 0.301 0.055 -0.081
## 11 12 13 14 15 16 17 18 19 20
## -0.089 -0.040 -0.099 -0.014 -0.113 -0.108 -0.006 0.116 -0.016 -0.018
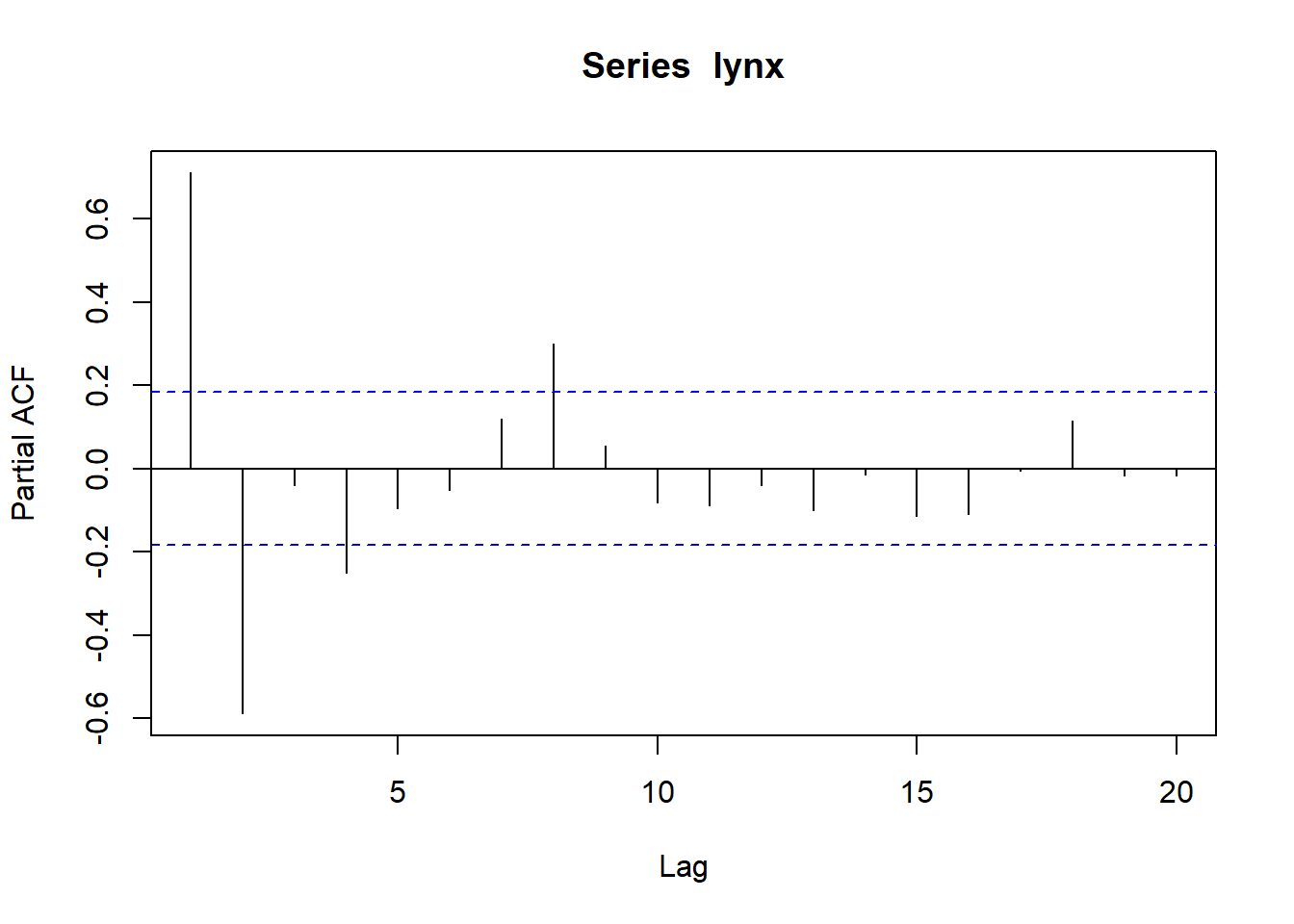
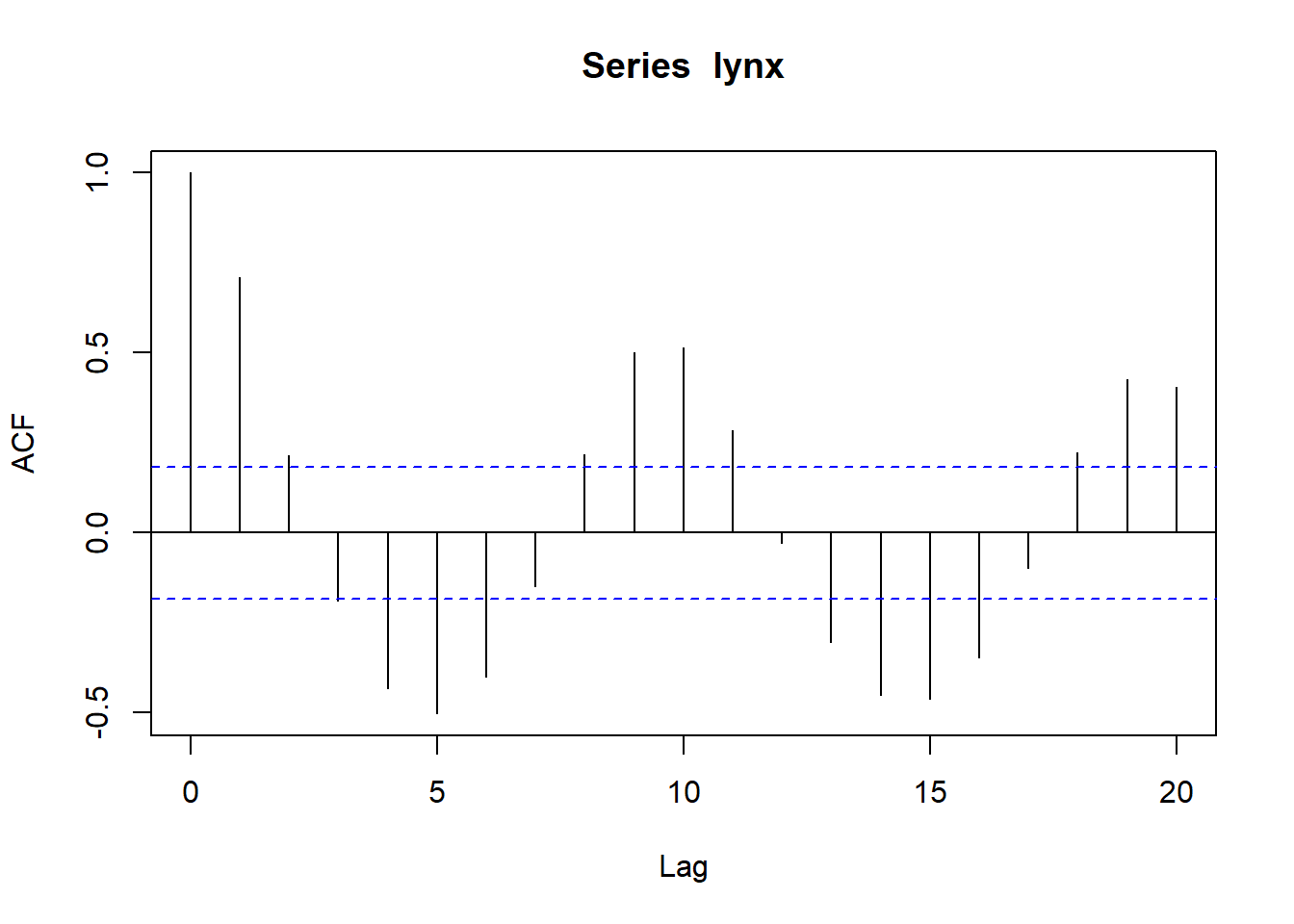
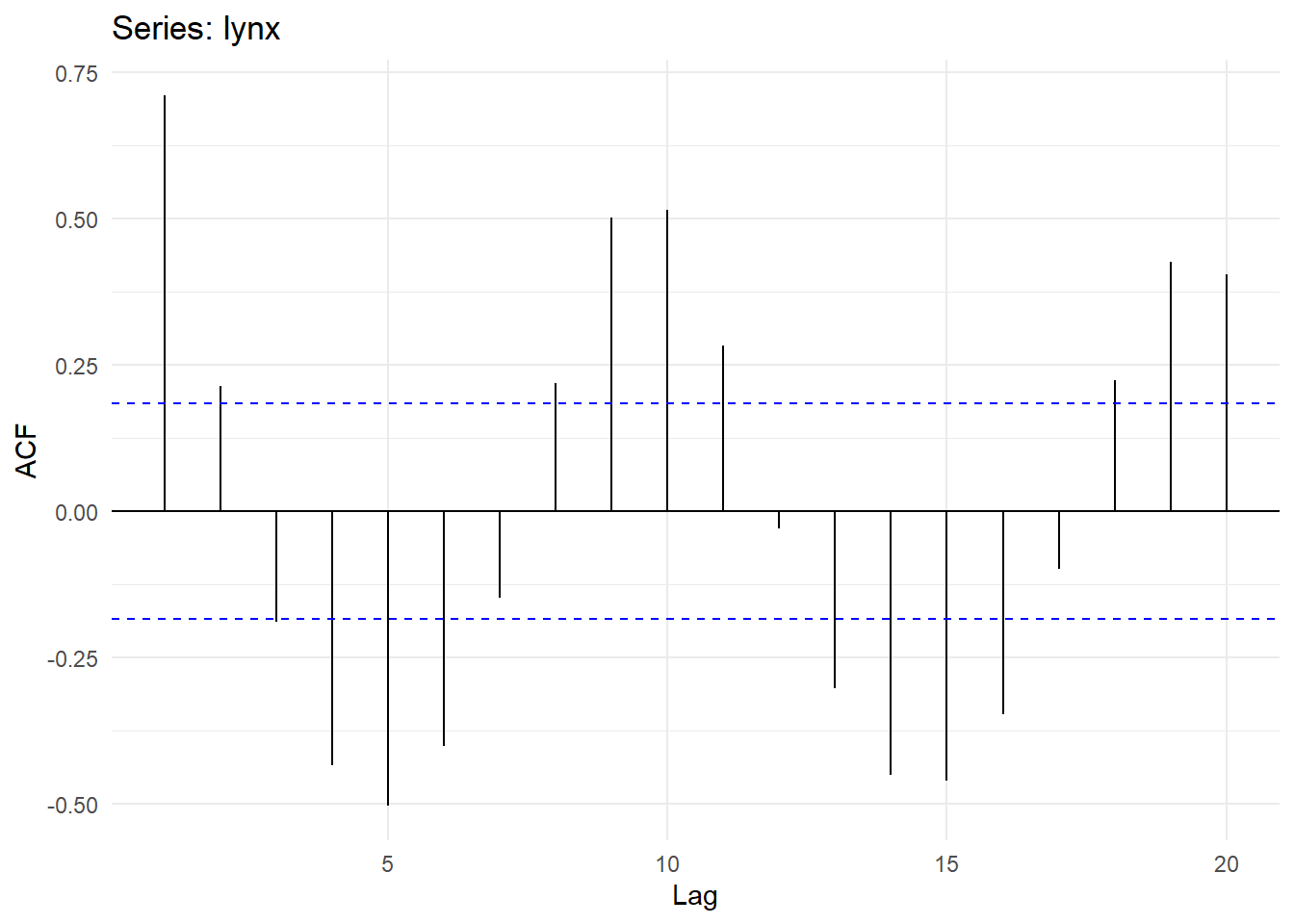
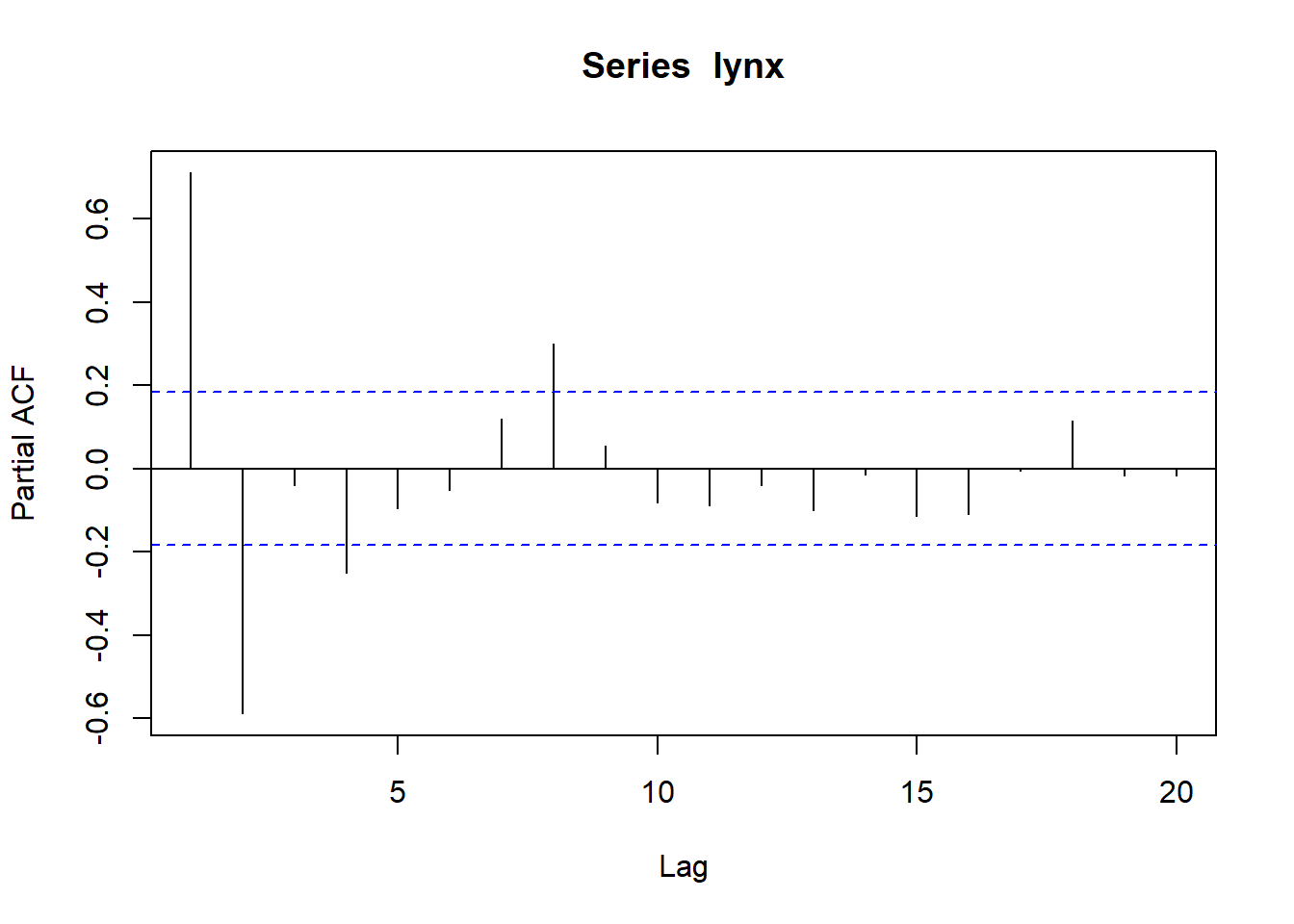
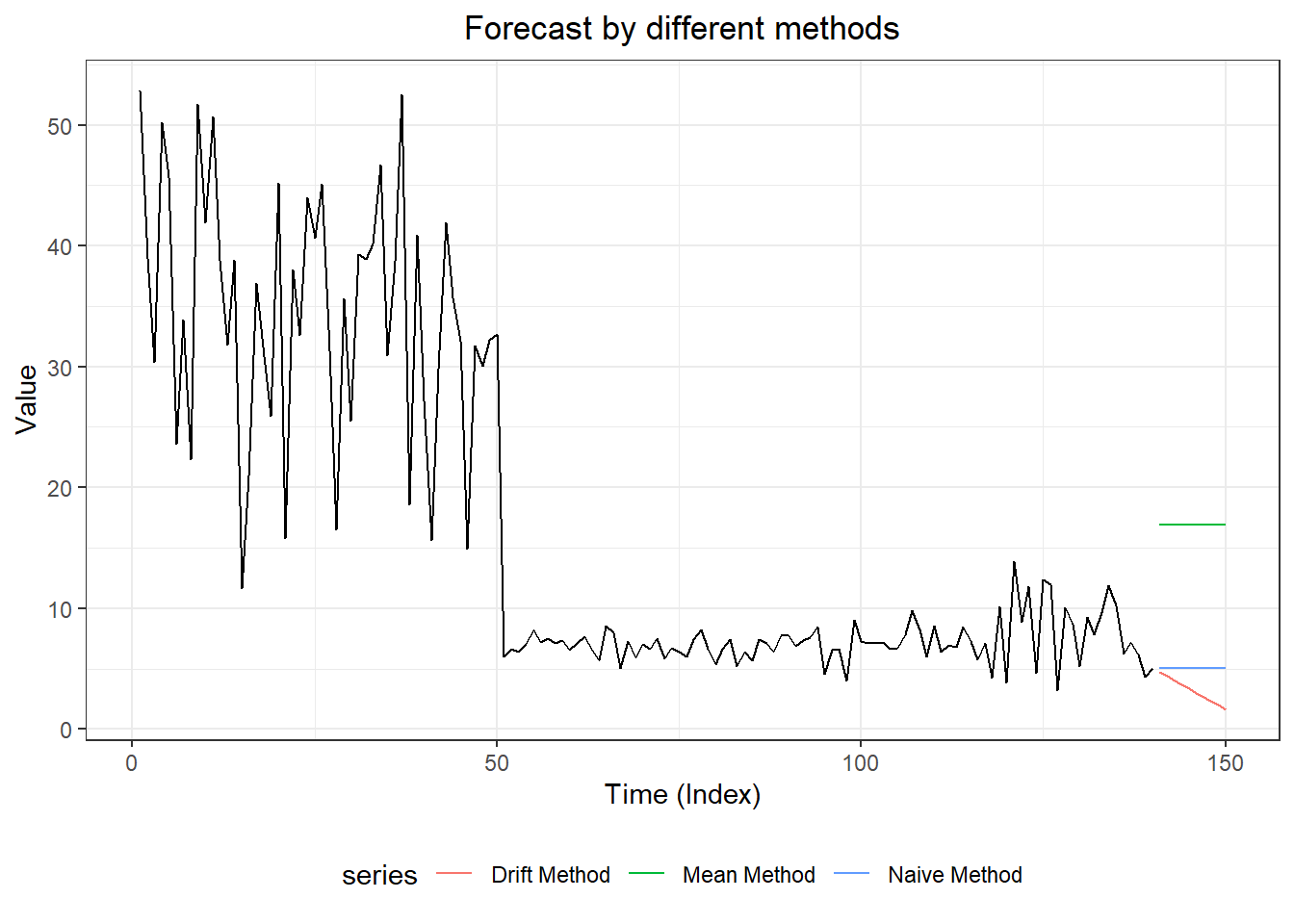
#2. three forecasting models
meanm <- meanf(myts, h = 10)
naivem <- naive(myts, h= 10)
driftm <- rwf(myts, h =10, drift = TRUE)
#3. plot forecasts from three forecasting models
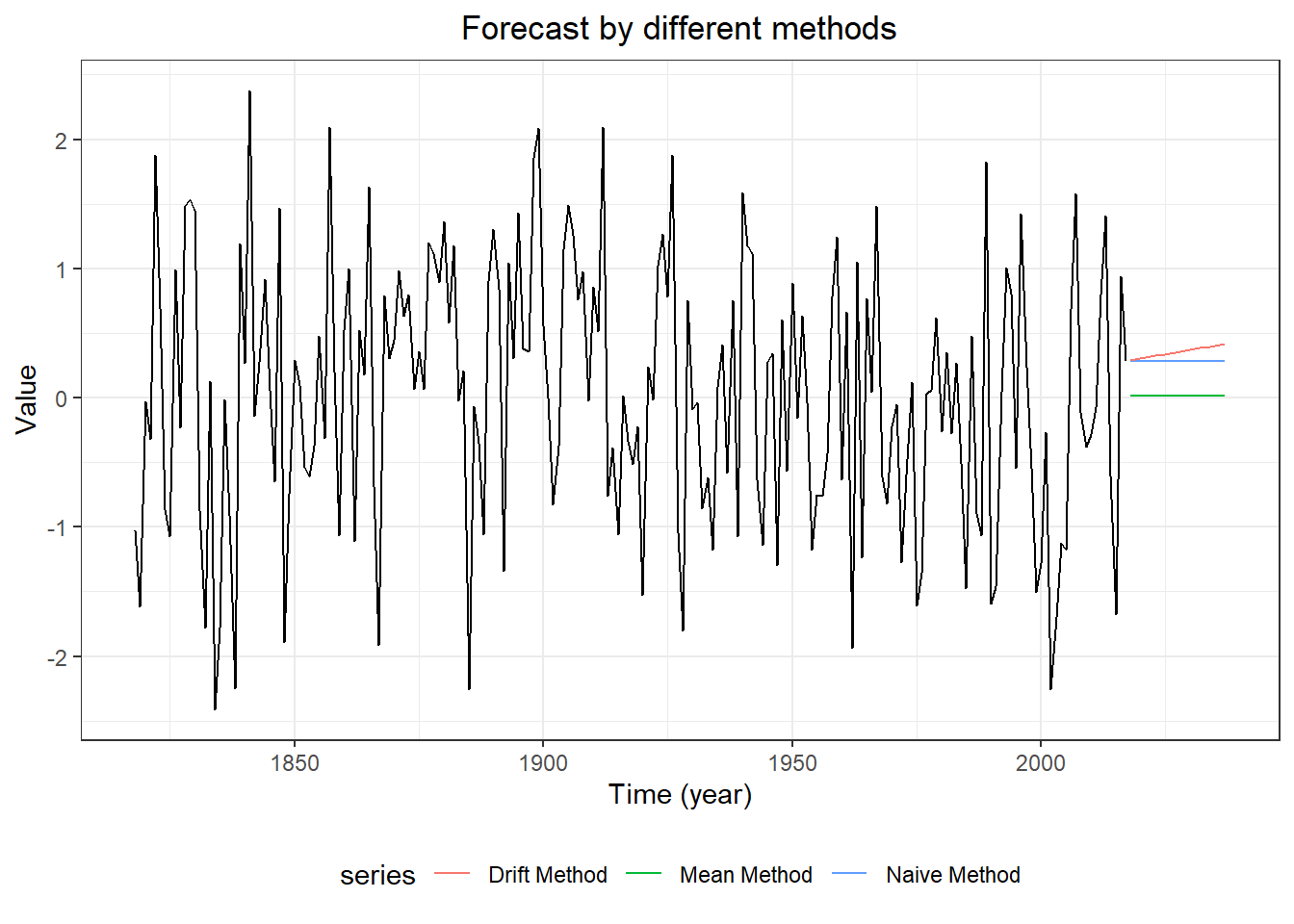
forecast::autoplot(myts,
PI = TRUE,
flwd = 2) +
autolayer(meanm$mean, series = "Mean Method", PI = TRUE) +
autolayer(naivem$mean, series = "Naive Method", PI = TRUE) +
autolayer(driftm$mean, series = "Drift Method", PI = TRUE) +
ggtitle("Forecast by different methods") +
xlab("Time (Index)") + ylab("Value") +
theme_bw() +
theme(plot.title = element_text(hjust = 0.5),
legend.position = "bottom")
## Warning: Ignoring unknown parameters: PI, flwd
## Warning: Ignoring unknown parameters: PI
## Warning: Ignoring unknown parameters: PI
## Warning: Ignoring unknown parameters: PI
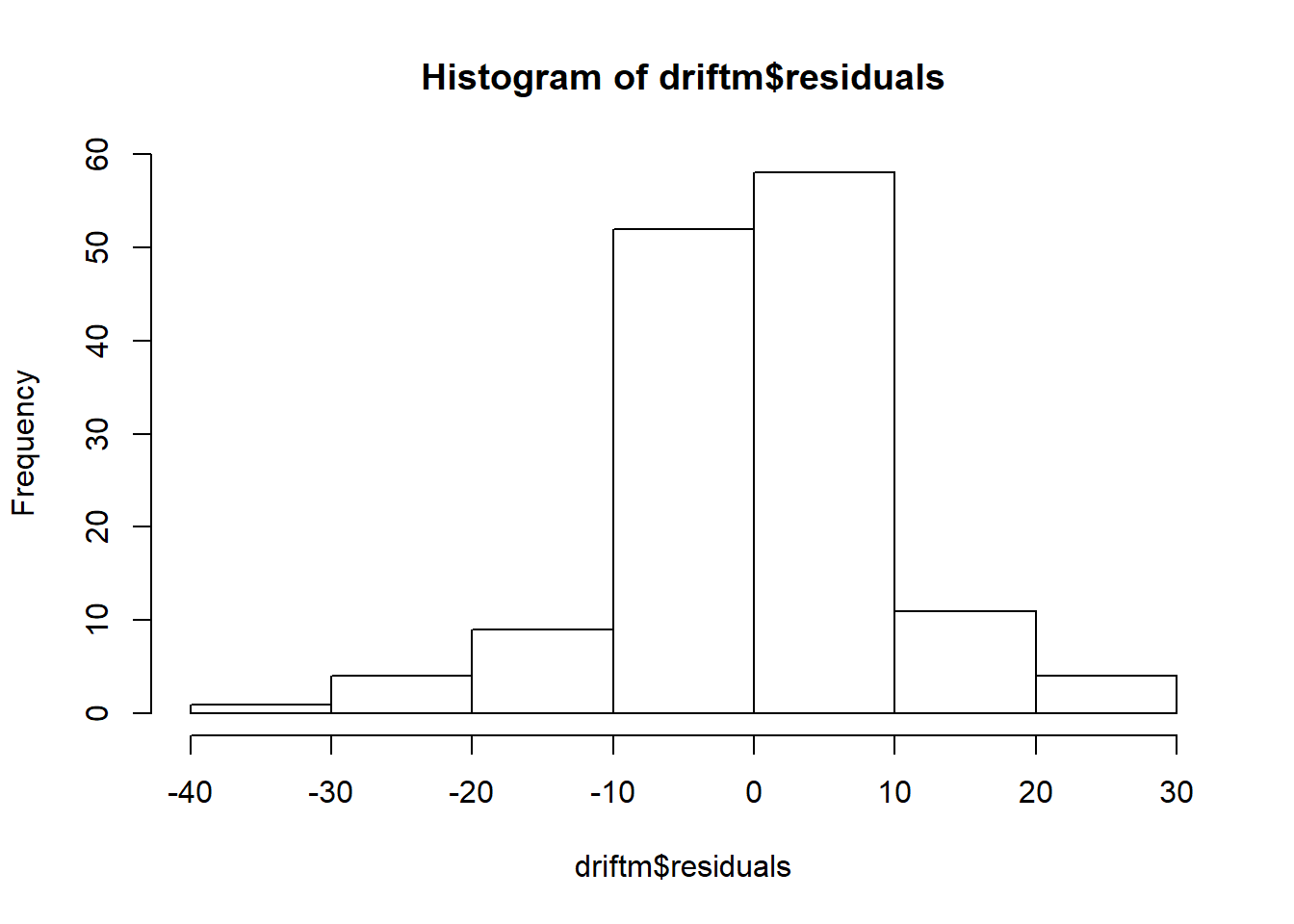
## [1] 211.0364
## [1] -1.591522e-15
## [1] NA
## [1] 84.53723
## [1] -0.3435748
## [1] 84.53723
## [1] 2.648343e-15
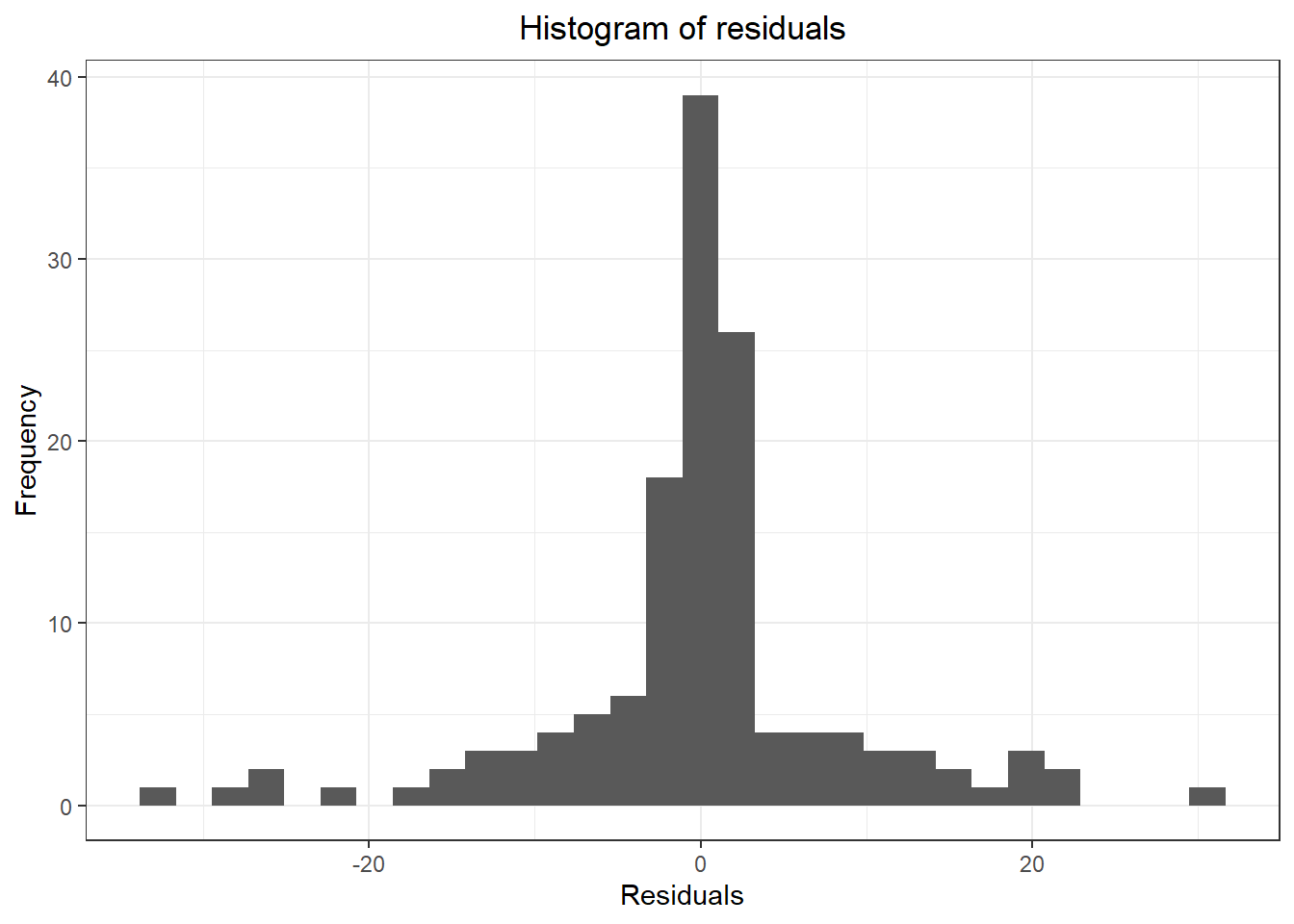
## Don't know how to automatically pick scale for object of type ts. Defaulting to continuous.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## Warning: Removed 1 rows containing non-finite values (stat_bin).
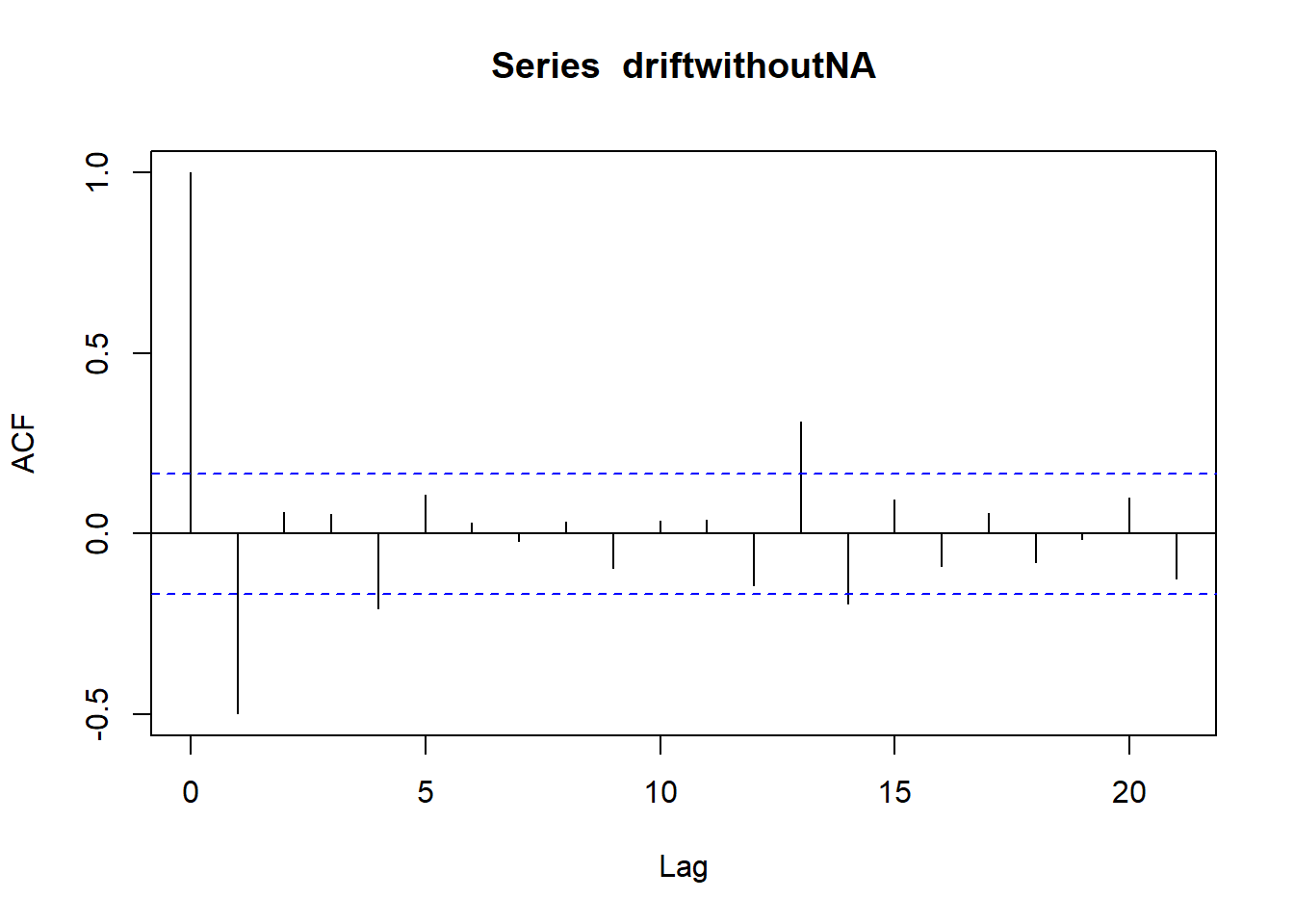
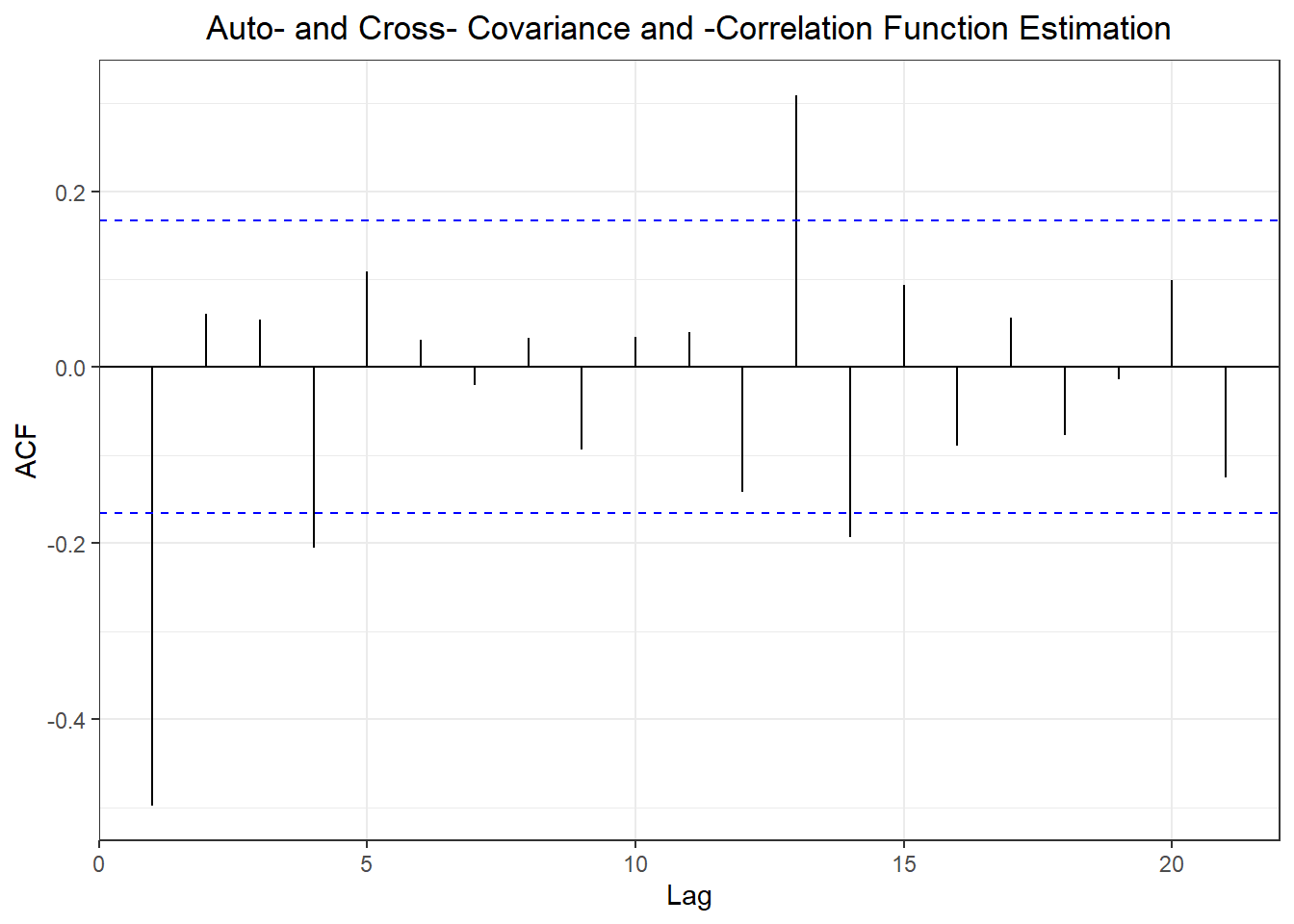
## ME RMSE MAE MPE MAPE MASE
## Training set -6.408719e-16 15.31467 13.94736 -84.86989 120.4406 2.231924
## Test set -1.125187e+01 11.61073 11.25187 -180.27778 180.2778 1.800578
## ACF1 Theil's U
## Training set 0.7632991 NA
## Test set -0.1703445 2.002248
## ME RMSE MAE MPE MAPE MASE
## Training set -0.4131666 10.024449 6.249032 -11.909758 33.36297 1.0000000
## Test set 0.9663084 3.022963 2.482045 -1.869095 33.51426 0.3971888
## ACF1 Theil's U
## Training set -0.4901263 NA
## Test set -0.1703445 0.6497522
## ME RMSE MAE MPE MAPE MASE
## Training set -1.624594e-17 10.015931 6.265918 -7.901602 33.29565 1.002702
## Test set 6.957224e+00 8.172915 6.974530 86.321252 86.72796 1.116098
## ACF1 Theil's U
## Training set -0.4901263 NA
## Test set 0.4327471 1.62159
##
## Shapiro-Wilk normality test
##
## data: naivem$residuals
## W = 0.89587, p-value = 2.061e-08
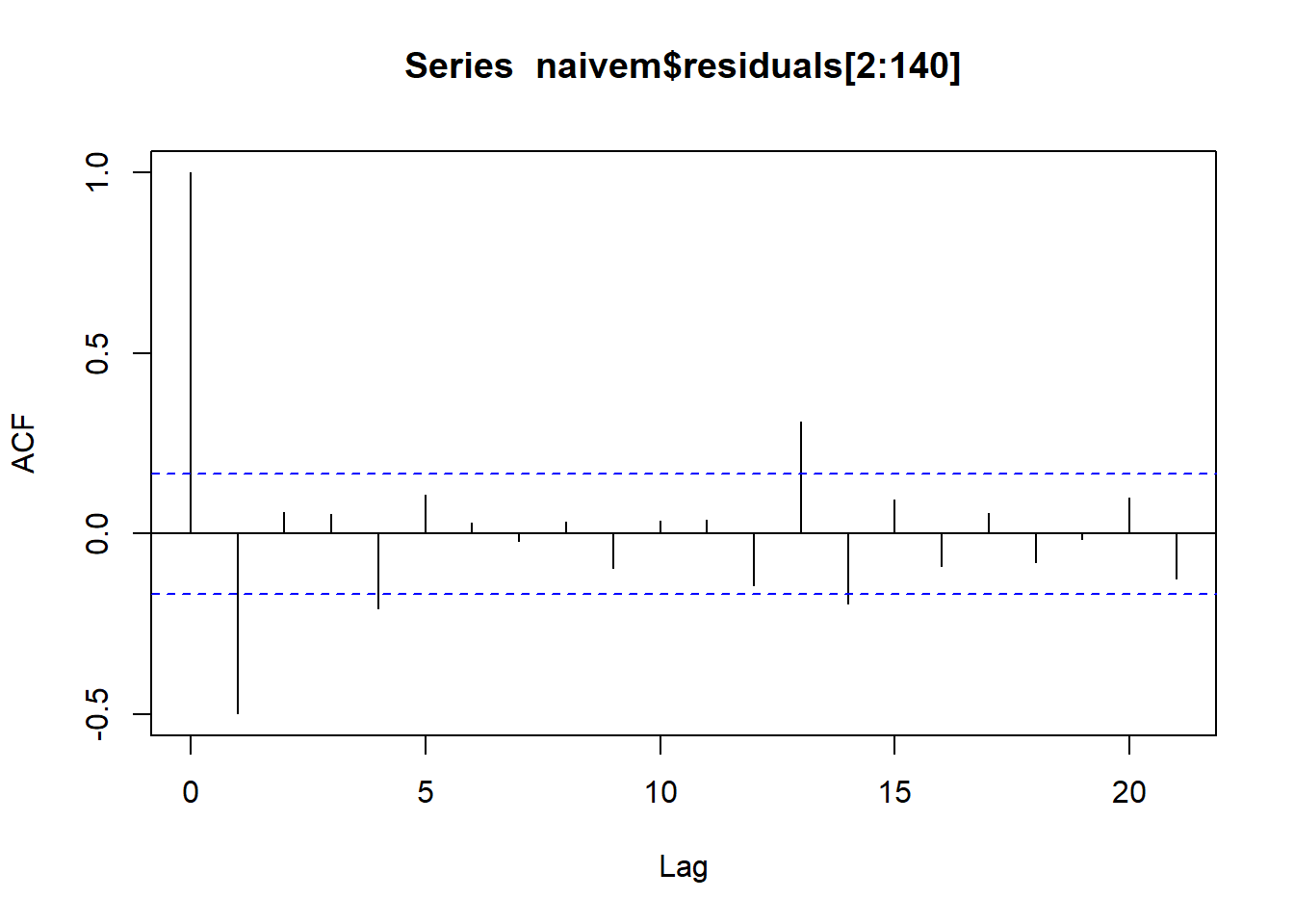
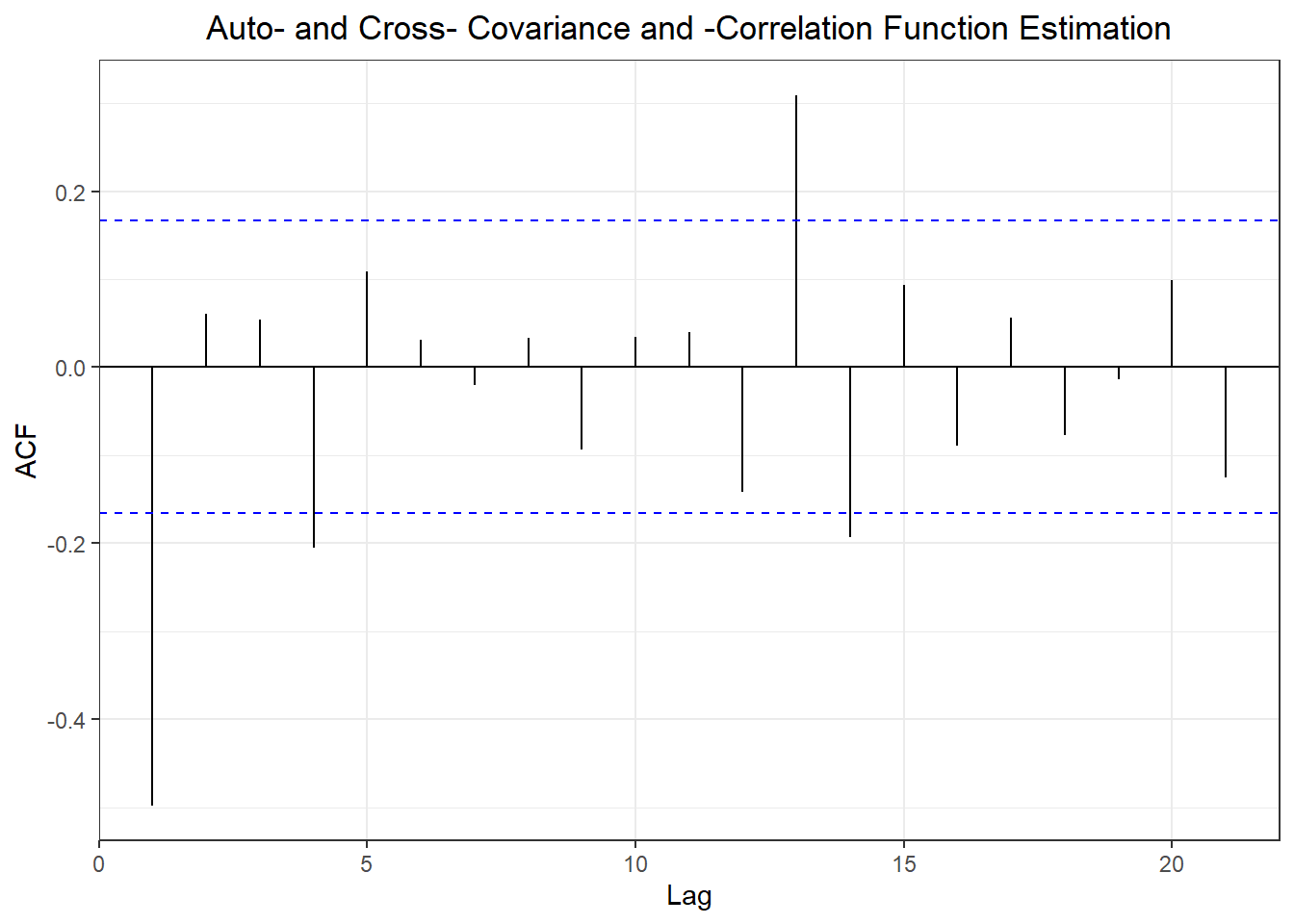
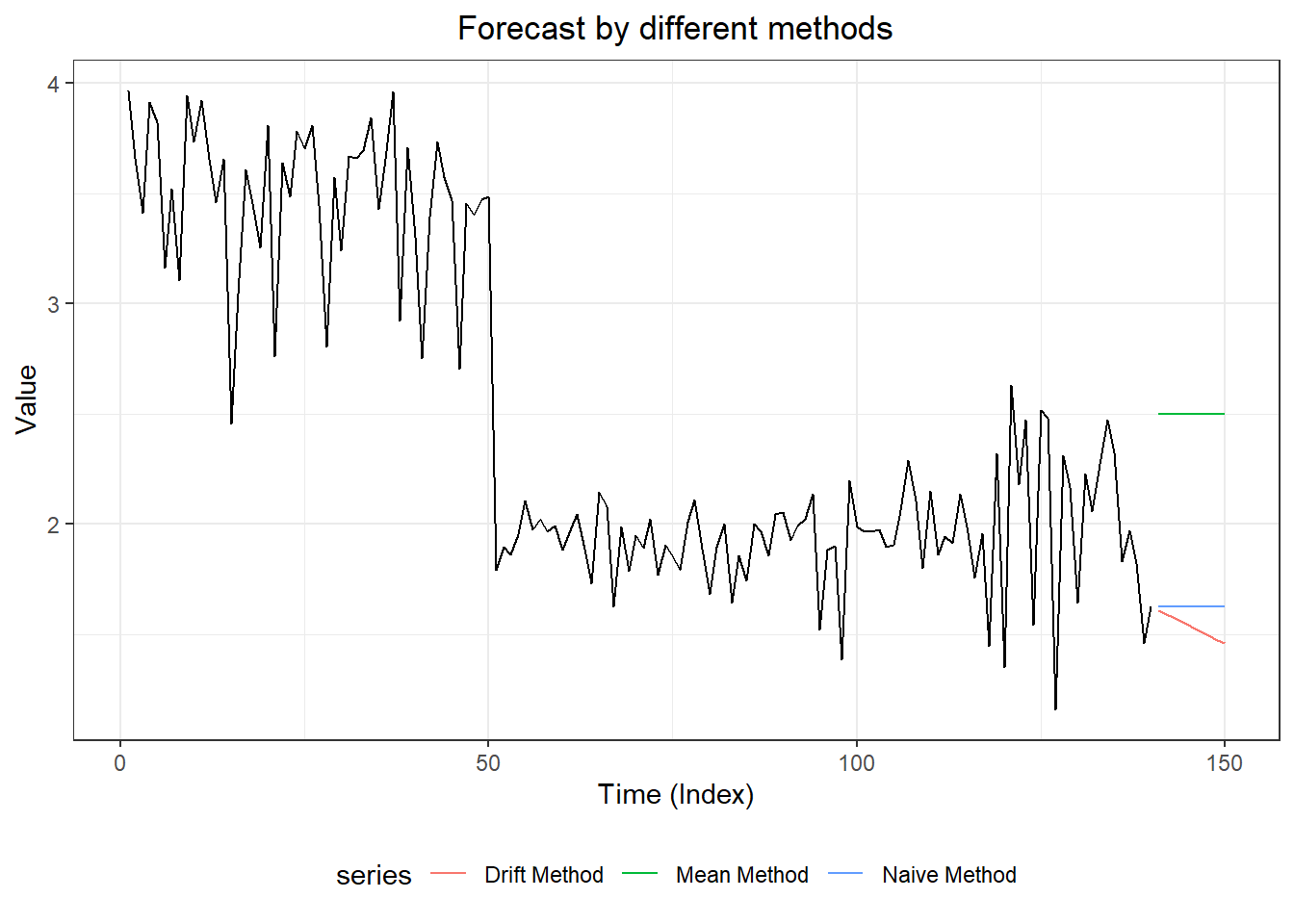
#2. three forecasting models
meanm <- meanf(myts, h = 10)
naivem <- naive(myts, h= 10)
driftm <- rwf(myts, h =10, drift = TRUE)
#3. plot forecasts from three forecasting models
forecast::autoplot(myts,
PI = TRUE,
flwd = 2) +
autolayer(meanm$mean, series = "Mean Method", PI = TRUE) +
autolayer(naivem$mean, series = "Naive Method", PI = TRUE) +
autolayer(driftm$mean, series = "Drift Method", PI = TRUE) +
ggtitle("Forecast by different methods") +
xlab("Time (Index)") + ylab("Value") +
theme_bw() +
theme(plot.title = element_text(hjust = 0.5),
legend.position = "bottom")
## Warning: Ignoring unknown parameters: PI, flwd
## Warning: Ignoring unknown parameters: PI
## Warning: Ignoring unknown parameters: PI
## Warning: Ignoring unknown parameters: PI
## [1] 0.6290444
## [1] 1.510731e-16
## [1] NA
## [1] 0.2067372
## [1] -0.01684688
## [1] 0.2067372
## [1] -4.472841e-17
## Don't know how to automatically pick scale for object of type ts. Defaulting to continuous.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## Warning: Removed 1 rows containing non-finite values (stat_bin).
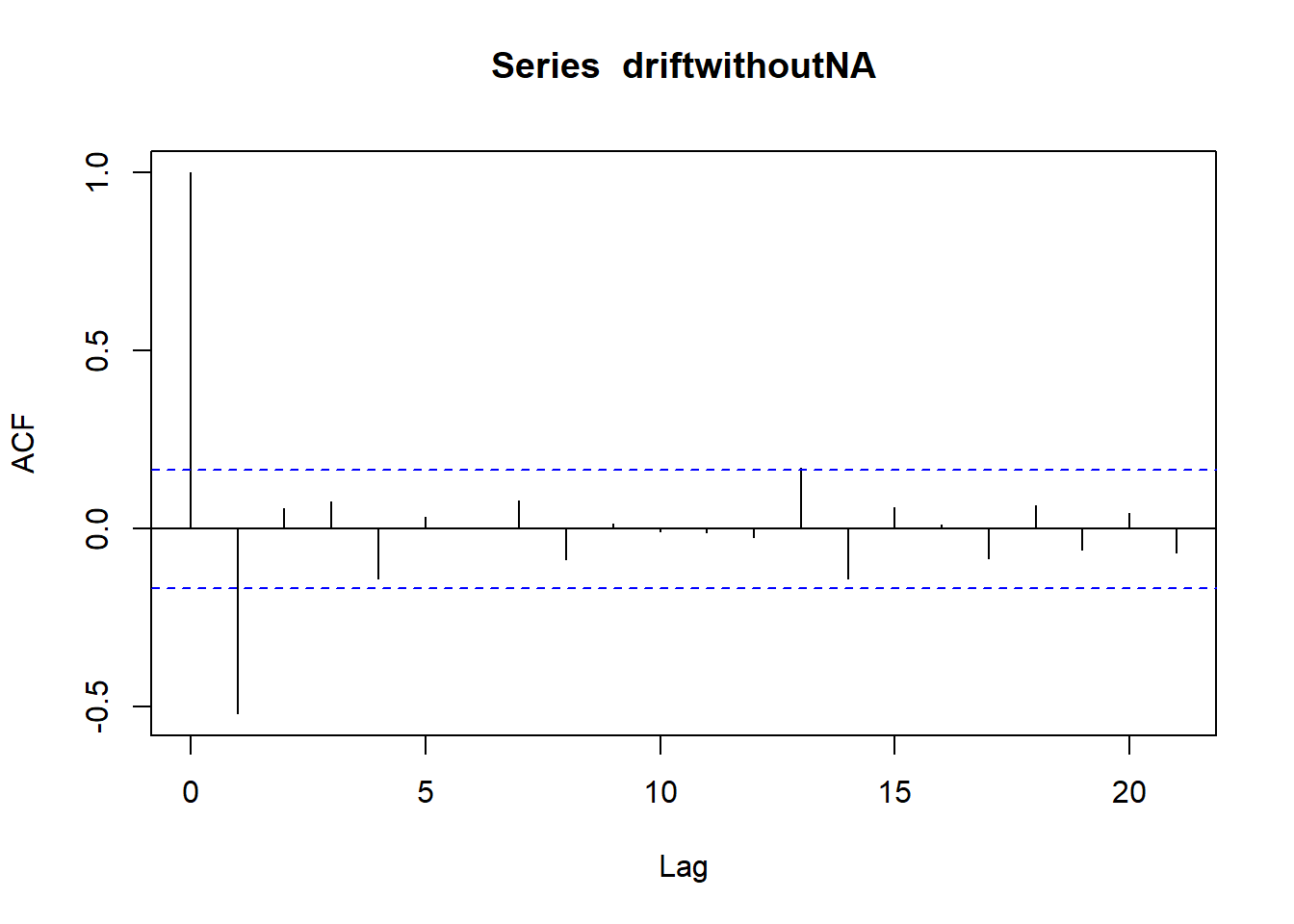
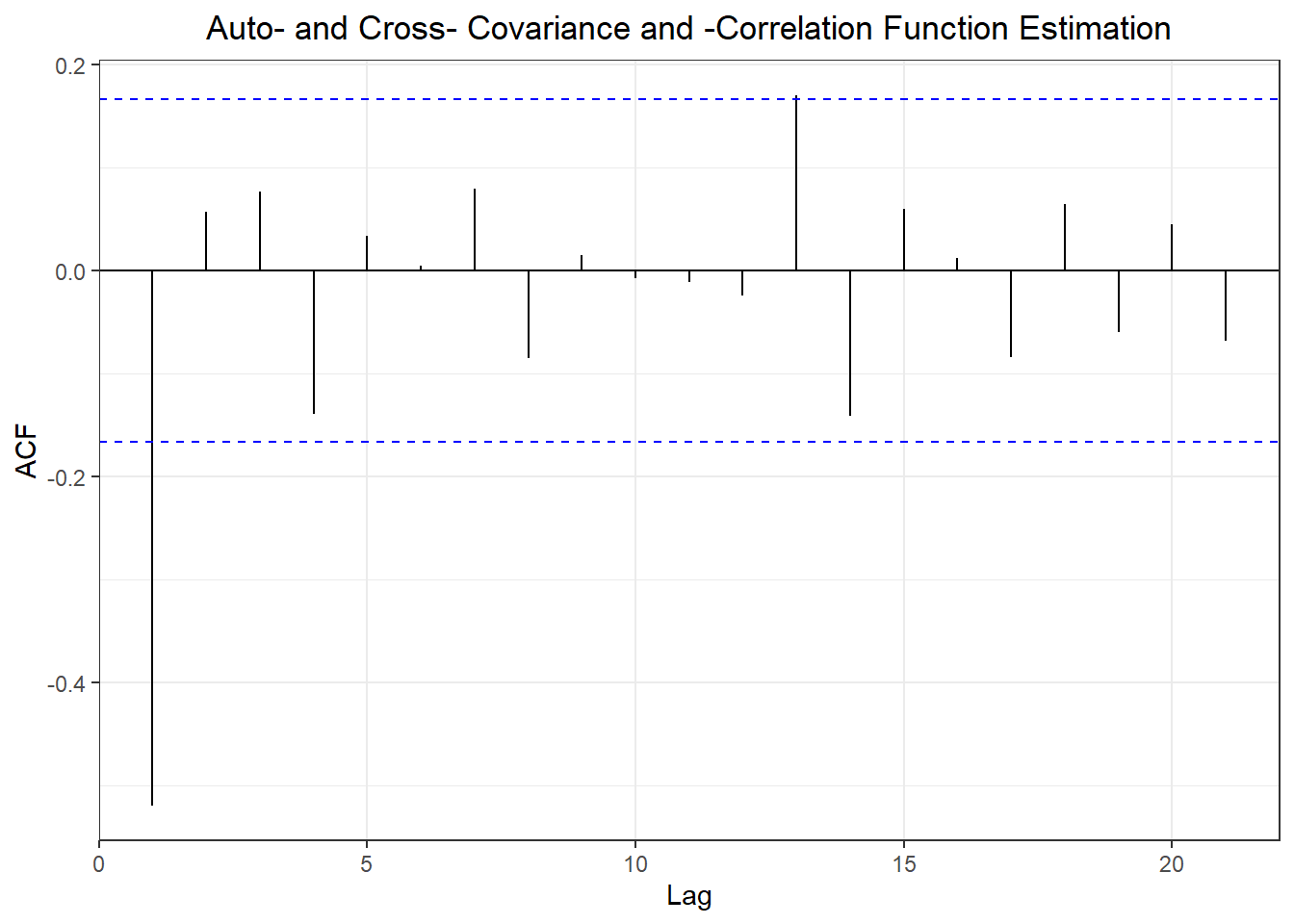
## ME RMSE MAE MPE MAPE
## Training set 1.846038e-16 0.8163346 0.7714004 -9.839234 31.23484
## Test set -6.198835e-01 0.7307518 0.6204553 -36.737960 36.75971
## MASE ACF1 Theil's U
## Training set 2.684607 0.8615304 NA
## Test set 2.159291 -0.2306541 0.9716032
## ME RMSE MAE MPE MAPE MASE
## Training set -0.01824047 0.4071185 0.2873421 -1.870232 11.33626 1.000000
## Test set 0.05893104 0.3914276 0.3316489 -1.328849 17.76316 1.154196
## ACF1 Theil's U
## Training set -0.4450652 NA
## Test set -0.2306541 0.577168
## ME RMSE MAE MPE MAPE MASE
## Training set -9.802537e-17 0.4067096 0.2876207 -1.103181 11.31901 1.000970
## Test set 3.234179e-01 0.5206086 0.4411263 12.524858 21.27240 1.535196
## ACF1 Theil's U
## Training set -0.4450652 NA
## Test set -0.1049056 0.7824451
##
## Shapiro-Wilk normality test
##
## data: naivem$residuals
## W = 0.961, p-value = 0.0005413
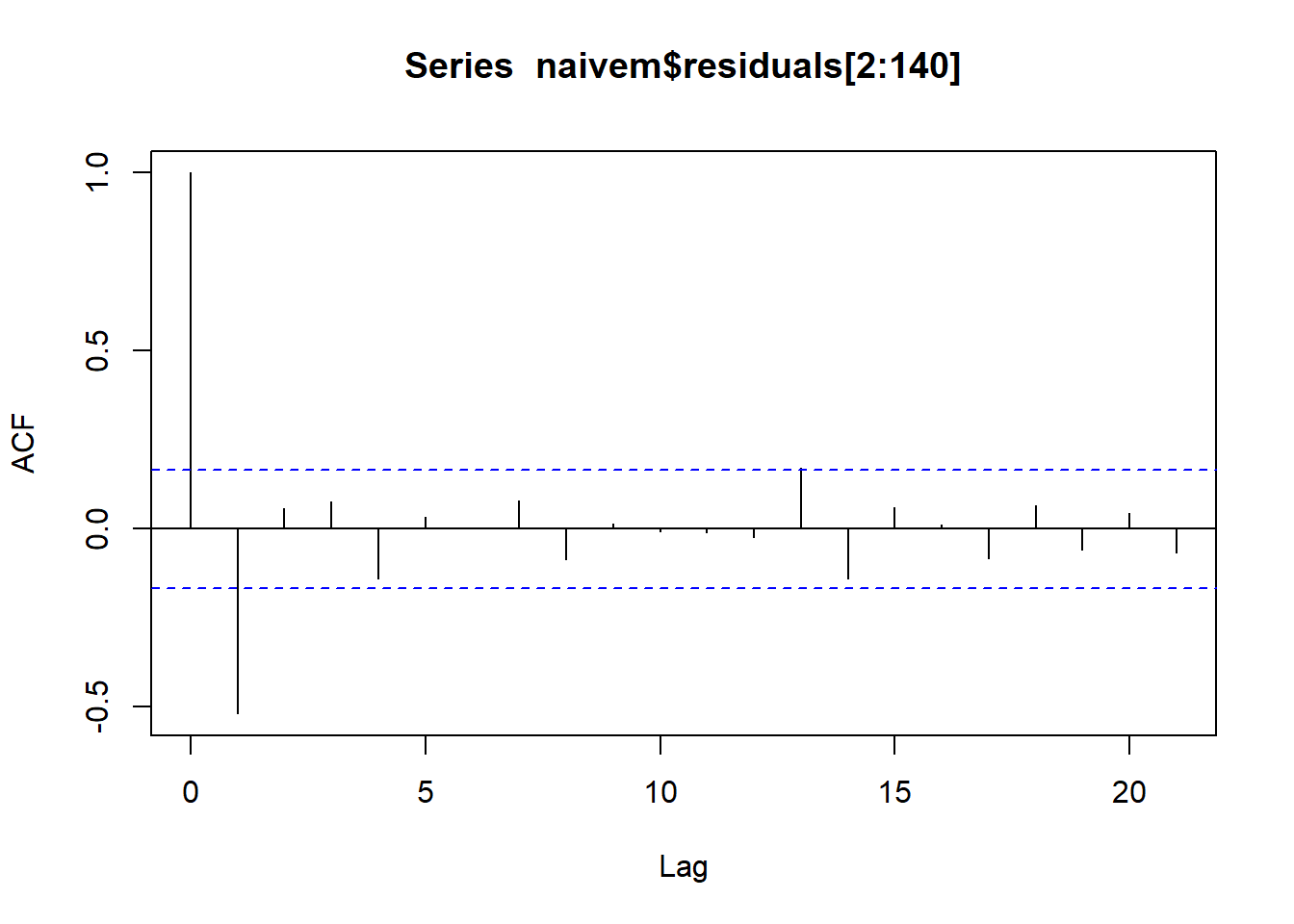
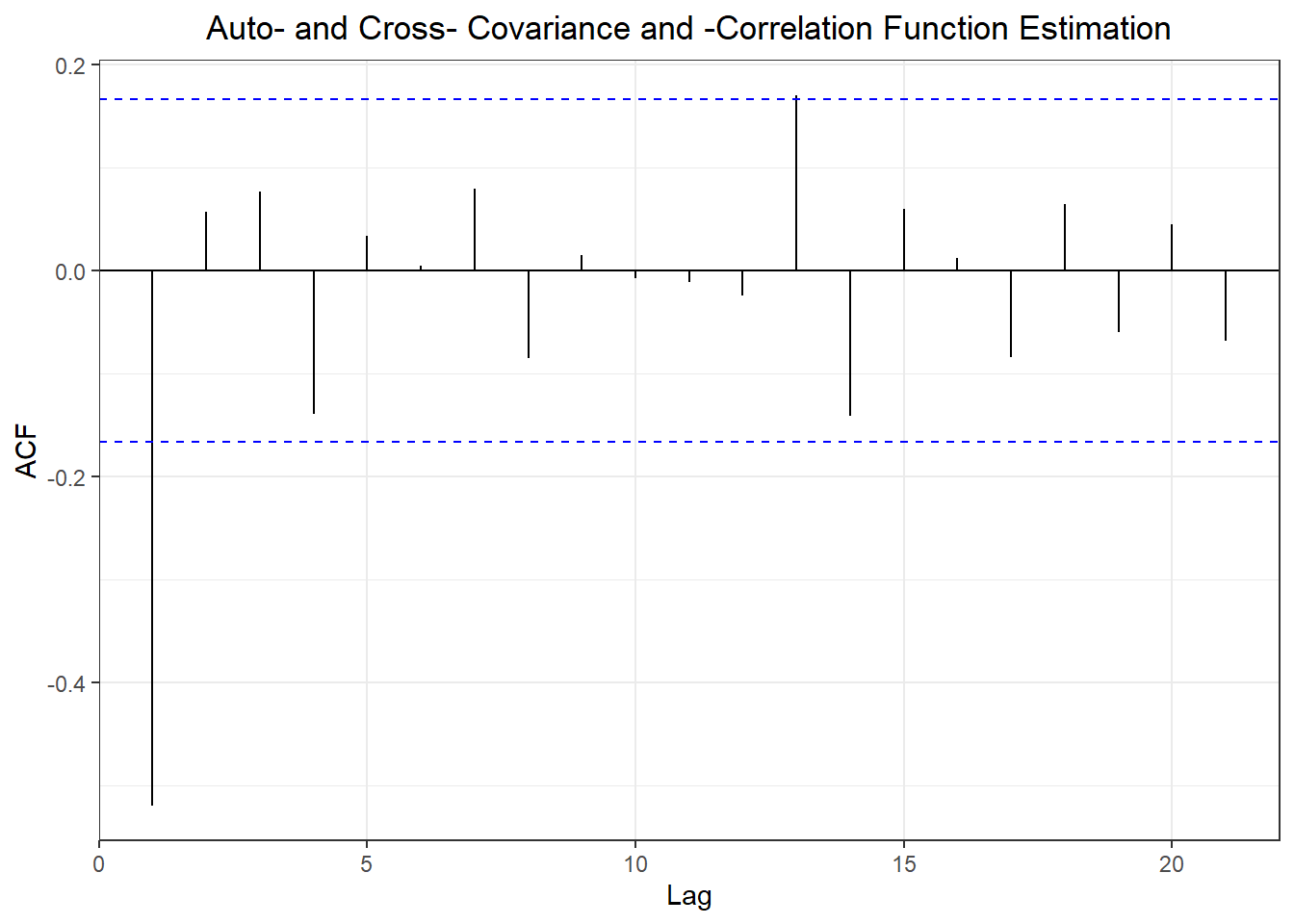
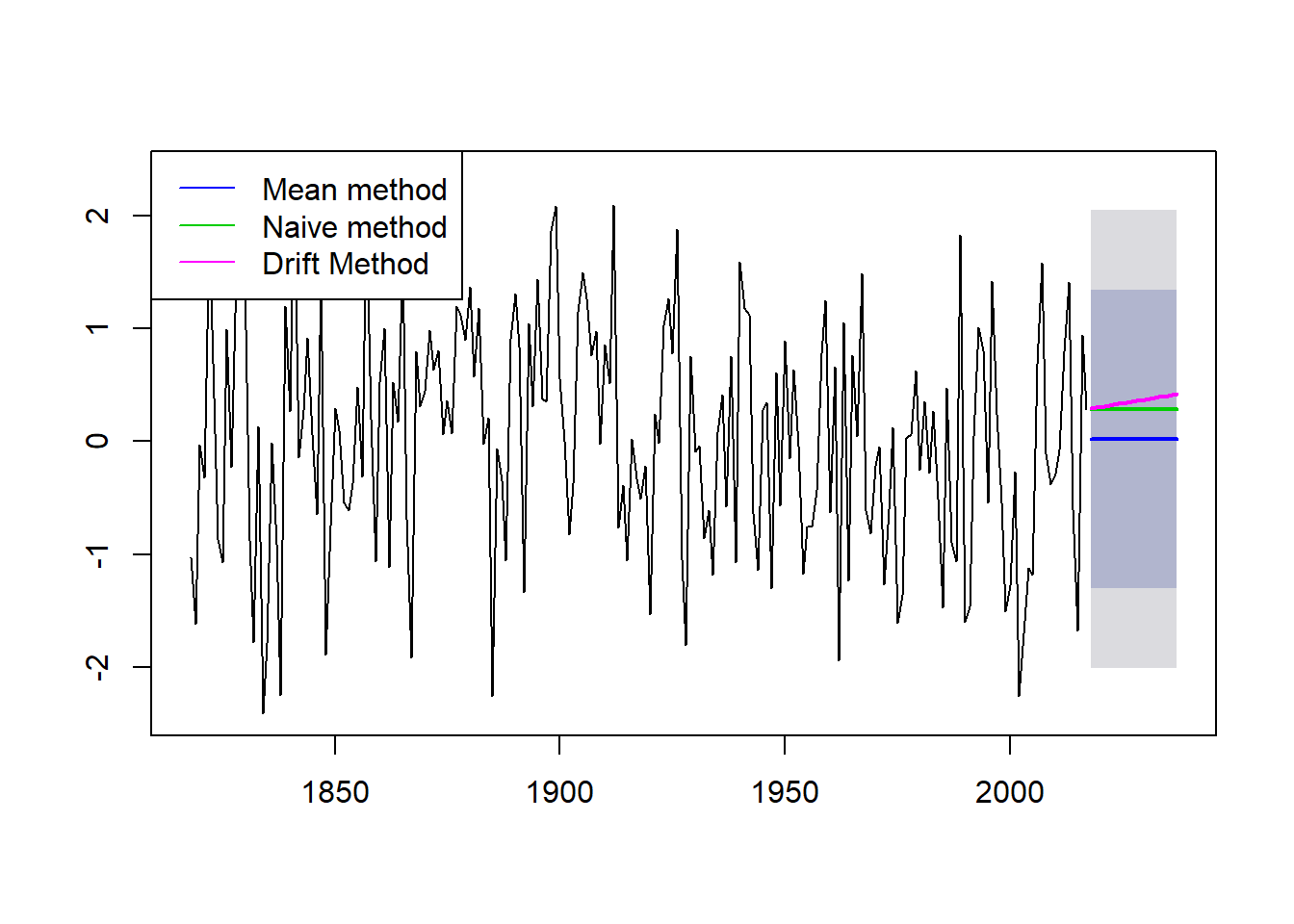
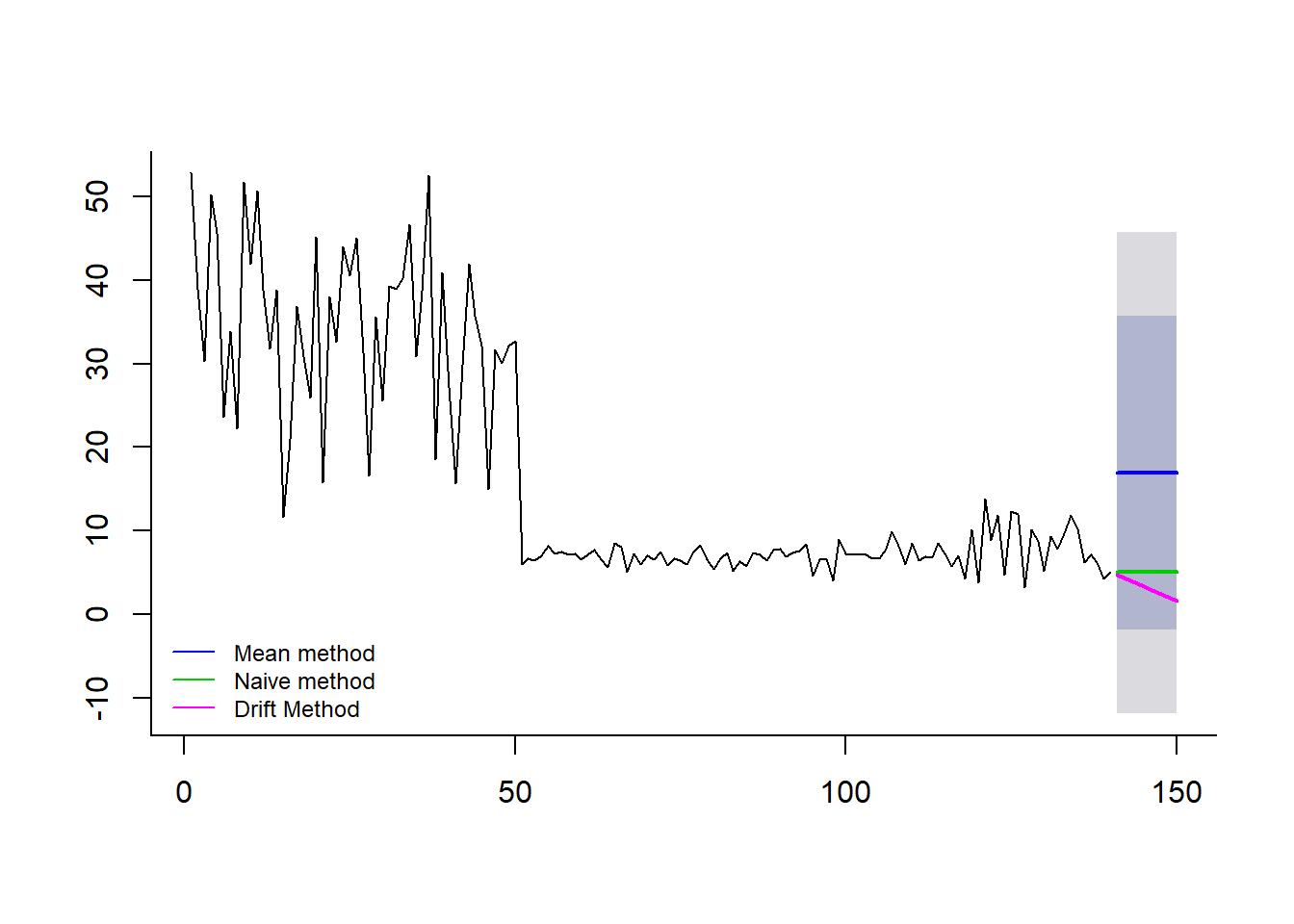
library(forecast)
meanm <- meanf(myts, h=10)
naivem <- naive(myts, h=10)
driftm <- rwf(myts, h=10, drift = T)
plot(meanm, main = "", bty = "l")
lines(naivem$mean, col=123, lwd = 2)
lines(driftm$mean, col=22, lwd = 2)
legend("bottomleft",lty=1,col=c(4,123,22), bty = "n", cex = 0.75,
legend=c("Mean method","Naive method","Drift Method"))
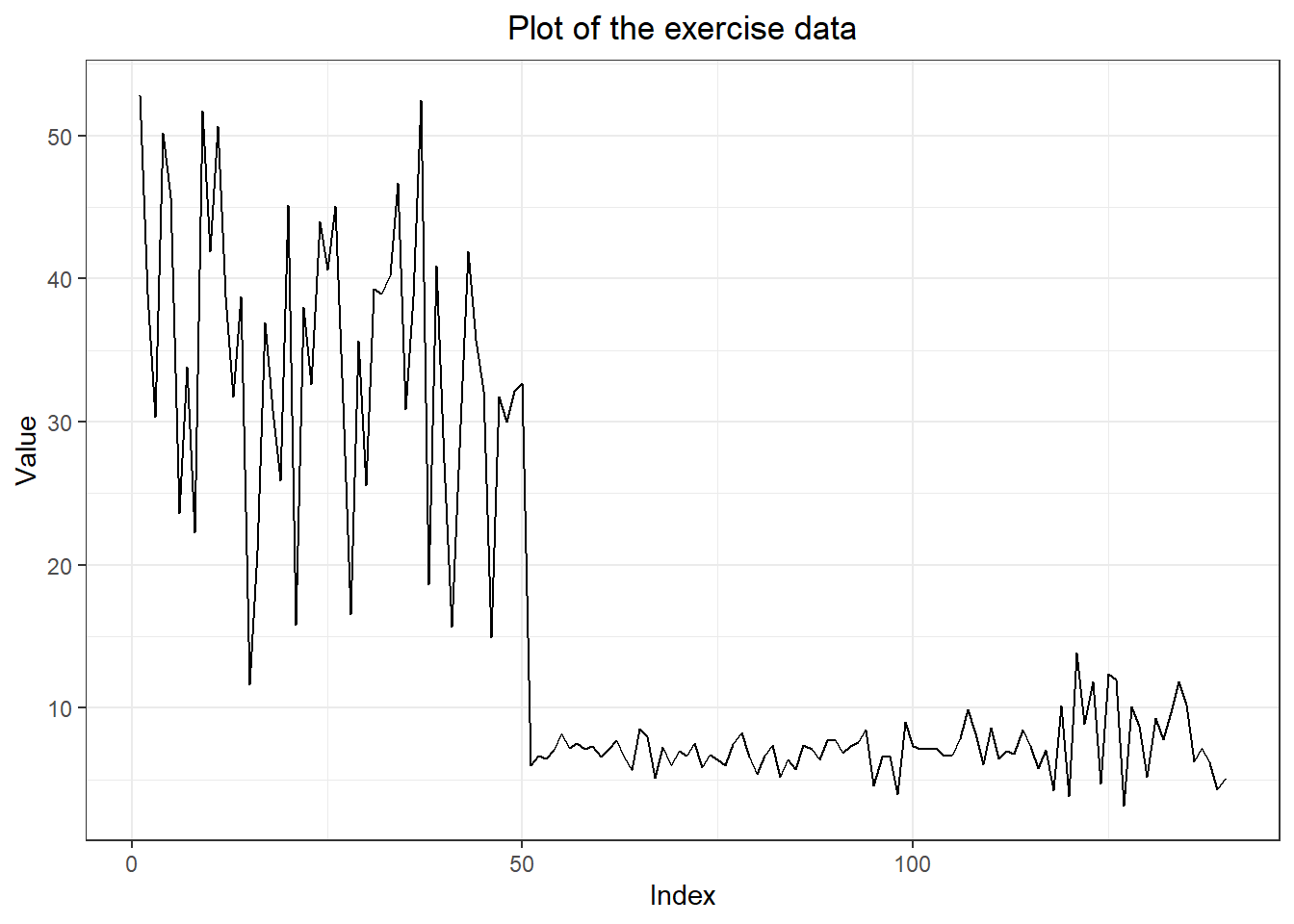
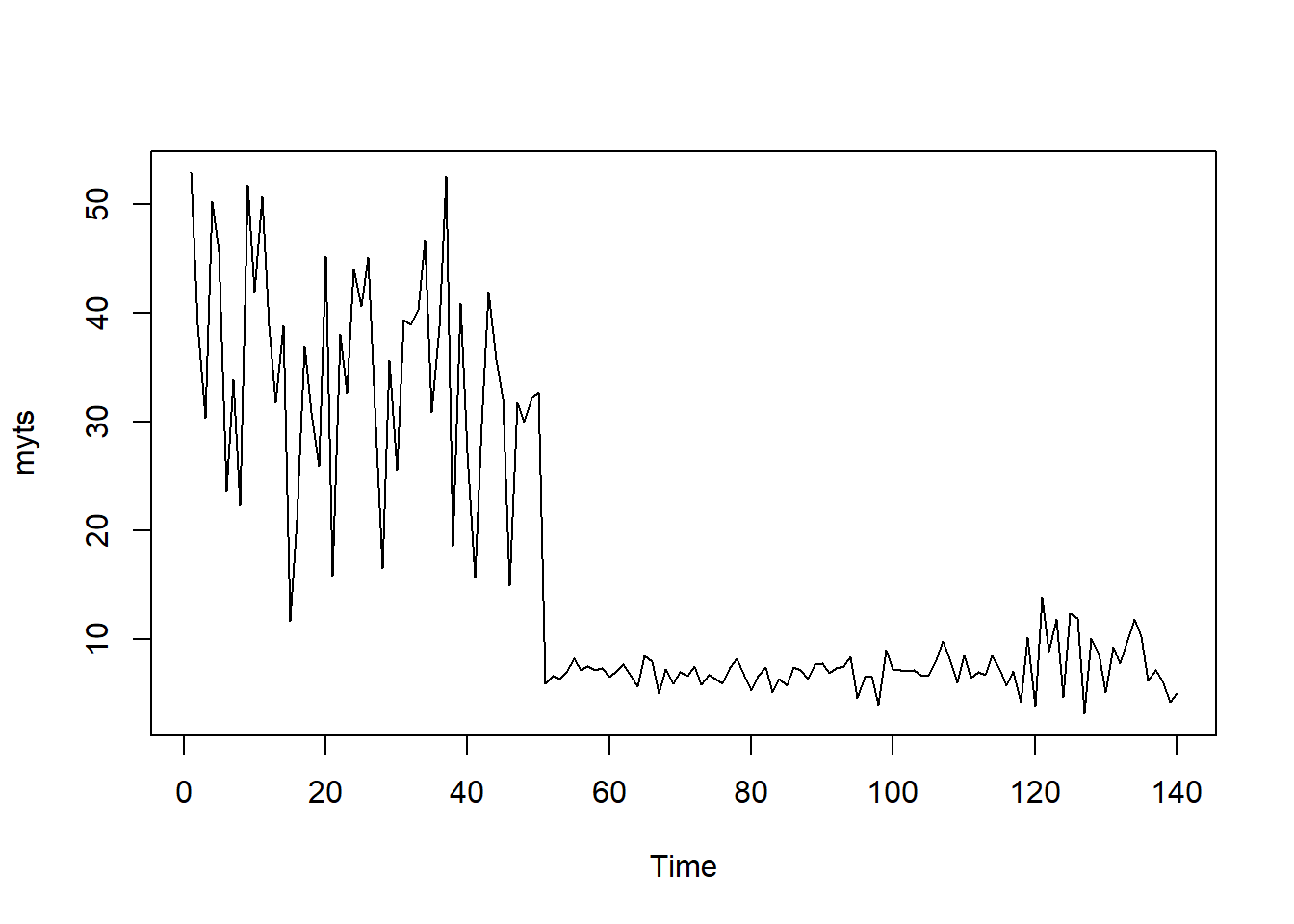
## [1] 140
## ME RMSE MAE MPE MAPE MASE
## Training set -6.408719e-16 15.31467 13.94736 -84.86989 120.4406 2.231924
## Test set -1.125187e+01 11.61073 11.25187 -180.27778 180.2778 1.800578
## ACF1 Theil's U
## Training set 0.7632991 NA
## Test set -0.1703445 2.002248
## ME RMSE MAE MPE MAPE MASE
## Training set -0.4131666 10.024449 6.249032 -11.909758 33.36297 1.0000000
## Test set 0.9663084 3.022963 2.482045 -1.869095 33.51426 0.3971888
## ACF1 Theil's U
## Training set -0.4901263 NA
## Test set -0.1703445 0.6497522
## ME RMSE MAE MPE MAPE MASE
## Training set -1.624594e-17 10.015931 6.265918 -7.901602 33.29565 1.002702
## Test set 6.957224e+00 8.172915 6.974530 86.321252 86.72796 1.116098
## ACF1 Theil's U
## Training set -0.4901263 NA
## Test set 0.4327471 1.62159
## [1] -0.3435748
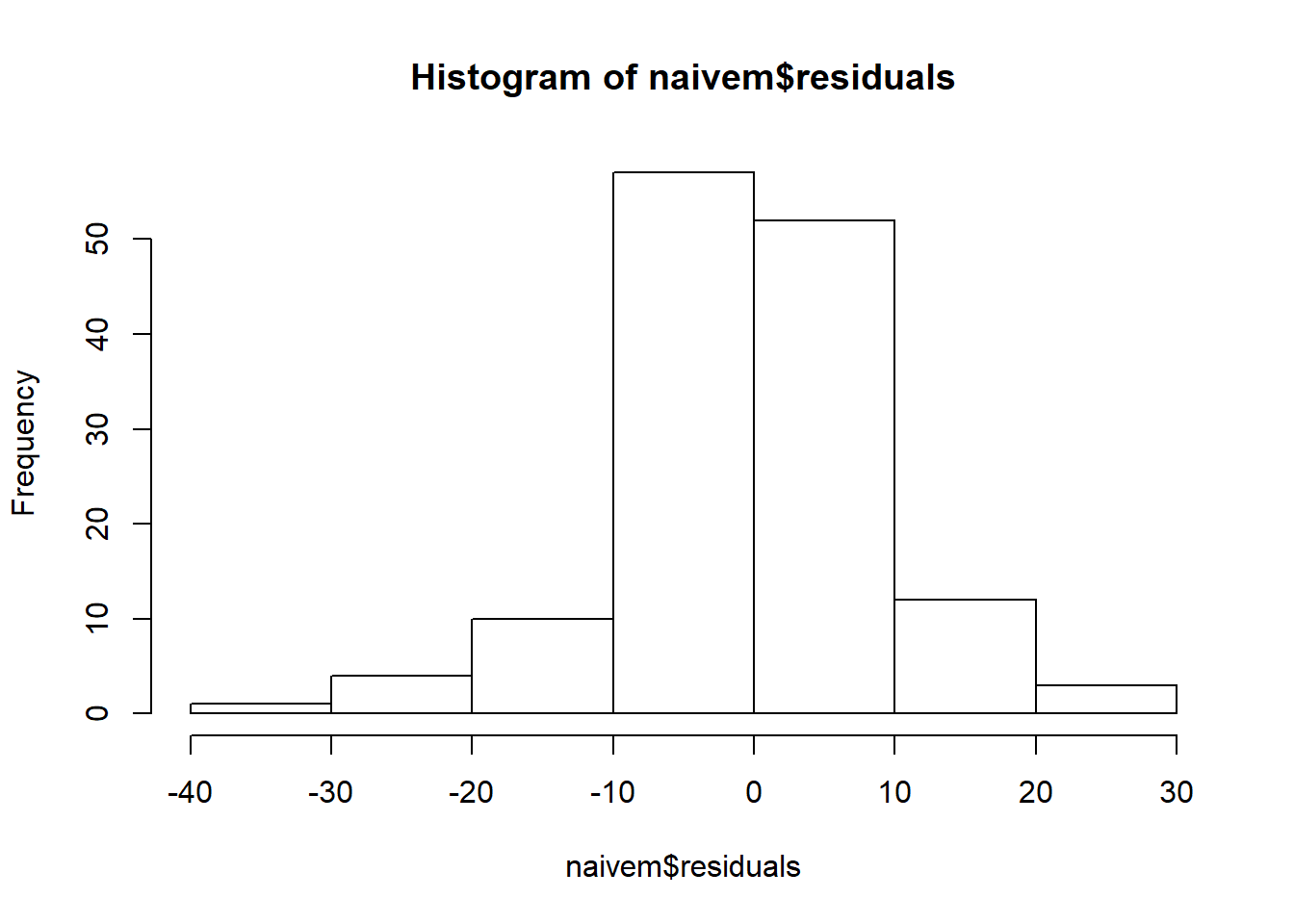
##
## Shapiro-Wilk normality test
##
## data: naivem$residuals
## W = 0.89587, p-value = 2.061e-08