Chapter 9 Microbial Genetics and Gene Regulation
The central dogma outlines the way in which genetic information is transferred to different levels of the organism (i.e., faithful replication). Yet, it is important to note that variations in the genetic code can still occur:
Figure 9.1: Central Dogma Flowchart
A mutation is a heritable change in an organism’s DNA sequence that is inherited by its offspring. Mutations can give rise to phenotypic variation and in some cases, even maximize the offsprings’ chance for survival.
9.1 Types of Mutations
Mutations can be phenotypic (i.e., they affect the phenotype of the organism).
There are also different types of mutations (know the table):
Figure 9.2: Types of Mutations
Furthermore, there are also many sources of mutations. Spontaneous mutations (i.e., mutations that occur due to random error) have a \(\frac{1}{1000000}\) chance of occuring!
Figure 9.3: Sources of Mutations
9.1.1 Penicilin counterselection for selecting mutants
Since penicilin kills growing cells, non-growing histidine auxotrophs will survive (i.e., these cells will be enriched).
Figure 9.4: Penicilin Selection
In this method, one merely identifies the His auxotrophs that they have enriched for!
9.1.2 Mutations by transposition
The general mechanism for transposition is as follows:
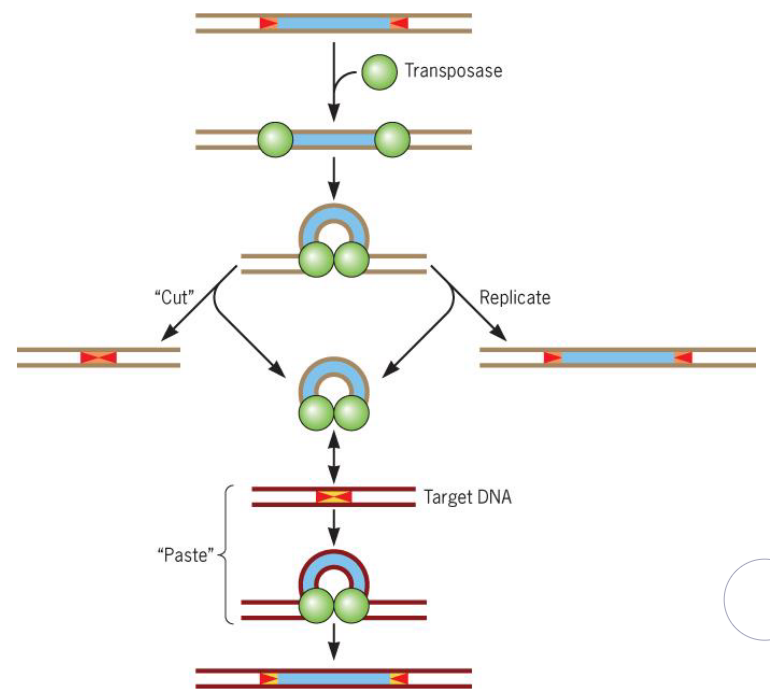
Figure 9.5: Tranposition in Action
There are also several transposable elements (know the table) and the following method for determining transposed elements:
Figure 9.6: Transposible Elements
9.2 Mechanisms of Gene Exchange in Prokaryotes
There are three different mechanisms of genetic exchange:
Figure 9.7: Mechanisms of Genetic Exchange
Prokaryotic genetic exchange doesn’t depend on reproduction, involves small amounts of DNA, and haploid organisms can directly express their alleles.
9.2.0.1 Griffith experiment
Observe the famous Griffith experiment:
Figure 9.8: Griffith Experiment
Natural transformation is often induced in dense cultures via quorum sensing (e.g., biofilms).
9.2.1 Genetic exchange via competence pili
Figure 9.9: Competence Pili
First, the pill bind to eDNA and translocates the DNA across the outer membrane via retraction. Then, the outer membrane’s secretin pore allows for DNA to diffuse across the outer membrane.
9.2.2 Life cycle of a phage
Observe the life cycle of a bacteriophage:
Figure 9.10: Bacteriophage Life Cycle
DNA in a bacteriophage can also be packaged in concatameric, rolling circle replications of phage genomes:
Figure 9.11: Concatameric Storage of DNA
There are cleavage at pac sites by phage enzymes (TerSL), and cleavage at similar pac sites in the host genome can also package non-viral DNA too!
Furthermore, there is also site-specific recombination and crossovers in bacteriophage DNA:
Figure 9.12: DNA Crossover in Bacteriophages
Gene transfer can also be transferred in one of two ways: specialized transduction or lateral transduction.
Figure 9.13: Types of Transductions
9.2.2.1 Lateral Transduction
Figure 9.14: Lateral Transduction
In step 1, there is the replication of the DNA. In step 2, there is the initiation of packaging from the pac site by ter.
All of the above occurs while prophage excision occurs and headfuls of host DNA are packaged!
9.2.2.2 Conjugation
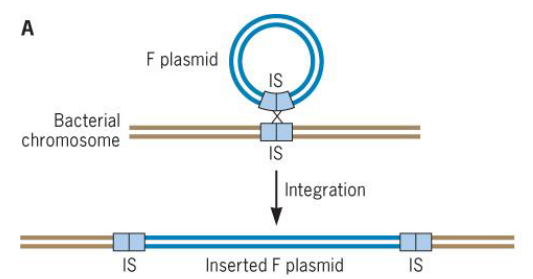
Figure 9.15: The F Plasmid
The integration of a F plasmid into a bacterium’s genome results in the creation of Hfr strains (i.e., strains with a high frequency of recombination).
Figure 9.16: Conjugation of an F plasmid
These Hfr strains, when mobilized, can drag along chromosomal DNA (which would take ~100 minutes to transfer to an entire E. coli genome).
Figure 9.17: Conjugation in E. faecalis
E. faecalis also has a conjugation mechanism (shown above), albeit it relies on pheromones and AS-mediated pumping.
9.2.3 CRISPR
Figure 9.18: CRISPR Schematic
CRISPR is a family of DNA sequences that are found in the genomes of prokaryotic organisms (e.g., bacteria and archaea) and are derived from the DNA fragments of bacteriophages that have previously infected the prokaryote.
9.3 Genes and Regulation of Cellular Processes
9.3.1 Evidence for coordinated regulation of metabolic reactions
E. coli cultured in minimal medium and 14C-glycerol from 14CO2 (as the carbon source) had 14C present in its macromolecules (including amino acids).
Similarly, E. coli cultured in minimal medium and histidine using the same carbon source had 14C present in all of its macromolecules but histidine!
9.3.2 Metabolic regulatory devices
Figure 9.19: Some Metabolic Regulatory Devices
Some other microbial themes include:
- Substrate limitations
- Short life span of mRNAs
- Operons and coordinated expressions
- Coupled tln and txns
9.3.2.1 Allostery
An allosteric site is a regulatory site that the effector binds to and is separate from the active site of the enzyme.
Note that the allosteric effector does not have to look anything like the substrate (or the product for that matter).
9.3.2.2 sRNA
Figure 9.20: sRNA Control of Protein Activity
The 6S RNA of E. coli regulates the transition to the stationary phase (of the cell cycle of the bacterium) by modifying the RNAP-$$70 housekeeping transcription activity (hence preventing the normal expression of genes in the exponential growth phase of the bacterium).
9.3.3 lac operon
Figure 9.21: lac operon
There are obviously many regulatory control points in the lac operon.
9.3.3.1 Positive transcriptional control
Figure 9.22: Positive Transcriptional Control
Here, the binding of a regulatory protein at a regulatory region on DNA promotes transcription initiation.
An inactive protein can be activated by an inducer and inactivated by an inhibitor.
9.3.3.2 Trp attenuation
Figure 9.23: Trp Attenuation
The trp operon is a group of genes that encode for biosynthetic enzymes for the amino acid tryptophan; these genes are also found in E. coli.
When tryptophan levels are low, the trp operon is turned on; otherwise, trytophan blocks the expression of the operon.
Such an amino acid synthesis is also regulated by attenutation (based off the coupling of transcription and translation).
9.3.3.3 Antisense sRNAs (in translation)
Figure 9.24: Antisense sRNAs
Antisense sRNAs have also been shown to exhibit regulatory effects on protein translation.
9.3.4 Two-component regulatory and phosphorelay systems
Many genes can be turned “on” or “off” in response to environmental conditions (e.g., temperature, osmolarity, O2 levels, etc). The regulatory proteins responsible for the latter are part of a two-component signalling system that links external events to the regulation of gene expression.
All three domains of life have these systems - bacteria and archaeal organisms have many two component signal transduction systems. Though, some of these systems regulate protein activity instead of gene transcription.
9.3.4.1 Sensor kinase
Figure 9.25: Sensor Kinase
These pathways span the entire cytoplasmic membrane and have an extracellular receptor for signals. They also play a role in intracellular communication pathways.
9.3.4.2 Response-regulator proteins
Figure 9.26: Response Regulator Proteins
These are activated by a sensor kinase and a DNA-binding protein. An activator enhances transcription whereas a repressor inhibits transcription (unless needed).
9.4 Global Regulations
Catabolite repression is the regulation of transcription by both repressors and activators.
Figure 9.27: Catabolite Repression
Diauxic growth has two patterns: a biphasic growth phase and a lag phase.
A biphasic growth pattern involves the preferential use of one carbon source over another when both sources are available in the environment.
As witnessed in the above graphic, lag occurs after a preferred substrate is exhausted - microbe growth then continues (using the second carbon source) after a brief moment of time.
Figure 9.28: Catabolite Repression Example
Catabolite repression plays a role in this pattern of growth!
9.4.1 Stringent response
Figure 9.29: Stringent Response
When cells are starved for amino acids, protein synthesis is unable to proceed (for obvious reasons).
Consequently, tRNA and rRNA production is decreased, and the protein ReIA downregulates the synthesis of (p)ppGpp - an alarmone. Cells also increase the biosynthesis of the amino acids needed.