Chapter 3 The Cell
Prokaryotic cells lack a true “nucleus” (i.e., their DNA are not bound by a membrane), but they are also much smaller in size than their eukaryotic counterparts (they are roughly as big as a mitochondrion).
Eukaryotic cells, on the contrary, have a membrane-bound nucleus and are more complex morphologically. They are also usually larger than prokaryotic cells.
NB: Prokaryotes grow much quicker than eukaryotes due to their large surface area to volume ratio!
3.1 Budding and Filamentous Bacteria
Caulobacter are found in freshwater environments and are used as a model organism for studying complex prokaryotic cell cycles.
Chloroflexus, on the other hand, are ancient bacteria that are also photosynthetic (though they do not produce O2).
3.2 Morphogenesis
We say that a bacterium is monomorphic if it only has one shape. The majority of bacteria are monomorphic.
However, we say that a bacterium is pleomorphic if it has multiple shapes. Pleomorphic bacterium not only change their shapes during their life cycle (generally becoming smaller when they become older), but they also respond to environmental cues and can sporulate in the event of limited nutrients.
3.3 Bacterial and Archaeal Membranes and Means of Transport
Bacterial membranes serve three purposes:
- a permeability barrier: this prevents leakages and functions as a gateway for the transport of nutrients into and out of the cell.
- Protein anchors: the membrane is also a site of many proteins involved in transport, bioenergetics, and chemotaxis.
- Energy conservation: energy is also generated here via the proton motive force (i.e., the mitochondria).
Furthermore, note the structural differences between the bi-layer in bacteria and archaea:
Figure 3.1: Bilayer in Bacteria and Archaea
Figure 3.2: Bilayer in Bacteria and Archaea
Also, know the following graphic:
Figure 3.3: Means of Membrane Transport
3.4 The Cell Wall
Gram-positive bacteria have a thick peptidoglycan cell wall and a thin membrane. This is in contrast to their gram-negative counterparts: a thin layer of peptidoglycan, an outer membrane of lipopolysaccharides and proteins, and a periplasm (i.e., intermembrane space).
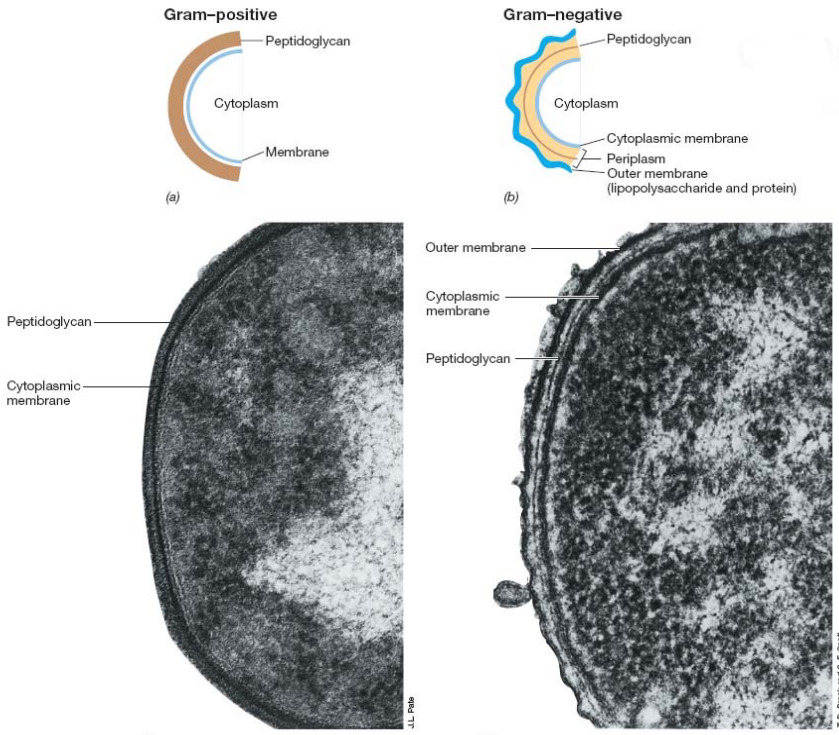
Figure 3.4: Cell Walls of Gram Positive and Negative Bacteria
Furthermore, gram positive bacteria have a peptide interbridge - a feature missing in gram-negative bacteria:
Figure 3.5: Peptide Cross Bridges in Gram Postiive and Gram Negative Bacterium
Nevertheless, peptidoglycan is comprised of amino acids and alternating units of NAM and NAG sugars linked via a \(\beta\)(1, 4) glycosidic bond.
It is also worth noting that cells placed in a sucrose and lysozyme solution will lead to the formation of protoplasts:
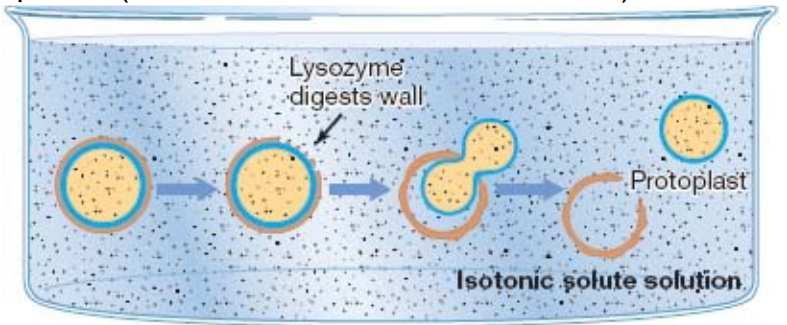
Figure 3.6: Protoplast Formation
3.5 Gram Staining
The gram stain is a technique invented by Christian Gram in 1884; the test serves to differentiate gram positive bacteria from gram negative ones.
In the gram stain, cells are first uniformly stained with crystal violet (a dye) before iodine (a mordant) is added to form an insoluble crystal violet-iodine complex.
The cell sample is then decolorized with alcohol before being counterstained with safranin (a dye); gram positive bacterium will appear purple (due to their thick peptidoglycan) whereas gram negative bacteria will appear pink (due to the safranin):
Figure 3.7: Gram Positive and Negative Bacterium
3.6 Pseudopeptidoglycan (murein) in Methanogenic Archaea
Pseudopeptidoglycan has NAT (N-Acetyl-talosaminuronic acid) residues in lieu of NAM residues in its chemical structure.
Further, NAG and NAT residues are linked via a \(\beta\)(1, 3) glycosidic bond, effectively preventing lysozyme attacks and making the archaean resistant to penicillin.
3.7 Teichoic Acids in Gram Positive Cells
These structures extend past the surface of the cell wall and grant the cell wall a negative charge.
Teichoic acids also provide flexibility to an otherwise rigid structure:
Figure 3.8: Teichoic Acids in Gram Positive Bacterium
3.8 Lipopolysaccharides (LPS) in Gram Negative Cell Walls
LPSs are an important barrier against host defenses.
Lipid A of the LPA is an endotoxin that is responsible for the bacterium’s pathogenicity. The O-specific polysaccharide of LPS varies from species to species.
The order of the LPS is as follows:
\[\begin{equation*} \text{Lipid A - ketodeoxyoctonate - core - O specific} \end{equation*}\]
3.9 Periplasmic Space
The periplasmic space is the space between the inner and outer membranes of a bacterium. It contains enzymes that participate in nutrient acquisition and metabolism.
3.9.1 For gram negative bacteria:
Chemolithotrophs have an extensive amount of transport proteins that extend to the outer membrane for uptaking and metabolizing inorganic ions.
Gram negative bacteria also have enzymes to oxidize toxic chemicals; these bacteria also have dioxygenase enzymes that are linked to the outer membrane.
3.9.2 For gram positive bacteria:
Exoenzymes are secreted by the bacterium to help transport nutrients into the cell (e.g., Bacillus secretes amylases to aid in mobilizing sugar).
3.10 Capsule and Slime Layers
The polysaccharide matrix of a bacterium is important for its survival, hydration, virulence, and attachment to surfaces.
Furthermore, India Ink (a dye) does not penetrate the capsule of bacteria (note the outline of the cells):
Figure 3.9: India Ink and Bacterial Capsules
3.11 Cellular Inclusions
There are many cellular inclusions in microorganisms; the ones examined in BS2002 are shown in the following sections.
3.11.1 Carbon inclusions
These take the form of poly \(\beta\) hydroxybutyrate in both archaea and bacteria:
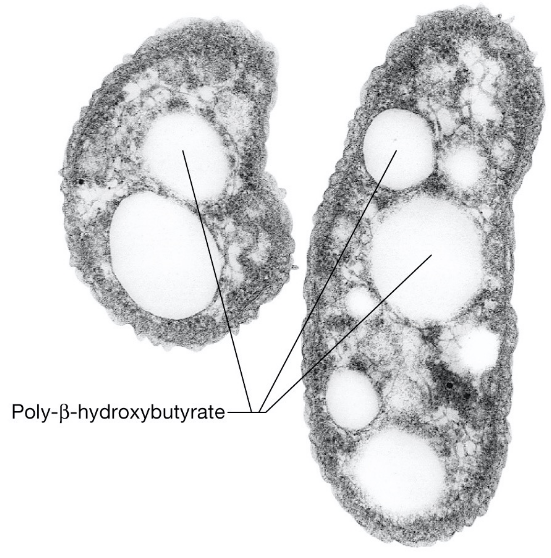
Figure 3.10: Carbon Inclusions in Archaea and Bacteria
3.11.2 Sulfur granules
Shown below are sulfur granules in the purple, photosynthetic bacterium Chromatium buderi:
Figure 3.11: Sulfur Granules in C. buderi
3.11.3 Magnetosomes
These are cellular inclusions that help bacteria orient and migate towards Earth’s geomagnetic fields.
Magenetosomes are often associated with high O2 concentrations.
3.11.4 Gas vesicles
These cellular inclusions are important for aquatic microorganisms. They allow the microorganism to float on water.
3.12 Flagella
There are serveral types of flagella in bacteria; they can be categorized into four types:
- Monotrichous = attached on one end.
- Amphitrichous = attached on both ends.
- Lophotrichous = a bundle on one end.
- Peritrichous = all around the cell’s surface.
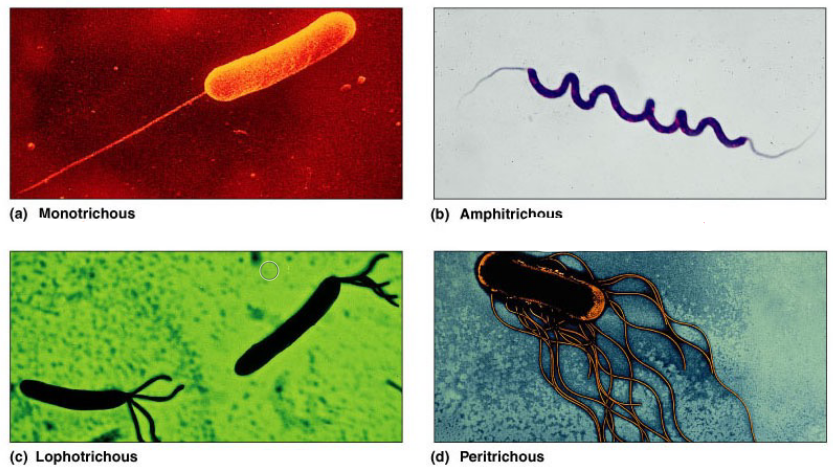
Figure 3.12: Types of Flagella
3.12.1 Flagella of gram negative bacteria
A typical gram negative bacterium’s flagella has three components:
- The filament (made from flagellin)
- The hook (made from a hook protein)
- The basal body (made from three rings)
The three rings mentioned in the last bullet point are the:
- MS ring (assembled in the cytoplasmic membrane)
- P ring (assembled in the periplasm)
- L ring (assembled in the LPS)
From there, the flagellin protein flows through the hook to form a filament.
Note that the motor of the flagella is driven by a proton motive force, not ATP!
3.12.2 Flagella of gram positive bacteria
A typical gram positive bacterium’s flagella has a filament-hook-base structure, is powered by the proton motive force, and is homologous.
However, only two rings: the MS and P ring are present.
3.12.3 Flagella of archaeans
Archaeal flagella are analogous to bacterial flagella, albeit they are powered by ATP and not the proton motive force.
The flagella also grows at the base and not the tip.
3.13 Swimming, Chemotaxis, and Phototaxis
There is a random run and tumble movement (of the microorganism) in the absence of an attractant.
The attractant causes an increased “run” time towards the attractant and a shorter “run” away from the attractant".
Figure 3.13: Swimming and Chemotaxis Demonstration
In the case of peritrichous flagella, the flagella rotate counterclockwise in a bundle:
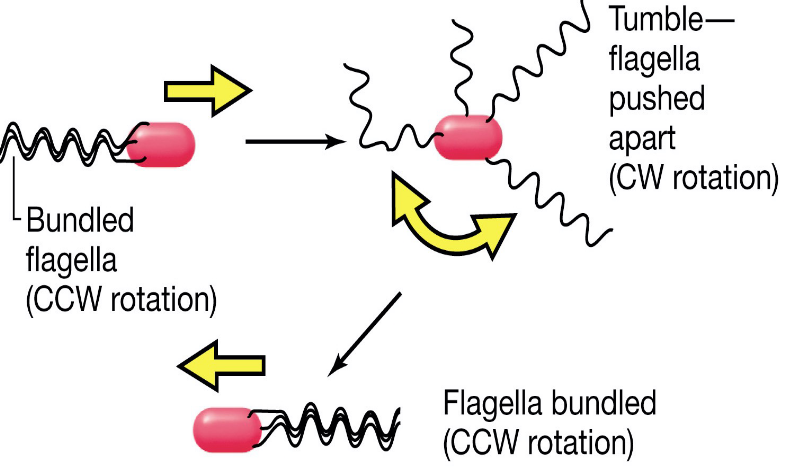
Figure 3.14: Peritrochous Flagella Movement
Polar flagella (i.e., flagella that are present on both ends of the microbe) change the direction of their movement via reversing their rotation:
Figure 3.15: Polar Flagella Movement
3.13.1 Capiliary tube assay
In a bacterial suspension involving three tubes: a blue tube (control), a red tube (repellent) and a green tube(attractant), the bacteria can be shown swimming towards the attractant:
Figure 3.16: A Capiliary Tube Assay
3.13.2 Phototaxis
Photosynthetic bacteria accumulate at the wavelength where pigments absorb. In a bacterial solution, bacteria swarm towards the light source.
3.13.3 Surface appendages
Pili cover the surface of bacteria and form conjugation bridges between bacterial cells (to transfer DNA).
3.14 Gram Positive Endospores
Sporulation occurs as a result of nutrient deprivation. It is not trigerred in response to environmental stress (not including nutrient limitations).
Nevertheless, spores are extreme resistant to environmental stresses; with a few exceptions, all spore-forming bacteria are gram-positive.
The sporulation process is controlled by a complex cascade of sigma factor gene expression events.
The different types of bacterial spores are shown below:
Figure 3.17: Different Types of Bacterial Spores
3.14.1 Returning spores to a vegetative state
First, the spore must be heated to a sublethal temperature.
Then, the heated spore must be placed in nutrient broth.
The above two steps result in very rapid germination and synthesis of RNA, protein, and DNA. The spore coat breaks and is discarded.
3.14.2 Dipicolinic acid
Dipicolinic acid constitutes 10% of a spore’s dry weight.
Dipicolinic acid molecules are cross-linked by Ca2+ ions:
Figure 3.18: Dipicolinic Acid in Bacterial Spores