Chapter 2 Principal figures (Descriptive Analysis)
2.1 Packages, data, and helpers
library(tidyverse); library(useful);library(epiDisplay);library(lubridate)
library(spdep); library(sf); library(sp); library(devtools); library(pkgload)
load("./_data/CLIM_MAL_basedata.RData")
devtools::source_gist("https://gist.github.com/gcarrascoe/89e018d99bad7d3365ec4ac18e3817bd")2.2 Figure 1 [COMPLETE] - Study Area
2.2.1 Figure 1A
# Preprocessing (final figure)
o_dist <- dt_final %>% group_by(NOMBDIST,year) %>%
summarise(viv=sum(vivax), fal=sum(falciparum),pop=mean(pop2015)) %>%
mutate(Pviv_1k=viv*1000/pop, Pfal_1k=fal*1000/pop) %>%
ungroup() %>%
group_by(NOMBDIST) %>%
summarise(avg_1viv = mean(Pviv_1k, na.rm=T),
avg_2fal = mean(Pfal_1k, na.rm=T))
o_LOR.sf <- area.sf %>%
left_join(o_dist, by="NOMBDIST") %>%
gather(spp, val, avg_1viv:avg_2fal)
(Fig_1b<- o_LOR.sf %>%
ggplot() +
geom_sf(aes(fill=log10(val+1)), size = 0.3, col = "black") +
scale_fill_distiller(palette = "RdYlGn", name = "Log10 \n API") +
theme_void() +
theme(panel.grid.major = element_line(color = "white"),
legend.position=c(0.9, 0.05), legend.justification = c(1, 0),
strip.text = element_text(size=15)) +
facet_wrap(.~spp, nrow = 1, ncol = 2, labeller = labeller(spp = c(avg_1viv="P. vivax", avg_2fal="P. falciparum"))))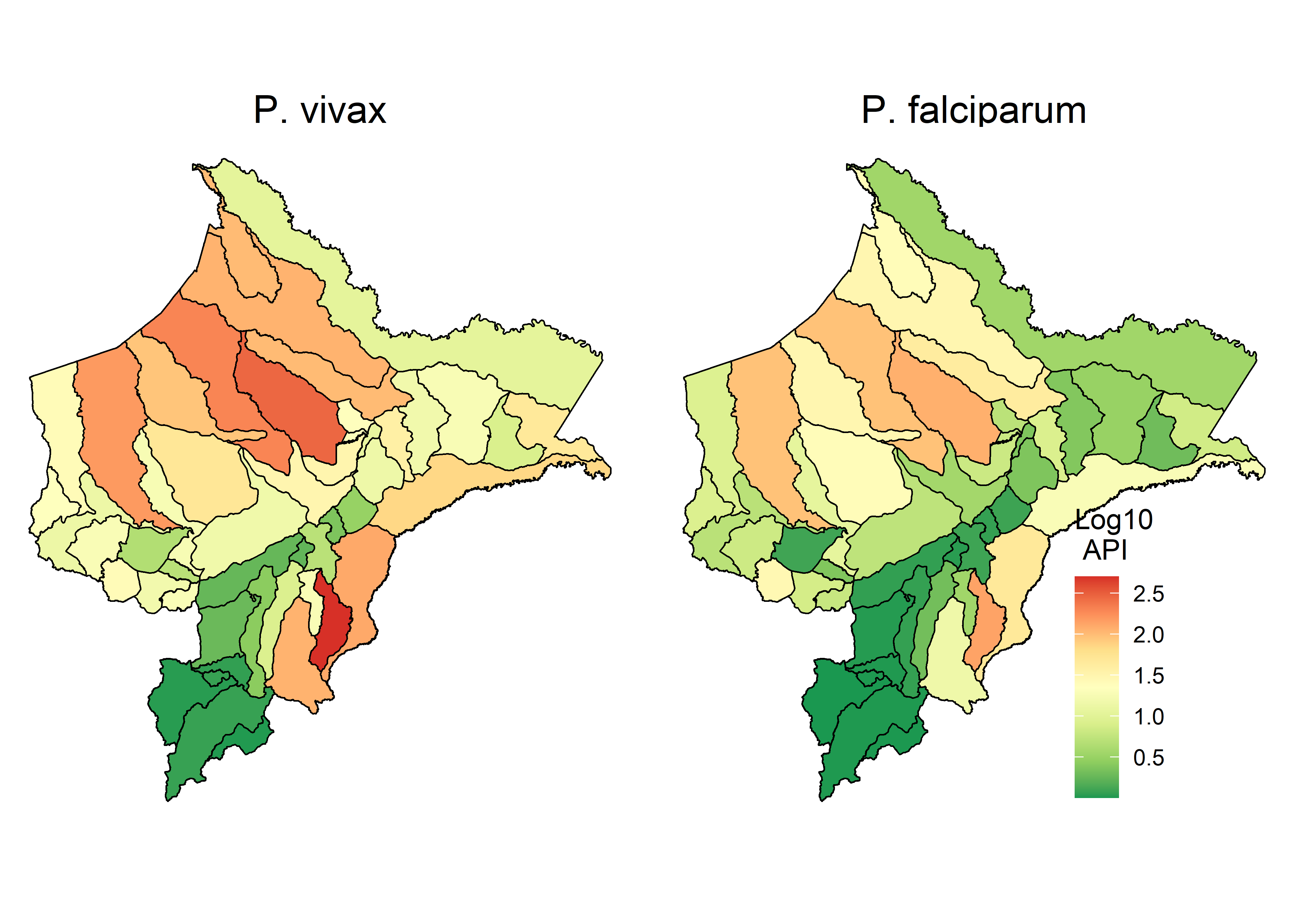
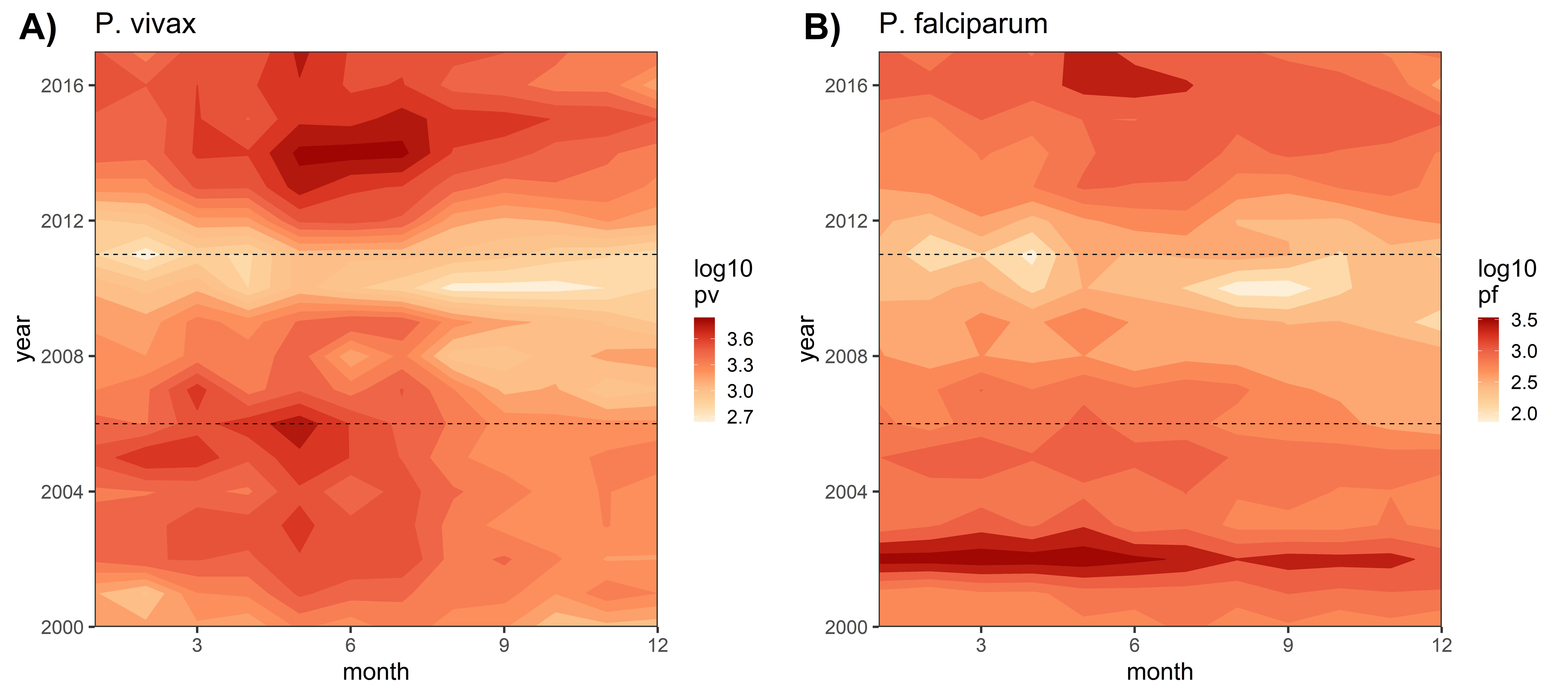
2.3 Figure 2 [COMPLETE] Seasonal and inter-annual trends of P.vivax and P.Falciparum
# Extra requirements
library(isoband)
library(ggisoband)
library(cowplot)
# Regional dataset
dt_region <- dat %>% group_by(year,month) %>%
summarise(pv=sum(vivax), pf=sum(falciparum), pop=sum(pop2015),
aet = mean(aet), prcp = mean(prcp), q = mean(q), soilm = mean(soilm), tmax = mean(tmax), tmin = mean(tmin), w.deff = mean(water_deficit),
loss = sum(loss), l.km2 = sum(loss_km2), cl.km2 = sum(cum_loss_km2))
f2a <- filled.contour.tidy(dt_region, month, year, pv, palette = "OrRd", dire = 1, hline = c(2006,2011), main= "P. vivax", b.size = 20)
f2b <- filled.contour.tidy(dt_region, month, year, pf, "OrRd", dire = 1, hline = c(2006,2011), main= "P. falciparum", b.size = 20)
# Final figure
fig2<-plot_grid(f2a,f2b, labels = c("A)","B)"), label_size=30)
fig2