PrioriTree: an Interactive Utility for Improving Geographic Phylodynamic Analyses in BEAST
Overview
PrioriTree is an interactive browser utility—distributed as an R package (see https://jsigao.shinyapps.io/prioritree/ for a demo of the utility)—designed to help researchers specify input files for—and process output files from—analyses of biogeographic history performed using the BEAST software package (Drummond et al. 2012; Suchard et al. 2018). The discrete-geographic models implemented in BEAST (Lemey et al. 2009; Edwards et al. 2011) contain many parameters that must be inferred from minimal information (the single geographic area in which each pathogen occurs); inferences under this approach are therefore inherently sensitive to the assumed priors on the model parameters. We recently demonstrated that the priors implemented as the defaults in BEAST—and used in the vast majority of published studies—are strongly informative and extremely unrealistic; these misinformative priors distort inferences of biogeographic history (see Gao et al. (2022) for details).
These considerations motivated our development of PrioriTree to help researchers: (1) interactively specify BEAST discrete-geographic phylodynamic analyses by providing graphical plots of specified priors; (2) specify input files for (and summarize output from) BEAST analyses to assess the prior sensitivity of biogeographic; (3) specify input files for (and summarize output from) BEAST analyses to assess the reative fit of candidate discrete-geographic (prior) models to the study data (by computing Bayes factors), and; (4) specify input files for (and summarize output from) BEAST analyses to assess the absolute fit (adequacy) of discrete-geographic (prior) models to the study data (by means of posterior-predictive simulation).
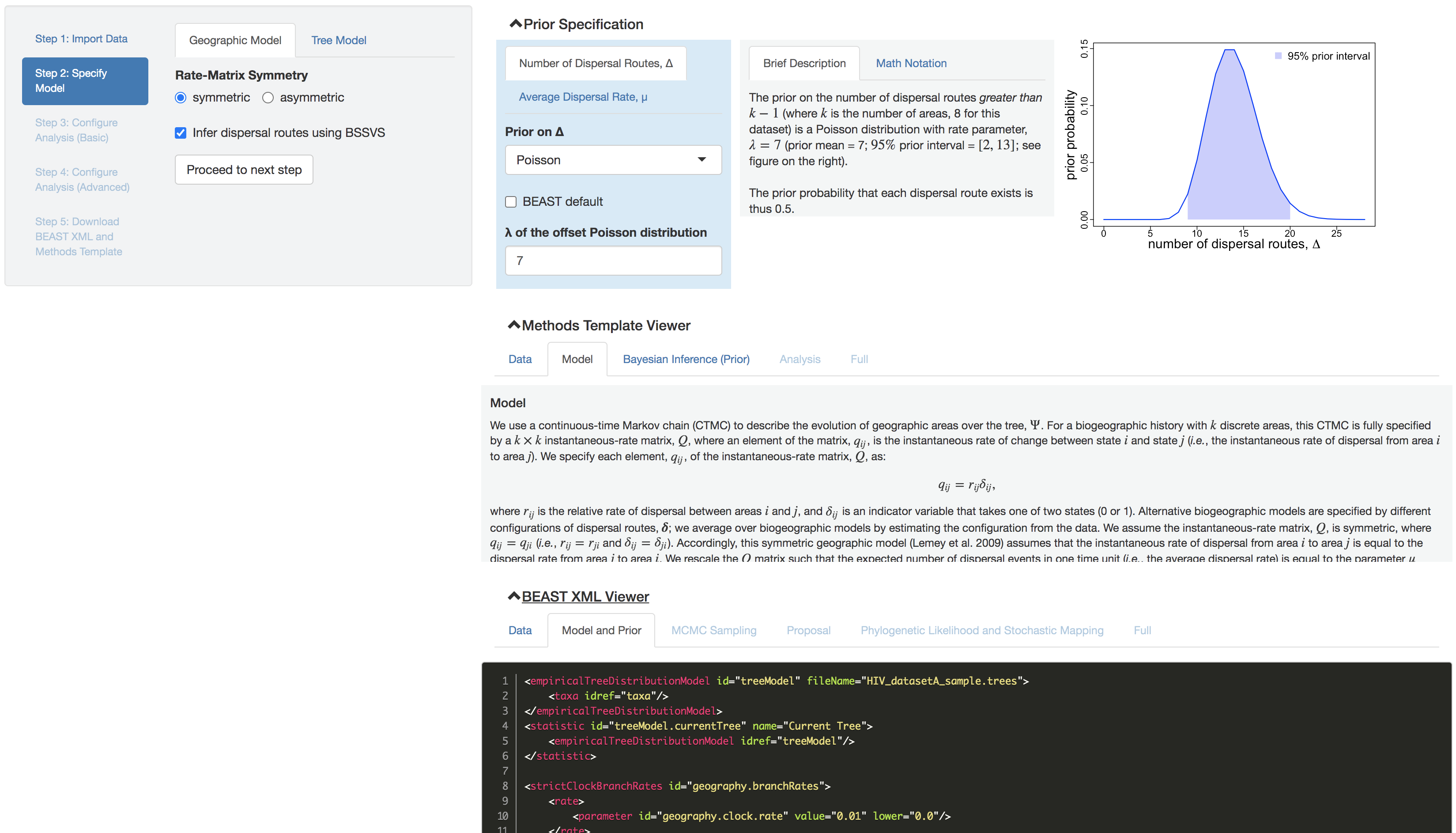
Figure 0.1: PrioriTree user interface. The main interface is divided into two panels: user input in the left panel, and the right panel dynamically renders the corresponding prior distributions, methods text, and BEAST XML script.
This manual consists of two major sections: (1) a quick-start guide provides an overview of the steps required to specify BEAST discrete-geographic phylodynamic analyses and to summarize the results using PrioriTree, and (2) a detailed guide that explains the functionalities provided by PrioriTree and the related theory in detail. Example input files can be found in this downloadable folder. More example files with real datasets (that are much larger) are available in this supplementary repository for Gao et al. (2022).