Chapter 3 Principal figures (Figure Models)
3.1 Packages, data, and helpers
library(tidyverse); library(useful);library(epiDisplay);library(lubridate)
library(spdep); library(sf); library(sp); library(devtools); library(pkgload); library(stringr)
load("./_data/CLIM_MAL_dat_20200826.RData")
devtools::source_gist("https://gist.github.com/gcarrascoe/89e018d99bad7d3365ec4ac18e3817bd")3.2 Figure 3 [COMPLETE] Fixed effects of baseline, linear and nonlinear models with seasonal-spatial and spatial effects for A) P.vivax and B) P Falciparum malaria
# Extra requirements
library(cowplot)
# P. Vivax
sf_6_pv <- test_pv_2[c(9,7,2,4,3,5)] %>%
plot_fixed.safe(title = "P. vivax",
filter=4,
legend = F,
ylim = 2.8,
angle = 0,
hjust = .5,
lim = c("enviro","n_eess", "nets", "cum_loss_km2", "prcp", "tmax"),
xlabs = c("enviro","hfd", "nets", "cum_loss_km2", "prcp", "tmax"),
breaks=c("1","2","3","4","5","6"),
lab_mod=c("baseline: spatial","baseline: seaonal-spatial",
"linear: spatial","linear: seaonal-spatial",
"nonlinear: spatial","nonlinear: seaonal-spatial"),
flip=T) +
theme(plot.margin = unit(c(3,3,3,3), "lines"))
# P. Falciparum
sf_6_pf <- test_pf_2[c(9,7,2,4,3,5)] %>%
plot_fixed.safe(title = "P. falciparum",
filter=4,
ylim = 1.1,
angle = 0,
hjust = .5,
lim = c("enviro","workers", "cum_loss_km2", "prcp", "tmax"),
breaks=c("1","2","3","4","5","6"),
lab_mod=c("baseline: spatial","baseline: seaonal-spatial",
"linear: spatial","linear: seaonal-spatial",
"nonlinear: spatial","nonlinear: seaonal-spatial"),
flip=T) +
theme(plot.margin = unit(c(3,3,3,3), "lines"))
# Final figure
(sf_6<-plot_grid(sf_6_pv, sf_6_pf, nrow = 1, ncol=2, labels = c("A)", "B)"), label_size=30, align = "hv", rel_widths = c(4.2,6)))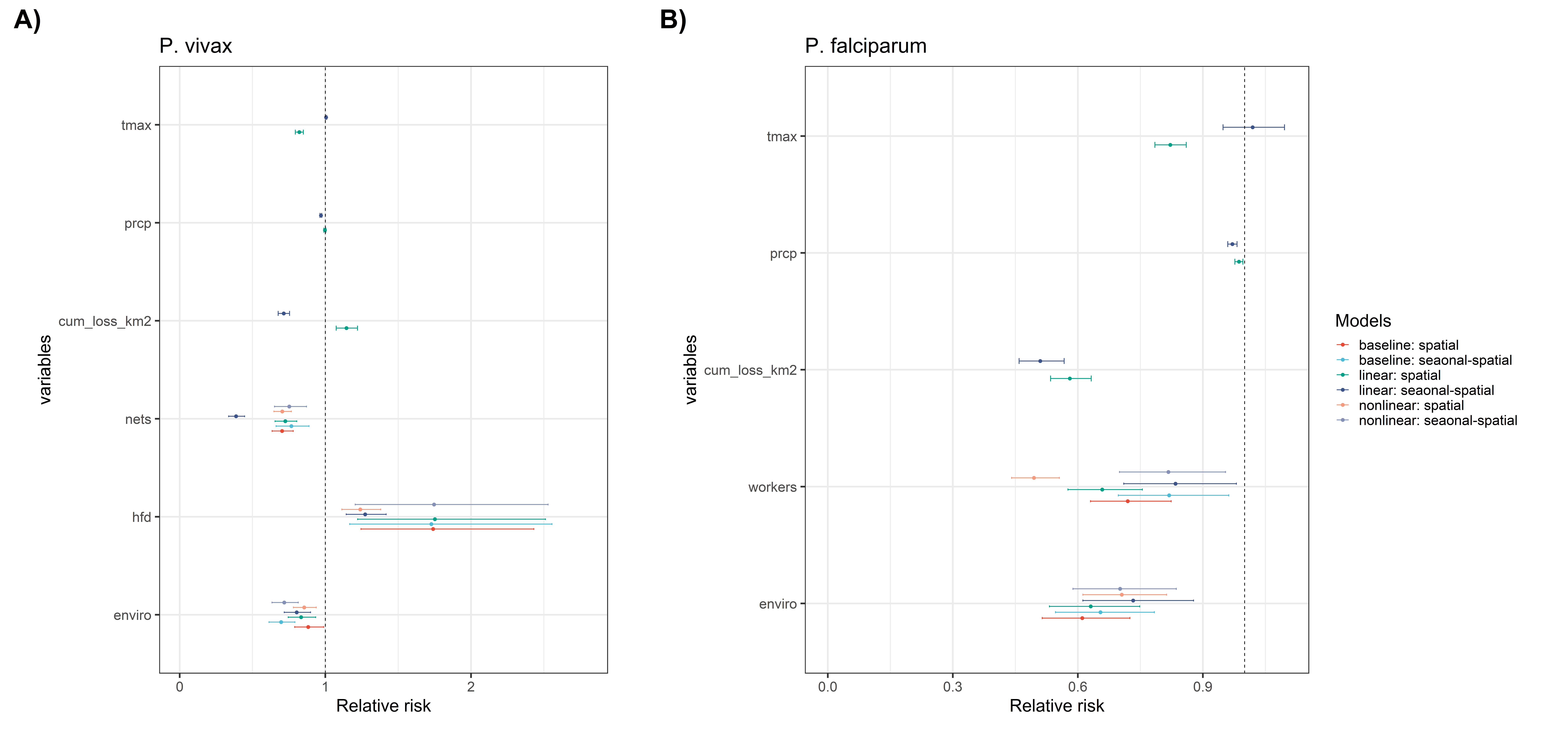
3.3 Figure 4 [COMPLETE] Spatial heterogeneity of covariates effects in terms of mean absolute error (MAE)
# Extra requirements
library(colorspace)
library(hrbrthemes)
library(sf)
library(ggthemes)
library(cowplot)
# ---------------
# Version 3
# ---------------
# Preprocessing figure A
f3_a<-plot_mae2(dat1 = dat, var = vivax, full = test_pv_2[[4]],
model1 = test_pv_2[[11]], model2 = test_pv_2[[10]], model3 = test_pv_2[[9]],
map = area.sf, id = NOMBDIST)
dat.f3a <- f3_a[[3]] %>%
dplyr::select(NOMBDIST, m1:m3, i1:i3) %>%
dplyr::rename(climate_val=m1, `malaria-control_val`=m2, deforestation_val=m3,
climate_i=i1, `malaria-control_i`=i2, deforestation_i=i3) %>%
gather(type, val, climate_val:deforestation_i) %>%
separate(type, into = c("type","i"), sep = "_") %>%
spread(i,val) %>%
mutate(i = as.numeric(i),
i = ifelse(i<0,NA,i), # only show added value
i = ifelse(i>100,NA,i)
) %>%
mutate(val = ifelse(val=="No Added", val, "Added"))
# Figure A
f3a <- dat.f3a %>%
ggplot() +
geom_sf(aes(fill=i)) +
scale_fill_continuous_sequential(palette="blues", na="white", trans="pseudo_log",
breaks = c(1,3,8,15,30), labels = c("No \nadded","3","8","15","30")) +
labs(fill = "Added value: MAE(diff) \n ") +
theme_few(base_size = 20) +
theme(legend.position = "top",
panel.spacing = unit(2, "lines"),
legend.text = element_text(size = 10)) +
facet_wrap(.~type)
# Preprocessing figure B
f3_b<-plot_mae2(dat1 = dat, var = falciparum, full = test_pf_2[[2]],
model1 = test_pf_2[[9]], model2 = test_pf_2[[8]], model3 = test_pf_2[[7]],
map = area.sf, id = NOMBDIST)
dat.f3b <- f3_b[[3]] %>%
dplyr::select(NOMBDIST, m1:m3, i1:i3) %>%
dplyr::rename(climate_val=m1, `malaria-control_val`=m2, deforestation_val=m3,
climate_i=i1, `malaria-control_i`=i2, deforestation_i=i3) %>%
gather(type, val, climate_val:deforestation_i) %>%
separate(type, into = c("type","i"), sep = "_") %>%
spread(i,val) %>%
mutate(i = as.numeric(i),
i = ifelse(i<0,NA,i), # only show added value
i = ifelse(i>100,NA,i)
) %>%
mutate(val = ifelse(val=="No Added", val, "Added"))
# Figure B
f3b <- dat.f3b %>%
ggplot() +
geom_sf(aes(fill=i)) +
scale_fill_continuous_sequential(palette="blues", na="white", trans="pseudo_log",
breaks = c(1,3,8,15,30), labels = c("No \nadded","3","8","15","30")) +
labs(fill = "Added value: MAE(diff) \n ") +
theme_few(base_size = 20) +
theme(legend.position = "top",
panel.spacing = unit(2, "lines"),
legend.text = element_text(size = 10)) +
facet_wrap(.~type)
# Final figure
plot_grid(f3a,f3b, nrow = 2, labels = c("A)","B)"), label_size=30) 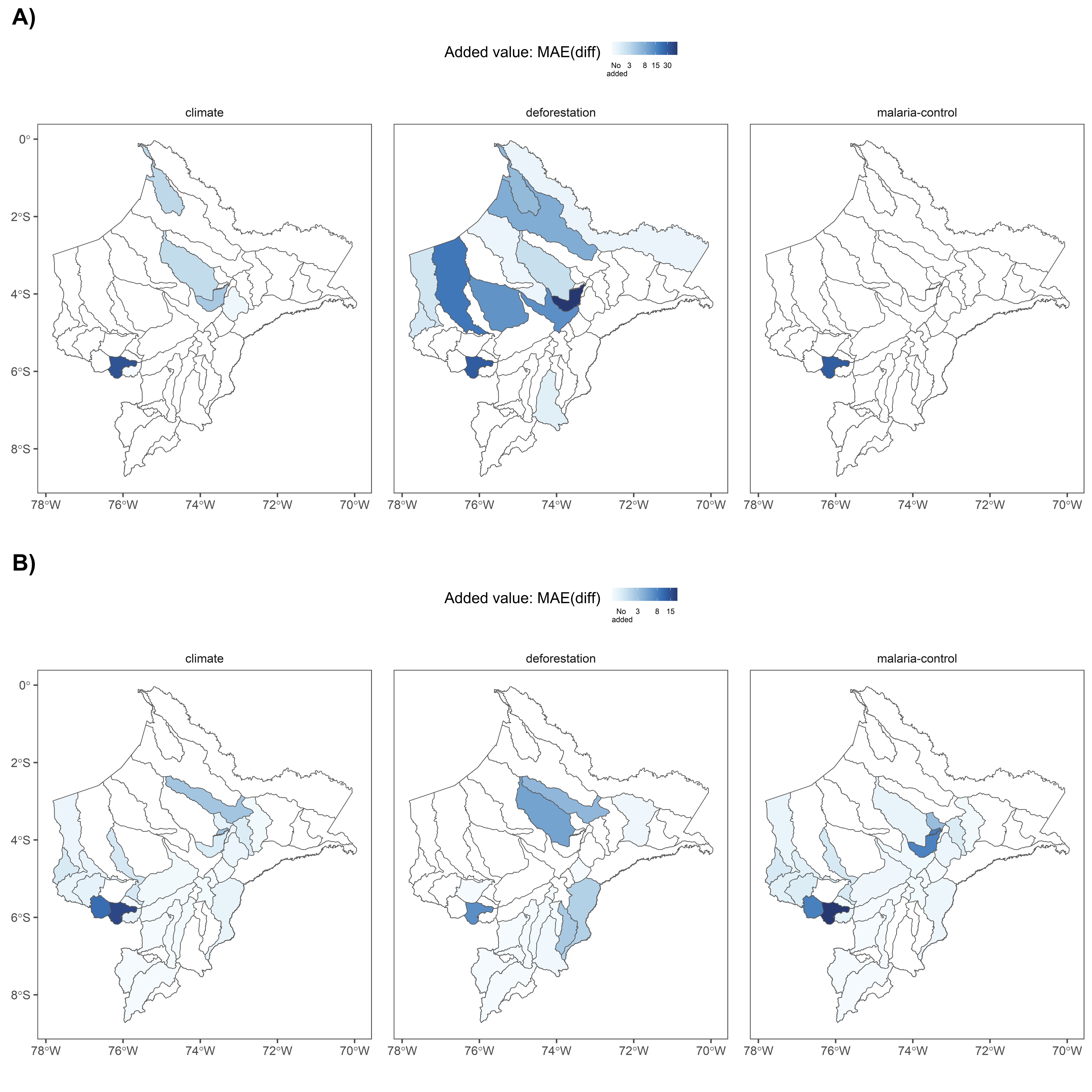
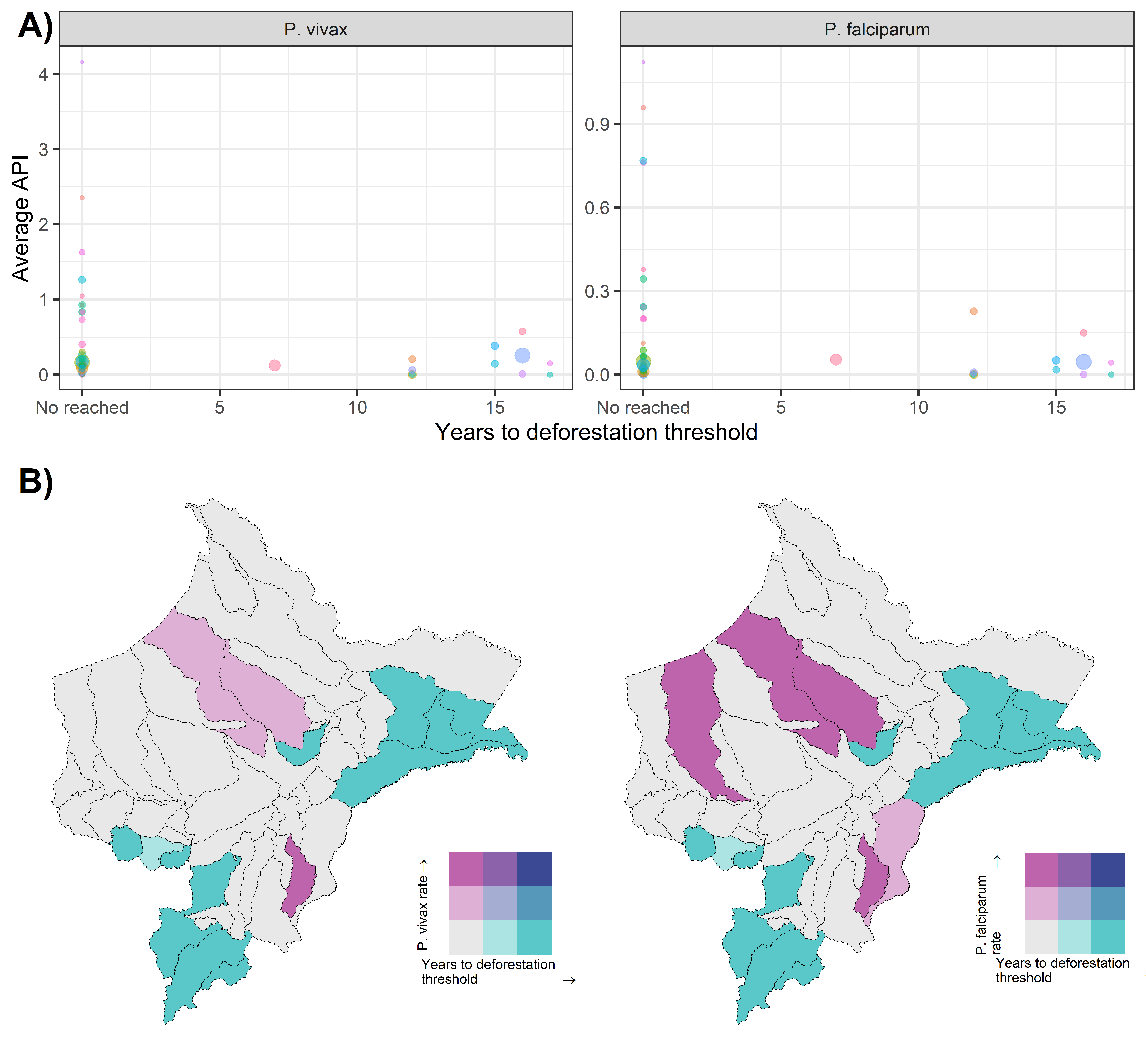
3.4 Figure 5 [COMPLETE] A)Scatter plot and B)map of time to cumulative deforestation threshold relative to P.vivax and P.falciparum
# Extra requirements
library(biscale)
library(cowplot)
#######################################################################
# PANEL A
#######################################################################
a <- dat %>%
group_by(NOMBDIST) %>%
mutate(defo = ifelse(cum_loss_km2<1.5,0,1),
i_def = defo - lag(defo)) %>%
filter(i_def == 1) %>%
mutate(year_def = year-2000) %>%
dplyr::select(NOMBDIST,year_def)
a2 <- dat %>%
group_by(NOMBDIST) %>%
mutate(pv_rate = 100*vivax/pop2015,
pf_rate = 100*falciparum/pop2015) %>%
summarise(pv_rate = mean(pv_rate),
pf_rate = mean(pf_rate),
pop = mean(pop2015)) %>%
full_join(a, by="NOMBDIST")
a2.sf <- area.sf %>%
full_join(a2, by="NOMBDIST") %>%
mutate(area = as.numeric(st_area(.)/1000000))
f4a <- a2.sf %>%
mutate(year_def = ifelse(is.na(year_def),0,year_def)) %>%
dplyr::select(NOMBDIST, year_def,pf_rate, pv_rate, pop) %>%
dplyr::rename(apv_rate = pv_rate, bpf_rate = pf_rate) %>%
gather(spp, rate, apv_rate:bpf_rate) %>%
ggplot(aes(x= year_def, y= rate)) +
geom_point(aes(size = pop, col=NOMBDIST), alpha = .5) +
labs(x = "Years to deforestation threshold", y = "Average API") +
guides(size=F, col=F) +
theme_bw(base_size = 20) +
scale_x_continuous(breaks = c(0,5,10,15), labels = c("No reached","5","10","15")) +
facet_wrap(.~spp, scales = "free_y",
labeller = labeller(spp = c(apv_rate = "P. vivax", bpf_rate = "P. falciparum"))
)
#######################################################################
# PANEL B
#######################################################################
map <- a2.sf %>%
mutate(year_def = ifelse(is.na(year_def),0,year_def)) %>%
bi_class(x = year_def, y = pv_rate, style = "equal", dim = 3) %>%
ggplot() +
geom_sf(mapping = aes(fill = bi_class), color = "black",
size = 0.3, show.legend = FALSE, linetype = "dashed") +
bi_scale_fill(pal = "DkBlue", dim = 3) +
bi_theme()
legend <- bi_legend(pal = "DkBlue",
dim = 3,
xlab = "Years to deforestation \nthreshold",
ylab = "P. vivax rate",
size = 12)
finalPlot <- ggdraw() +
draw_plot(map, 0, 0, 1, 1) +
draw_plot(legend, 0.7, 0.1, 0.3, 0.3)
map2 <- a2.sf %>%
mutate(year_def = ifelse(is.na(year_def),0,year_def)) %>%
bi_class(x = year_def, y = pf_rate, style = "equal", dim = 3) %>%
ggplot() +
geom_sf(mapping = aes(fill = bi_class), color = "black",
size = 0.3, show.legend = FALSE, linetype = "dashed") +
bi_scale_fill(pal = "DkBlue", dim = 3) +
bi_theme()
legend2 <- bi_legend(pal = "DkBlue",
dim = 3,
xlab = "Years to deforestation \nthreshold",
ylab = "P. falciparum \nrate",
size = 12)
finalPlot2 <- ggdraw() +
draw_plot(map2, 0, 0, 1, 1) +
draw_plot(legend2, 0.7, 0.1, 0.3, 0.3)
f4b <- plot_grid(finalPlot, finalPlot2)
# Final figure
(f4 <- plot_grid(f4a, f4b, ncol = 1, labels = c("A)","B)"), rel_heights = c(3,4), label_size=30))