Chapter 5 New Fidings
5.1 Finding the best model
5.1.1 Falciparum
library(tidyverse); library(useful);library(epiDisplay);library(lubridate); library(tictoc);library(usethis)
library(spdep); library(sf); library(sp); library(devtools); library(pkgload);library(readr);
library(stringr);library("viridis")
library(cowplot)
library(INLA)
library("devtools")
library("haven")
library("tidyverse")
library(Hmisc)
devtools::source_gist("https://gist.github.com/gcarrascoe/89e018d99bad7d3365ec4ac18e3817bd")
load("./CLIM_MAL_dat_20200826.RData")
dat<-dat %>%
mutate(q_std =((q - mean(q,na.rm=T)) / sd(q,na.rm=T)),
aet_std =((aet - mean(aet,na.rm=T)) / sd(aet,na.rm=T)),
soilm_std =((soilm - mean(soilm,na.rm=T)) / sd(soilm,na.rm=T)),
prcp_day_std =(((prcp/30) - mean((prcp/30),na.rm=T)) / sd((prcp/30),na.rm=T)),
tmax_std =((tmax - mean(tmax,na.rm=T)) / sd(tmax,na.rm=T)),
tmin_std =((tmin - mean(tmin,na.rm=T)) / sd(tmin,na.rm=T)),
cum_loss_km2_std =((cum_loss_km2 - mean(cum_loss_km2,na.rm=T)) / sd(cum_loss_km2,na.rm=T)),
water_deficit_std =((water_deficit - mean(water_deficit,na.rm=T)) / sd(water_deficit,na.rm=T)))
dat<- dat %>%
group_by(NOMBDIST) %>%
mutate(workers_lag_12=Lag(workers,+12),
nets_lag_12=Lag(nets,+12),
n_eess_lag_12=Lag(n_eess,+12),
enviro_lag_12=Lag(enviro,+12),
cum_loss_km2_std_lag_12=Lag(cum_loss_km2_std,+12),
#Lag1
soilm_std_lag1=Lag(soilm_std,+1),
q_std_lag1=Lag(q_std,+1),
aet_std_lag1=Lag(aet_std,+1),
tmax_std_lag1=Lag(tmax_std,+1),
tmin_std_lag1=Lag(tmin_std,+1),
prcp_day_std_lag1=Lag(prcp_day_std,+1),
water_deficit_std_lag1=Lag(water_deficit_std,+1),
#lag2
soilm_std_lag2=Lag(soilm_std,+2),
q_std_lag2=Lag(q_std,+2),
aet_std_lag2=Lag(aet_std,+2),
tmax_std_lag2=Lag(tmax_std,+2),
tmin_std_lag2=Lag(tmin_std,+2),
prcp_day_std_lag2=Lag(prcp_day_std,+2),
water_deficit_std_lag2=Lag(water_deficit_std,+2),
#lag3
soilm_std_lag3=Lag(soilm_std,+3),
q_std_lag3=Lag(q_std,+3),
aet_std_lag3=Lag(aet_std,+3),
tmax_std_lag3=Lag(tmax_std,+3),
tmin_std_lag3=Lag(tmin_std,+3),
prcp_day_std_lag3=Lag(prcp_day_std,+3),
water_deficit_std_lag3=Lag(water_deficit_std,+3)
)
dat.rev.vf<-dat %>%
dplyr::select(NOMBDIST,vivax,falciparum,index,year,month2,
prcp_day_std,tmax_std,
prcp_day_std_lag1,tmax_std_lag1,
prcp_day_std_lag2,tmax_std_lag2,
prcp_day_std_lag3,tmax_std_lag3,
cum_loss_km2_std,
n_eess,workers,enviro,nets,
n_eess_lag_12, workers_lag_12, enviro_lag_12, nets_lag_12,
pop2015,
water_deficit_std,water_deficit_std_lag1,
water_deficit_std_lag2,water_deficit_std_lag3,
soilm_std,aet_std,q_std,tmin_std,
soilm_std_lag1,aet_std_lag1,q_std_lag1,tmin_std_lag1,
soilm_std_lag2,aet_std_lag2,q_std_lag2,tmin_std_lag2,
soilm_std_lag3,aet_std_lag3,q_std_lag3,tmin_std_lag3
)
dim(dat.rev.vf)
dat.rev.vf.cplt<-dat.rev.vf[complete.cases(dat.rev.vf),]
dim(dat.rev.vf.cplt) #9996
##############################################################################
pf_explore <- c(
## models_pf_7
# 1
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
workers + enviro,
# 2
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12,
# 3
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std_lag1 + q_std_lag1 + aet_std_lag1,
# 4
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std + q_std + aet_std,
# 5
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 6
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 7
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std_lag1 + q_std_lag1 + aet_std_lag1,
# 8
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std + q_std + aet_std,
# 9
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 10
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 11
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std_lag1 + q_std_lag1 + aet_std_lag1,
# 12
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std + q_std + aet_std,
# 13
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 14
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 15
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std_lag1 + q_std_lag1 + aet_std_lag1,
# 16
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std + q_std + aet_std,
# 17
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 18
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 19
formula = falciparum ~ 1,
## models_pf_7_1
# 1
formula = falciparum ~ 1,
# 2
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 3
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1"),
# 4
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1"),
# 5
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1"),
# 6
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1"),
# 7
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1"),
# 8
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1"),
# 9
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 10
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 11
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 12
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 13
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 14
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 15
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 16
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 17
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 18
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 19
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 20
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 21
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 22
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 23
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 24
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 25
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 26
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 27
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
## models_pf_7.3
# 1
formula = falciparum ~ 1,
# 2
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 3
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 4
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 5
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 6
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 7
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 8
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 9
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1") +
f(inla.group(water_deficit), model = "rw1"),
# 10
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 11
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1") +
f(inla.group(water_deficit), model = "rw1"),
# 12
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 13
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 14
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 15
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 16
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 17
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 18
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 19
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 20
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 21
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 22
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 23
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 24
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 25
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 26
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 27
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
## models_pf_7_book
# 1
formula = falciparum ~ 1,
# 2
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 3
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
workers + enviro,
# 4
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12,
# 5
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
aet_std_lag1,
## models_pf_8
# 1
formula = falciparum ~ 1,
# 2
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1"),
# 3
formula = falciparum ~ 1 + f(index, model = "besag", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1"),
# 4
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2"),
# 5
formula = falciparum ~ 1 + f(index, model = "besag", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2"),
# 6
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2"),
# 7
formula = falciparum ~ 1 + f(index, model = "besag", graph = "map.graph") +
f(year, model = "rw1") + f(month2, model = "rw2"),
# 8
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "rw1") + f(month2, model = "rw2"),
## models_pf_8.1.2
# 1
formula = falciparum ~ 1,
# 2
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 3
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1"),
# 4
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1"),
# 5
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1"),
# 6
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag2), model = "rw1"),
# 7
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag2), model = "rw1"),
# 8
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag2), model = "rw1"),
# 9
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 10
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 11
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 12
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag2), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 13
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag2),
model = "rw1"),
# 14
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag2), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 15
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag2),
model = "rw1"),
# 16
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag2), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 17
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag2),
model = "rw1"),
# 18
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag2), model = "rw1") + f(inla.group(q_std_lag2),
model = "rw1"),
# 19
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag2), model = "rw1") + f(inla.group(aet_std_lag2),
model = "rw1"),
# 20
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag2), model = "rw1") + f(inla.group(aet_std_lag2),
model = "rw1"),
# 21
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag2), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 22
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag2),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 23
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag2), model = "rw1"),
# 24
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag2), model = "rw1") + f(inla.group(q_std_lag2),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 25
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag2), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag2), model = "rw1"),
# 26
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag2),
model = "rw1") + f(inla.group(aet_std_lag2), model = "rw1"),
# 27
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag2), model = "rw1") + f(inla.group(q_std_lag2),
model = "rw1") + f(inla.group(aet_std_lag2), model = "rw1"),
## models_pf_8.2
# 1
formula = falciparum ~ 1,
# 2
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 3
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1"),
# 4
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1"),
# 5
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1"),
# 6
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1"),
# 7
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1"),
# 8
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1"),
# 9
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 10
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 11
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 12
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 13
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 14
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 15
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 16
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 17
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 18
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 19
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 20
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 21
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 22
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 23
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 24
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 25
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 26
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 27
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1")
)
names(pf_explore)<-pf_explore
##############################################################################
inla.batch.cplt<-function(formula, dat1 = dat.rev.vf.cplt) {
result = inla(formula, data=dat1, family="nbinomial", offset=log(pop2015), verbose = F,
#control.inla=list(strategy="gaussian"),
control.inla=list(strategy="adaptive"),
control.compute=list(config=F, dic=T, cpo=T, waic=T,
return.marginals = FALSE),
control.fixed = list(correlation.matrix=T),
control.predictor=list(link=1,compute=TRUE)
)
return(result)
}
inla.batch.cplt.safe <- possibly(inla.batch.cplt, otherwise = NA_real_)
##############################################################################
# Running our models
tic()
INLA:::inla.dynload.workaround()
test_pf_explore<-pf_explore %>% purrr::map(~inla.batch.cplt.safe(formula = .))
toc()
test_pf_explore
saveRDS(test_pf_explore, file= "./_data/models_pf_explore.rds")
##############################################################################
# Choosing our best model
pf_explore.s <- pf_explore_check %>%
purrr::map(~Rsq.batch.safe(model = ., dic.null = pf_explore_check[[1]]$dic, n = n)) %>%
bind_rows(.id = "formula") %>% mutate(id = row_number())
pf_explore.s %>%
plot_score()5.1.2 Vivax
library(tidyverse); library(useful);library(epiDisplay);library(lubridate); library(tictoc);library(usethis)
library(spdep); library(sf); library(sp); library(devtools); library(pkgload);library(readr);
library(stringr);library("viridis")
library(cowplot)
library(INLA)
library("devtools")
library("haven")
library("tidyverse")
library(Hmisc)
devtools::source_gist("https://gist.github.com/gcarrascoe/89e018d99bad7d3365ec4ac18e3817bd")
load("./CLIM_MAL_dat_20200826.RData")
dat<-dat %>%
mutate(q_std =((q - mean(q,na.rm=T)) / sd(q,na.rm=T)),
aet_std =((aet - mean(aet,na.rm=T)) / sd(aet,na.rm=T)),
soilm_std =((soilm - mean(soilm,na.rm=T)) / sd(soilm,na.rm=T)),
prcp_day_std =(((prcp/30) - mean((prcp/30),na.rm=T)) / sd((prcp/30),na.rm=T)),
tmax_std =((tmax - mean(tmax,na.rm=T)) / sd(tmax,na.rm=T)),
tmin_std =((tmin - mean(tmin,na.rm=T)) / sd(tmin,na.rm=T)),
cum_loss_km2_std =((cum_loss_km2 - mean(cum_loss_km2,na.rm=T)) / sd(cum_loss_km2,na.rm=T)),
water_deficit_std =((water_deficit - mean(water_deficit,na.rm=T)) / sd(water_deficit,na.rm=T)))
dat<- dat %>%
group_by(NOMBDIST) %>%
mutate(workers_lag_12=Lag(workers,+12),
nets_lag_12=Lag(nets,+12),
n_eess_lag_12=Lag(n_eess,+12),
enviro_lag_12=Lag(enviro,+12),
cum_loss_km2_std_lag_12=Lag(cum_loss_km2_std,+12),
#Lag1
soilm_std_lag1=Lag(soilm_std,+1),
q_std_lag1=Lag(q_std,+1),
aet_std_lag1=Lag(aet_std,+1),
tmax_std_lag1=Lag(tmax_std,+1),
tmin_std_lag1=Lag(tmin_std,+1),
prcp_day_std_lag1=Lag(prcp_day_std,+1),
water_deficit_std_lag1=Lag(water_deficit_std,+1),
#lag2
soilm_std_lag2=Lag(soilm_std,+2),
q_std_lag2=Lag(q_std,+2),
aet_std_lag2=Lag(aet_std,+2),
tmax_std_lag2=Lag(tmax_std,+2),
tmin_std_lag2=Lag(tmin_std,+2),
prcp_day_std_lag2=Lag(prcp_day_std,+2),
water_deficit_std_lag2=Lag(water_deficit_std,+2),
#lag3
soilm_std_lag3=Lag(soilm_std,+3),
q_std_lag3=Lag(q_std,+3),
aet_std_lag3=Lag(aet_std,+3),
tmax_std_lag3=Lag(tmax_std,+3),
tmin_std_lag3=Lag(tmin_std,+3),
prcp_day_std_lag3=Lag(prcp_day_std,+3),
water_deficit_std_lag3=Lag(water_deficit_std,+3)
)
dat.rev.vf<-dat %>%
dplyr::select(NOMBDIST,vivax,falciparum,index,year,month2,
prcp_day_std,tmax_std,
prcp_day_std_lag1,tmax_std_lag1,
prcp_day_std_lag2,tmax_std_lag2,
prcp_day_std_lag3,tmax_std_lag3,
cum_loss_km2_std,
n_eess,workers,enviro,nets,
n_eess_lag_12, workers_lag_12, enviro_lag_12, nets_lag_12,
pop2015,
water_deficit_std,water_deficit_std_lag1,
water_deficit_std_lag2,water_deficit_std_lag3,
soilm_std,aet_std,q_std,tmin_std,
soilm_std_lag1,aet_std_lag1,q_std_lag1,tmin_std_lag1,
soilm_std_lag2,aet_std_lag2,q_std_lag2,tmin_std_lag2,
soilm_std_lag3,aet_std_lag3,q_std_lag3,tmin_std_lag3
)
dim(dat.rev.vf)
dat.rev.vf.cplt<-dat.rev.vf[complete.cases(dat.rev.vf),]
dim(dat.rev.vf.cplt) #9996
##############################################################################
pv_explore <- c(
## models_pv_7
# 1
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess + enviro + nets,
# 2
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12,
# 3
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std_lag1 + q_std_lag1 + aet_std_lag1,
# 4
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std + q_std + aet_std,
# 5
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 6
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 7
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std_lag1 + q_std_lag1 + aet_std_lag1,
# 8
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std + q_std + aet_std,
# 9
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 10
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 11
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std_lag1 + q_std_lag1 + aet_std_lag1,
# 12
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std + q_std + aet_std,
# 13
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 14
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 15
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std_lag1 + q_std_lag1 + aet_std_lag1,
# 16
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
soilm_std + q_std + aet_std,
# 17
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 18
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 19
formula = vivax ~ 1,
## models_pv_7_1
# 1
formula = vivax ~ 1,
# 2
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 3
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1"),
# 4
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1"),
# 5
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1"),
# 6
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1"),
# 7
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1"),
# 8
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1"),
# 9
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 10
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 11
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 12
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 13
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 14
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 15
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 16
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 17
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 18
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 19
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 20
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 21
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 22
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 23
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 24
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 25
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 26
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 27
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
## models_pv_7_2
# 1
formula = vivax ~ 1,
# 2
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 3
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1"),
# 4
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1"),
# 5
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1"),
# 6
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1"),
# 7
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1"),
# 8
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1"),
# 9
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 10
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 11
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 12
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 13
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 14
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 15
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 16
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 17
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 18
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 19
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 20
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 21
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 22
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 23
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 24
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 25
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 26
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 27
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
## models_pv_7_3
# 1
formula = vivax ~ 1,
# 2
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 3
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 4
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 5
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 6
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 7
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 8
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 9
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1") + f(inla.group(water_deficit),
model = "rw1"),
# 10
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 11
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1") + f(inla.group(water_deficit),
model = "rw1"),
# 12
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 13
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 14
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 15
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 16
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 17
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 18
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 19
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 20
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1") + f(inla.group(water_deficit_std_lag1),
model = "rw1"),
# 21
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 22
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 23
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 24
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 25
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 26
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
# 27
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(water_deficit_std_lag1), model = "rw1"),
## models_pv_7_book
# 1
formula = vivax ~ 1,
# 2
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 3
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
nets + n_eess + enviro,
# 4
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12,
# 5
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
aet_std_lag1,
## models_pv_8_2
# 1
formula = vivax ~ 1,
# 2
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 3
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1"),
# 4
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1"),
# 5
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1"),
# 6
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1"),
# 7
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1"),
# 8
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1"),
# 9
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 10
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 11
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 12
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 13
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 14
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 15
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 16
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 17
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 18
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 19
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 20
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 21
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 22
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 23
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 24
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 25
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 26
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 27
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
## models_pv_8_2_2
# 1
formula = vivax ~ 1,
# 2
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 3
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1"),
# 4
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1"),
# 5
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1"),
# 6
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag3), model = "rw1"),
# 7
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag3), model = "rw1"),
# 8
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag3), model = "rw1"),
# 9
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 10
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 11
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 12
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag3), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 13
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag3),
model = "rw1"),
# 14
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag3), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 15
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag3),
model = "rw1"),
# 16
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag3), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 17
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag3),
model = "rw1"),
# 18
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag3), model = "rw1") + f(inla.group(q_std_lag3),
model = "rw1"),
# 19
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag3), model = "rw1") + f(inla.group(aet_std_lag3),
model = "rw1"),
# 20
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag3), model = "rw1") + f(inla.group(aet_std_lag3),
model = "rw1"),
# 21
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag3), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 22
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag3),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 23
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag3), model = "rw1"),
# 24
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag3), model = "rw1") + f(inla.group(q_std_lag3),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 25
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag3), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag3), model = "rw1"),
# 26
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag3),
model = "rw1") + f(inla.group(aet_std_lag3), model = "rw1"),
# 27
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag3), model = "rw1") + f(inla.group(q_std_lag3),
model = "rw1") + f(inla.group(aet_std_lag3), model = "rw1"),
## models_pv_8_3
# 1
formula = vivax ~ 1,
# 2
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 3
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1"),
# 4
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1"),
# 5
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1"),
# 6
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1"),
# 7
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1"),
# 8
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1"),
# 9
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 10
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 11
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 12
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 13
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 14
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 15
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 16
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 17
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 18
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 19
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 20
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 21
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 22
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 23
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 24
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 25
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 26
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 27
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
## models_pv_8_4
# 1
formula = vivax ~ 1,
# 2
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 3
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1"),
# 4
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1"),
# 5
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1"),
# 6
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1"),
# 7
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1"),
# 8
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1"),
# 9
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 10
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 11
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 12
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 13
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 14
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 15
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 16
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 17
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 18
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 19
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 20
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 21
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 22
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 23
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 24
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 25
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 26
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 27
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
## models_pv_8_6
# 1
formula = vivax ~ 1,
# 2
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 3
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1"),
# 4
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1"),
# 5
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1"),
# 6
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1"),
# 7
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1"),
# 8
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1"),
# 9
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 10
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 11
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 12
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 13
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 14
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 15
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 16
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 17
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 18
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 19
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 20
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 21
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 22
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 23
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 24
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 25
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 26
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 27
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
## models_pv_8_9
# 1
formula = vivax ~ 1,
# 2
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 3
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1"),
# 4
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std), model = "rw1"),
# 5
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1"),
# 6
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1"),
# 7
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1"),
# 8
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1"),
# 9
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 10
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 11
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 12
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 13
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 14
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std),
model = "rw1"),
# 15
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 16
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# 17
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 18
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1"),
# 19
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 20
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
# 21
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 22
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 23
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 24
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std), model = "rw1"),
# 25
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 26
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1"),
# 27
formula = vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmin_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(soilm_std_lag1), model = "rw1") + f(inla.group(q_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1")
)
names(pv_explore)<-pv_explore
##############################################################################
inla.batch.cplt<-function(formula, dat1 = dat.rev.vf.cplt) {
result = inla(formula, data=dat1, family="nbinomial", offset=log(pop2015), verbose = F,
#control.inla=list(strategy="gaussian"),
control.inla=list(strategy="adaptive"),
control.compute=list(config=F, dic=T, cpo=T, waic=T,
return.marginals = FALSE),
control.fixed = list(correlation.matrix=T),
control.predictor=list(link=1,compute=TRUE)
)
return(result)
}
inla.batch.cplt.safe <- possibly(inla.batch.cplt, otherwise = NA_real_)
##############################################################################
# Running our models
tic()
INLA:::inla.dynload.workaround()
test_pv_explore<-pv_explore %>% purrr::map(~inla.batch.cplt.safe(formula = .))
toc()
test_pv_explore
saveRDS(test_pv_explore, file= "./_data/models_pv_explore.rds")
##############################################################################
# Choosing our best model
pv_explore.s <- pv_explore_check %>%
purrr::map(~Rsq.batch.safe(model = ., dic.null = pv_explore_check[[1]]$dic, n = n)) %>%
bind_rows(.id = "formula") %>% mutate(id = row_number())
pv_explore.s %>%
plot_score()5.2 Final Models
5.2.1 Falciparum
1. Spatio-temporal model
\[ \begin{aligned} Falciparum = &\alpha + \beta_{t(m(rw1))} + \gamma_{t(a)} +\phi_{s} +\nu_{s} \\ \end{aligned} \]
2. Climate model
\[ \begin{aligned} Falciparum = &\alpha + \beta_{t(m(rw1))} + \gamma_{t(a)} + \phi_{s} + \nu_{s} \\ &+f(prcp) +f(t_{max,lag1}) +f(runoff_{lag1}) +f(aet_{lag1}) \end{aligned} \]
3. Malaria-control model
\[ \begin{aligned} Falciparum = &\alpha + \beta_{t(m(rw1))} + \gamma_{t(a)} + \phi_{s} + \nu_{s} \\ &+f(prcp) +f(t_{max,lag1}) +f(runoff_{lag1}) +f(aet_{lag1}) \\ &+enviro_{lag12} + nets_{lag12} + hfd_{lag12} + workers_{lag12} \end{aligned} \]
4. Full model
\[ \begin{aligned} Falciparum = &\alpha + \beta_{t(m(rw1))} + \gamma_{t(a)} + \phi_{s} + \nu_{s} \\ &+f({cumloss\ km^2}) +f(prcp) +f(t_{max,lag1}) +f(runoff_{lag1}) +f(aet_{lag1}) \\ &+enviro_{lag12} + nets_{lag12} + hfd_{lag12} + workers_{lag12} \end{aligned} \]
# P. Falciparum
# 1. Spatio-temporal model
falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1")
# 2. Climate model
falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(q_std_lag1), model = "rw1")
# 3. Malaria-control model
falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(q_std_lag1), model = "rw1") + n_eess_lag_12 +
workers_lag_12 + enviro_lag_12 + nets_lag_12
# 4. Full model
falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1")5.2.2 Vivax
1. Spatio-temporal model
\[ \begin{aligned} Vivax = &\alpha + \beta_{t(m(rw2))} + \gamma_{t(a)} +\phi_{s} +\nu_{s} \\ \end{aligned} \]
2. Climate model
\[ \begin{aligned} Vivax = &\alpha + \beta_{t(m(rw2))} + \gamma_{t(a)} +\phi_{s} +\nu_{s} \\ &+f(prcp_{lag1}) +f(t_{max}) +f(aet_{lag1}) +f(runoff) \end{aligned} \]
3. Malaria-control model
\[ \begin{aligned} Vivax = &\alpha + \beta_{t(m(rw2))} + \gamma_{t(a)} +\phi_{s} +\nu_{s} \\ &+f(prcp_{lag1}) +f(t_{max}) +f(aet_{lag1}) +f(runoff) \\ &+enviro_{lag12} + nets_{lag12}+ hfd_{lag12} + workers_{lag12} \end{aligned} \]
4. Full model
\[ \begin{aligned} Vivax = &\alpha + \beta_{t(m(rw2))} + \gamma_{t(a)} +\phi_{s} +\nu_{s} \\ & +f({cumloss\ km^2}) +f(prcp_{lag1}) +f(t_{max}) +f(aet_{lag1}) +f(runoff) \\ &+enviro_{lag12} + nets_{lag12}+ hfd_{lag12} + workers_{lag12} \end{aligned} \]
# P. Vivax
# 1. Spatio-temporal model
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2")
# 2. Climate model
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(q_std), model = "rw1")
# 3. Malaria-control model
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + n_eess_lag_12 + workers_lag_12 + enviro_lag_12 +
nets_lag_12 + f(inla.group(aet_std_lag1), model = "rw1") +
f(inla.group(q_std), model = "rw1")
# 4. Full model
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1")5.3 Selection Model
pf_check7.s<-test_pf_7_back_3[c(2,3,4,5)]%>%
purrr::map(~Rsq.batch.safe(model = ., dic.null = test_pf_7_back_3[[1]]$dic, n = n)) %>%
bind_rows(.id = "formula") %>% mutate(id = row_number())
pv_check7.s<-test_pv_8.3_back_2[c(2,3,4,5)]%>%
purrr::map(~Rsq.batch.safe(model = ., dic.null = test_pv_8.3_back_2[[1]]$dic, n = n)) %>%
bind_rows(.id = "formula") %>% mutate(id = row_number())
a.falciparum<-pf_check7.s %>%
gather(key, value, -formula)%>%
ungroup() %>%
group_by(key) %>%
filter(!(key %in% c("Rsq", "log score","id","pD","waic pD")))
a.falciparum_new<- a.falciparum %>%
group_by(key) %>%
mutate(position = rank(-value))
a.vivax<-pv_check7.s %>%
gather(key, value, -formula)%>%
ungroup() %>%
group_by(key) %>%
filter(!(key %in% c("Rsq", "log score","id","pD","waic pD")))
a.vivax_new<- a.vivax %>%
group_by(key) %>%
mutate(position = rank(-value))
graf.falciparum<- a.falciparum_new %>% ggplot(aes(x=key, y=value, fill=reorder(formula, -value), group = position)) +
geom_bar(stat="identity", position="dodge") +
theme(axis.title.y=element_blank(),
axis.title.x=element_blank(),
axis.ticks.x=element_blank(),
panel.background =element_blank(),
panel.grid.major=element_blank(),
panel.grid.minor=element_blank(),
strip.background =element_blank(),
strip.text = element_text(face="bold",size=12),
plot.title = element_text(face = "bold"))+
coord_cartesian(ylim=c(44000,46500))+
ggtitle("Falciparum: Selection model ", subtitle = "by dic and waic information criteria") +
scale_fill_manual(values=c("#86BB8D", "#4c4c4c", "#68a4bd", "#ff9900"),
labels = c("Spatio-temporal model", "Climate model", "Malaria-control model", "Full model") ,name="Model")
graf.vivax<- a.vivax_new %>% ggplot(aes(x=key, y=value, fill=reorder(formula, -value), group = position)) +
geom_bar(stat="identity", position="dodge") +
theme(axis.title.y=element_blank(),
axis.title.x=element_blank(),
axis.ticks.x=element_blank(),
panel.background =element_blank(),
panel.grid.major=element_blank(),
panel.grid.minor=element_blank(),
strip.background =element_blank(),
strip.text = element_text(face="bold",size=12),
plot.title = element_text(face = "bold"))+
coord_cartesian(ylim=c(65000,72000))+
ggtitle("Vivax: Selection model ", subtitle = "by dic and waic information criteria") +
scale_fill_manual(values=c("#86BB8D", "#4c4c4c", "#68a4bd", "#ff9900"),
labels = c("Spatio-temporal model", "Climate model", "Malaria-control model", "Full model") ,name="Model")
library(cowplot)
(sf_5<-plot_grid(graf.vivax,graf.falciparum, nrow = 1, ncol=2, labels = c("A)", "B)"), label_size=15, align = "hv", rel_widths = 1))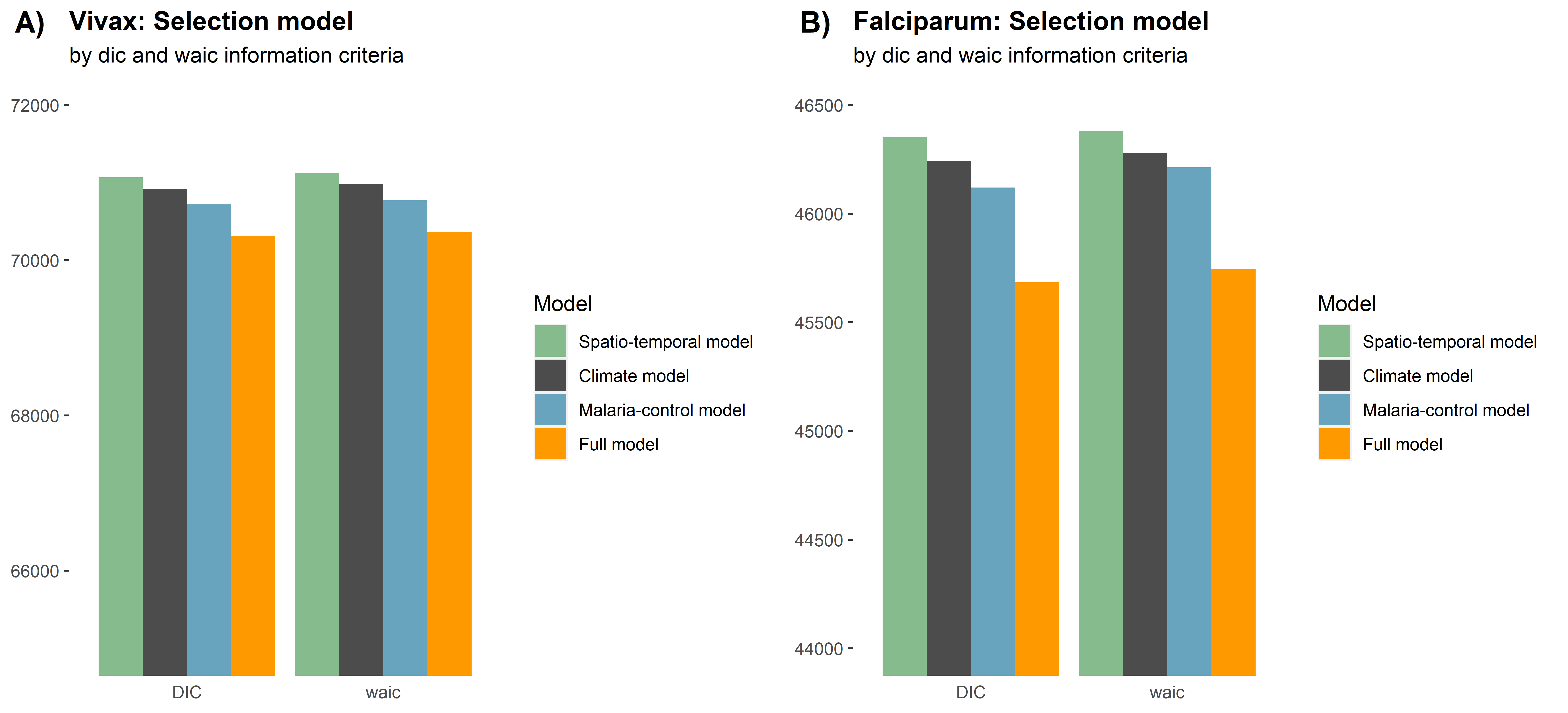
5.4 Fixed Effects
5.4.1 Nonlinear model
# Antiguo
# P. Vivax
sv_7_pv <- test_pv_8.3_back_2[c(5)] %>%
plot_fixed.safe(title = "P. Vivax",
filter=5,
legend = F,
ylim = 6,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12"),
xlab = c("Lag of workers","Lag of nets","Lag of hfd","Lag of enviro"),
breaks = c("1","2"),
lab_mod = c("Full model", ""),
flip=T)+
ylab("Incidence Rate Ratio (IRR)")+
theme(plot.margin = unit(c(1,1,1,1), "lines"))
# P. Falciparum
sf_7_pf <- test_pf_7_back_3[c(5)] %>%
plot_fixed.safe(title = "P. Falciparum",
filter=5,
legend = F,
ylim = 6,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12"),
xlab = c("Lag of workers","Lag of nets","Lag of hfd","Lag of enviro"),
breaks = c("1","2"),
lab_mod = c("Full model", ""),
flip=T)+
ylab("Incidence Rate Ratio (IRR)")+
theme(plot.margin = unit(c(1,1,1,1), "lines"))
library(cowplot)
(sf_6<-plot_grid(sv_7_pv, sf_7_pf, nrow = 1, ncol=2, labels = c("A)", "B)"), label_size=30, align = "hv", rel_widths = 1))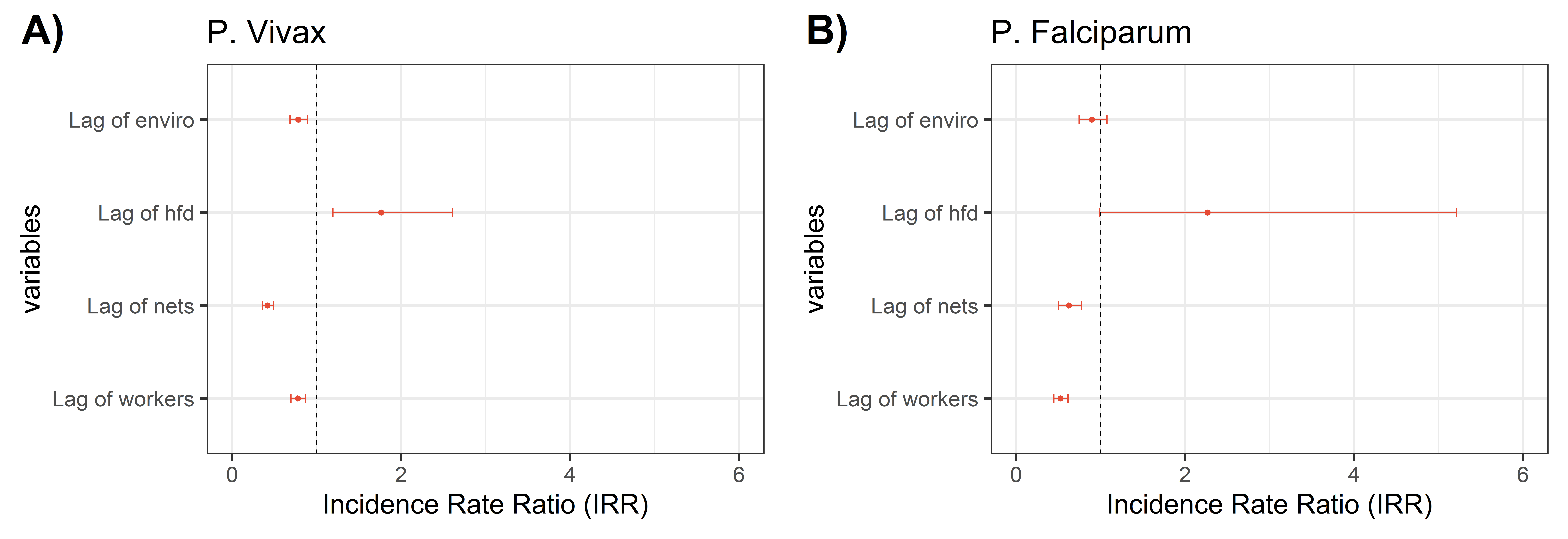
#Requerimientos
library(tidyverse)
library(INLA)
#Graficos sin exponenciar
plot.log.fixed<-function(model, label = id, col1="black", dire=-1, breaks, lab_mod, lim, title, legend=NULL, filter, ylab = "Relative risk", ylim = 10, angle=45, hjust = 1, xlabs=lim, b.size=20, flip=F) {
library(ggsci)
label <- enquo(label)
extract_dat <- function(model) {
mydata <- model$summary.fixed %>%
rownames_to_column(var = "var")
return(mydata)
}
dat_na <- data.frame(var=NA)
extract_dat.safe <- possibly(extract_dat, otherwise = dat_na)
fixed_log_plot <- model %>% purrr::map(~extract_dat.safe(model=.)) %>%
bind_rows(.id = "formula") %>%
mutate(id = match(formula, unique(formula))) %>%
#mutate(id = group_indices(., factor(formula, levels = unique(formula)))) %>%
filter(!is.na(`0.025quant`), `0.5quant`<filter) %>%
ggplot(aes(x = var, y = `0.5quant`, col=factor(id))) +
geom_point(position = position_dodge(0.3)) +
geom_errorbar(aes(ymin = `0.025quant`, ymax = `0.975quant`), width=.1, position = position_dodge(0.3)) +
geom_hline(aes(yintercept = 0), col=col1, linetype="dashed", size=.4) +
#scale_color_discrete(name="Models", breaks=breaks, labels=lab_mod) +
ggsci::scale_color_npg(name="Models", breaks=breaks, labels=lab_mod) +
scale_x_discrete(limits=lim, labels = xlabs) +
ylim(-3, ylim) +
{if(!is.null(legend)) {guides(color = legend)}} +
labs(title=title, x = "variables") +
theme_bw(base_size = b.size) +
labs(y=ylab) +
{if(isTRUE(flip)) {coord_flip()}} +
theme(axis.text.x = element_text(angle = angle, hjust = hjust))
return(fixed_log_plot)
}
plot.log.fixed.safe <- possibly(plot.log.fixed, otherwise = NA_real_)
# P. Vivax
sv_log_pv <- test_pv_8.3_back_2[c(5)] %>%
plot.log.fixed.safe(title = "P. Vivax",
filter=8,
legend = F,
ylim = 3,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12"),
xlab = c("Lag of workers","Lag of nets","Lag of hfd","Lag of enviro"),
breaks = c("1","2"),
lab_mod = c("Full model", ""),
flip=T)+
ylab("Log (IR)")+
theme(plot.margin = unit(c(1,1,1,1), "lines"))
# P. Falciparum
sf_log_pf <- test_pf_7_back_3[c(5)] %>%
plot.log.fixed.safe(title = "P. Falciparum",
filter=8,
legend = F,
ylim = 3,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12"),
xlab = c("Lag of workers","Lag of nets","Lag of hfd","Lag of enviro"),
breaks = c("1","2"),
lab_mod = c("Full model", ""),
flip=T)+
ylab("Log (IR)")+
theme(plot.margin = unit(c(1,1,1,1), "lines"))
library(cowplot)
(sf_6<-plot_grid(sv_log_pv, sf_log_pf, nrow = 1, ncol=2, labels = c("A)", "B)"), label_size=30, align = "hv", rel_widths = 1))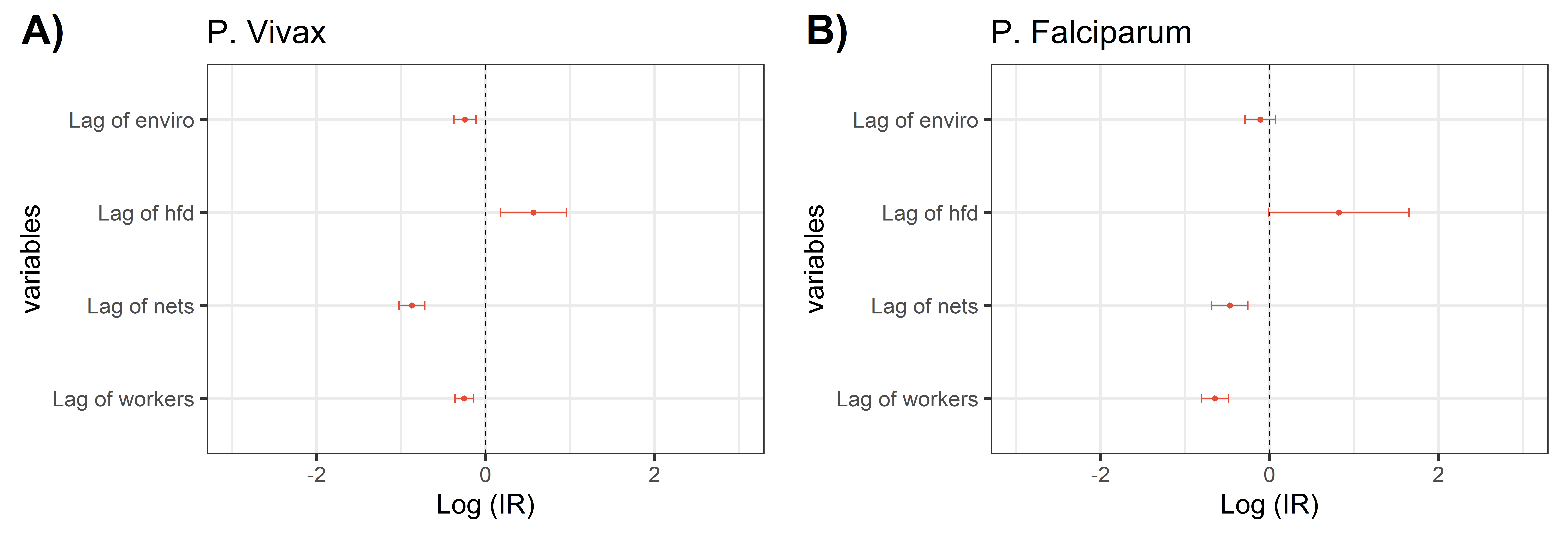
5.4.2 Linear model
#Antiguo
# P. Vivax
sv_7_pv <- test_pv_backlineal[c(5)] %>%
plot_fixed.safe(title = "P. Vivax",
filter=5,
legend = F,
ylim = 6,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12",
"prcp_day_std_lag1","tmax_std",
"cum_loss_km2_std","q_std",
"aet_std_lag1"),
xlab = c("Lag of workers","Lag of nets",
"Lag of hfd","Lag of enviro",
"Lag precipitation","Max temperature",
"Cum loss","Runoff",
"Lag aet"),
breaks = c("1","2"),
lab_mod = c("Full model", ""),
flip=T)+
ylab("Incidence Rate Ratio (IRR)")+
theme(plot.margin = unit(c(1,1,1,1), "lines"))
# P. Falciparum
sf_7_pf <- test_pf_backlineal[c(5)] %>%
plot_fixed.safe(title = "P. Falciparum",
filter=5,
legend = F,
ylim = 6,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12",
"prcp_day_std","tmax_std_lag1",
"cum_loss_km2_std","q_std_lag1",
"aet_std_lag1"),
xlab = c("Lag of workers","Lag of nets",
"Lag of hfd","Lag of enviro",
"Precipitation","Lag of max temperature",
"Cum loss","Lag of runoff",
"Lag of aet"),
breaks = c("1","2"),
lab_mod = c("Full model", ""),
flip=T)+
ylab("Incidence Rate Ratio (IRR)")+
theme(plot.margin = unit(c(1,1,1,1), "lines"))
library(cowplot)
(sf_6<-plot_grid(sv_7_pv, sf_7_pf, nrow = 1, ncol=2, labels = c("A)", "B)"), label_size=30, align = "hv", rel_widths = 1))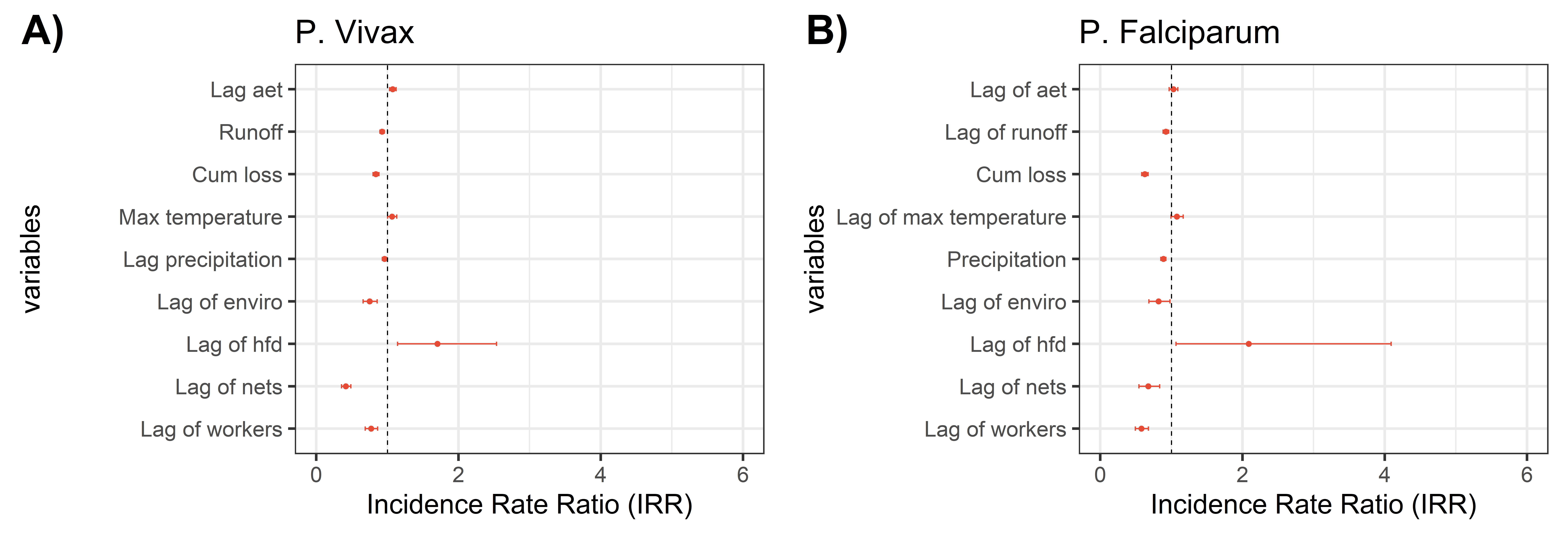
#Requerimientos
library(tidyverse)
library(INLA)
#Graficos sin exponenciar
plot.log.fixed<-function(model, label = id, col1="black", dire=-1, breaks, lab_mod, lim, title, legend=NULL, filter, ylab = "Relative risk", ylim = 10, angle=45, hjust = 1, xlabs=lim, b.size=20, flip=F) {
library(ggsci)
label <- enquo(label)
extract_dat <- function(model) {
mydata <- model$summary.fixed %>%
rownames_to_column(var = "var")
return(mydata)
}
dat_na <- data.frame(var=NA)
extract_dat.safe <- possibly(extract_dat, otherwise = dat_na)
fixed_log_plot <- model %>% purrr::map(~extract_dat.safe(model=.)) %>%
bind_rows(.id = "formula") %>%
mutate(id = match(formula, unique(formula))) %>%
#mutate(id = group_indices(., factor(formula, levels = unique(formula)))) %>%
filter(!is.na(`0.025quant`), `0.5quant`<filter) %>%
ggplot(aes(x = var, y = `0.5quant`, col=factor(id))) +
geom_point(position = position_dodge(0.3)) +
geom_errorbar(aes(ymin = `0.025quant`, ymax = `0.975quant`), width=.1, position = position_dodge(0.3)) +
geom_hline(aes(yintercept = 0), col=col1, linetype="dashed", size=.4) +
#scale_color_discrete(name="Models", breaks=breaks, labels=lab_mod) +
ggsci::scale_color_npg(name="Models", breaks=breaks, labels=lab_mod) +
scale_x_discrete(limits=lim, labels = xlabs) +
ylim(-3, ylim) +
{if(!is.null(legend)) {guides(color = legend)}} +
labs(title=title, x = "variables") +
theme_bw(base_size = b.size) +
labs(y=ylab) +
{if(isTRUE(flip)) {coord_flip()}} +
theme(axis.text.x = element_text(angle = angle, hjust = hjust))
return(fixed_log_plot)
}
plot.log.fixed.safe <- possibly(plot.log.fixed, otherwise = NA_real_)
# P. Vivax
sv_log_pv <- test_pv_backlineal[c(5)] %>%
plot.log.fixed.safe(title = "P. Vivax",
filter=8,
legend = F,
ylim = 3,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12",
"prcp_day_std_lag1","tmax_std",
"cum_loss_km2_std","q_std",
"aet_std_lag1"),
xlab = c("Lag of workers","Lag of nets",
"Lag of hfd","Lag of enviro",
"Lag precipitation","Max temperature",
"Cum loss","Runoff",
"Lag aet"),
breaks = c("1","2"),
lab_mod = c("Full model", ""),
flip=T)+
ylab("Log (IR)")+
theme(plot.margin = unit(c(1,1,1,1), "lines"))
# P. Falciparum
sf_log_pf <- test_pf_backlineal[c(5)] %>%
plot.log.fixed.safe(title = "P. Falciparum",
filter=8,
legend = F,
ylim = 3,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12",
"prcp_day_std","tmax_std_lag1",
"cum_loss_km2_std","q_std_lag1",
"aet_std_lag1"),
xlab = c("Lag of workers","Lag of nets",
"Lag of hfd","Lag of enviro",
"Precipitation","Lag of max temperature",
"Cum loss","Lag of runoff",
"Lag of aet"),
breaks = c("1","2"),
lab_mod = c("Full model", ""),
flip=T)+
ylab("Log (IR)")+
theme(plot.margin = unit(c(1,1,1,1), "lines"))
library(cowplot)
(sf_6<-plot_grid(sv_log_pv, sf_log_pf, nrow = 1, ncol=2, labels = c("A)", "B)"), label_size=30, align = "hv", rel_widths = 1))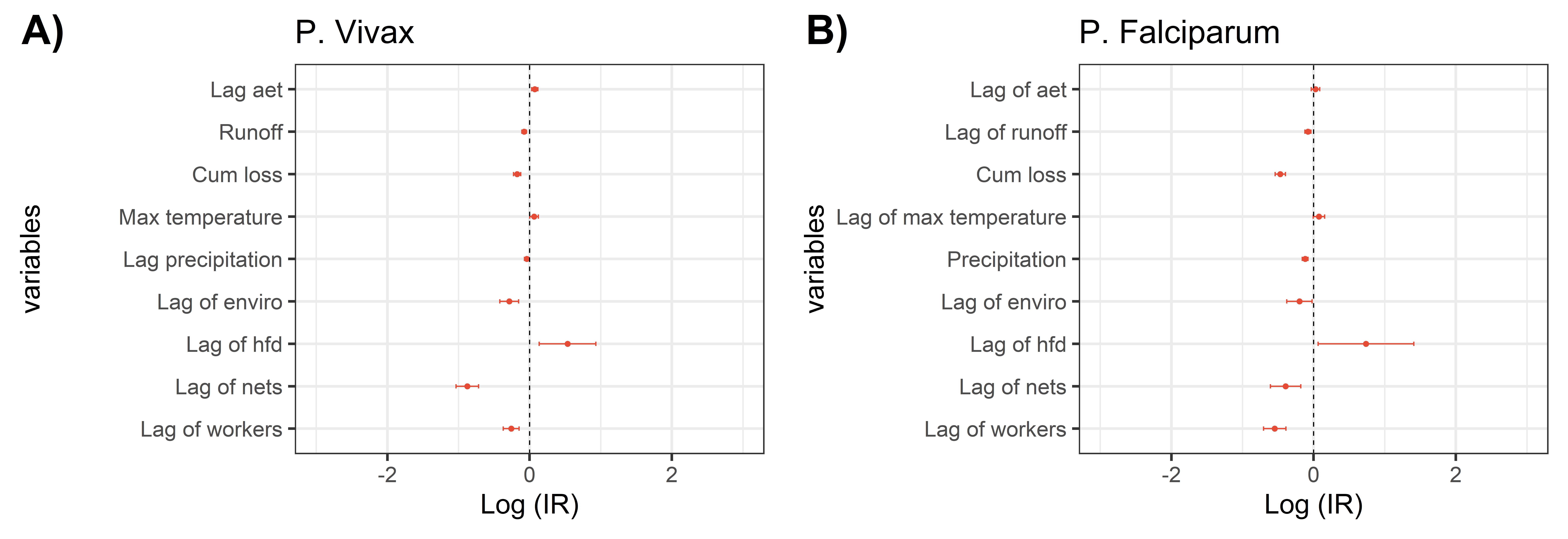
5.5 Figure 4 Spatial heterogeneity of covariates effects in terms of mean absolute error
library(colorspace)
library(sf)
library(ggthemes)
library(Hmisc)
dat<-dat %>%
mutate(q_std =((q - mean(q,na.rm=T)) / sd(q,na.rm=T)),
aet_std =((aet - mean(aet,na.rm=T)) / sd(aet,na.rm=T)),
soilm_std =((soilm - mean(soilm,na.rm=T)) / sd(soilm,na.rm=T)),
prcp_day_std =(((prcp/30) - mean((prcp/30),na.rm=T)) / sd((prcp/30),na.rm=T)),
tmax_std =((tmax - mean(tmax,na.rm=T)) / sd(tmax,na.rm=T)),
tmin_std =((tmin - mean(tmin,na.rm=T)) / sd(tmin,na.rm=T)),
cum_loss_km2_std =((cum_loss_km2 - mean(cum_loss_km2,na.rm=T)) / sd(cum_loss_km2,na.rm=T)),
water_deficit_std =((water_deficit - mean(water_deficit,na.rm=T)) / sd(water_deficit,na.rm=T)))
dat<- dat %>%
group_by(NOMBDIST) %>%
mutate(workers_lag_12=Lag(workers,+12),
nets_lag_12=Lag(nets,+12),
n_eess_lag_12=Lag(n_eess,+12),
enviro_lag_12=Lag(enviro,+12),
cum_loss_km2_std_lag_12=Lag(cum_loss_km2_std,+12),
soilm_std_lag1=Lag(soilm_std,+1),
q_std_lag1=Lag(q_std,+1),
aet_std_lag1=Lag(aet_std,+1),
tmax_std_lag1=Lag(tmax_std,+1),
tmin_std_lag1=Lag(tmin_std,+1),
prcp_day_std_lag1=Lag(prcp_day_std,+1),
water_deficit_std_lag1=Lag(water_deficit_std,+1))
dat.rev.vf<-dat %>%
dplyr::select(NOMBDIST,vivax,falciparum,index,year,month2,
prcp_day_std,prcp_day_std_lag1,tmax_std,tmax_std_lag1,
cum_loss_km2_std,
n_eess,workers,enviro,nets,
n_eess_lag_12, workers_lag_12, enviro_lag_12,
nets_lag_12,pop2015,water_deficit_std,water_deficit_std_lag1,
soilm_std,aet_std,q_std,tmin_std,
soilm_std_lag1,aet_std_lag1,q_std_lag1,tmin_std_lag1)
dat.rev.vf.cplt<-dat.rev.vf[complete.cases(dat.rev.vf),]
###########################################
# P. Vivax
###########################################
pv_8.ma<- c(
# clima
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12,
# control-malaria
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# deforestacion
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# full
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1")
)
###########################################
# P. Falciparum
###########################################
pf_7.ma<-c(
#Full model
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
#clima
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(cum_loss_km2_std), model = "rw1") + n_eess_lag_12 +
workers_lag_12 + enviro_lag_12 + nets_lag_12,
#controles malaria
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
#deforestacion
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + n_eess_lag_12 + workers_lag_12 + enviro_lag_12 +
nets_lag_12 + f(inla.group(q_std_lag1), model = "rw1") +
f(inla.group(aet_std_lag1), model = "rw1")
)
# P. Vivax#######################################################################
f3_a<-plot_mae2(dat1 = dat.rev.vf.cplt, var = vivax, full = test_pv_8_mae[[4]],
model1 = test_pv_8_mae[[1]], model2 = test_pv_8_mae[[2]], model3 = test_pv_8_mae[[3]],
map = area.sf, id = NOMBDIST)
dat.f3a <- f3_a[[3]] %>%
dplyr::select(NOMBDIST, m1:m3, i1:i3) %>%
dplyr::rename(climate_val=m1, `malaria-control_val`=m2, deforestation_val=m3,
climate_i=i1, `malaria-control_i`=i2, deforestation_i=i3) %>%
gather(type, val, climate_val:deforestation_i) %>%
separate(type, into = c("type","i"), sep = "_") %>%
spread(i,val) %>%
mutate(i = as.numeric(i),
i = ifelse(i<0,NA,i),
i = ifelse(i>100,NA,i)
) %>%
mutate(val = ifelse(val=="No Added", val, "Added"))
f3a <- dat.f3a %>%
ggplot() +
geom_sf(aes(fill=i)) +
scale_fill_continuous_sequential(palette="blues", na="white", trans="pseudo_log",
breaks = c(1,3,8,15,30), labels = c("No \nadded","3","8","15","30")) + labs(fill = "Added value: MAE(diff) \n ") +
theme_few(base_size = 20) +
theme(legend.position = "top",
panel.spacing = unit(2, "lines"),
legend.text = element_text(size = 10)) +
facet_wrap(.~type)
# P. Falciparum###################################################
f3_b<-plot_mae2(dat1 = dat.rev.vf.cplt, var = falciparum, full = test_pf_7_mae[[1]],
model1 = test_pf_7_mae[[2]], model2 = test_pf_7_mae[[3]], model3 = test_pf_7_mae[[4]],
map = area.sf, id = NOMBDIST)
dat.f3b <- f3_b[[3]] %>%
dplyr::select(NOMBDIST, m1:m3, i1:i3) %>%
dplyr::rename(climate_val=m1, `malaria-control_val`=m2, deforestation_val=m3,
climate_i=i1, `malaria-control_i`=i2, deforestation_i=i3) %>%
gather(type, val, climate_val:deforestation_i) %>%
separate(type, into = c("type","i"), sep = "_") %>%
spread(i,val) %>%
mutate(i = as.numeric(i),
i = ifelse(i<0,NA,i),
i = ifelse(i>100,NA,i)
) %>%
mutate(val = ifelse(val=="No Added", val, "Added"))
f3b <- dat.f3b %>%
ggplot() +
geom_sf(aes(fill=i)) +
scale_fill_continuous_sequential(palette="blues", na="white", trans="pseudo_log",
breaks = c(1,3,8,15,30), labels = c("No \nadded","3","8","15","30")) + labs(fill = "Added value: MAE(diff) \n ") +
theme_few(base_size = 20) +
theme(legend.position = "top",
panel.spacing = unit(2, "lines"),
legend.text = element_text(size = 10)) +
facet_wrap(.~type)
library(cowplot)
(figure4 <- plot_grid(f3a,f3b, nrow = 2, labels = c("A)","B)"), label_size=30))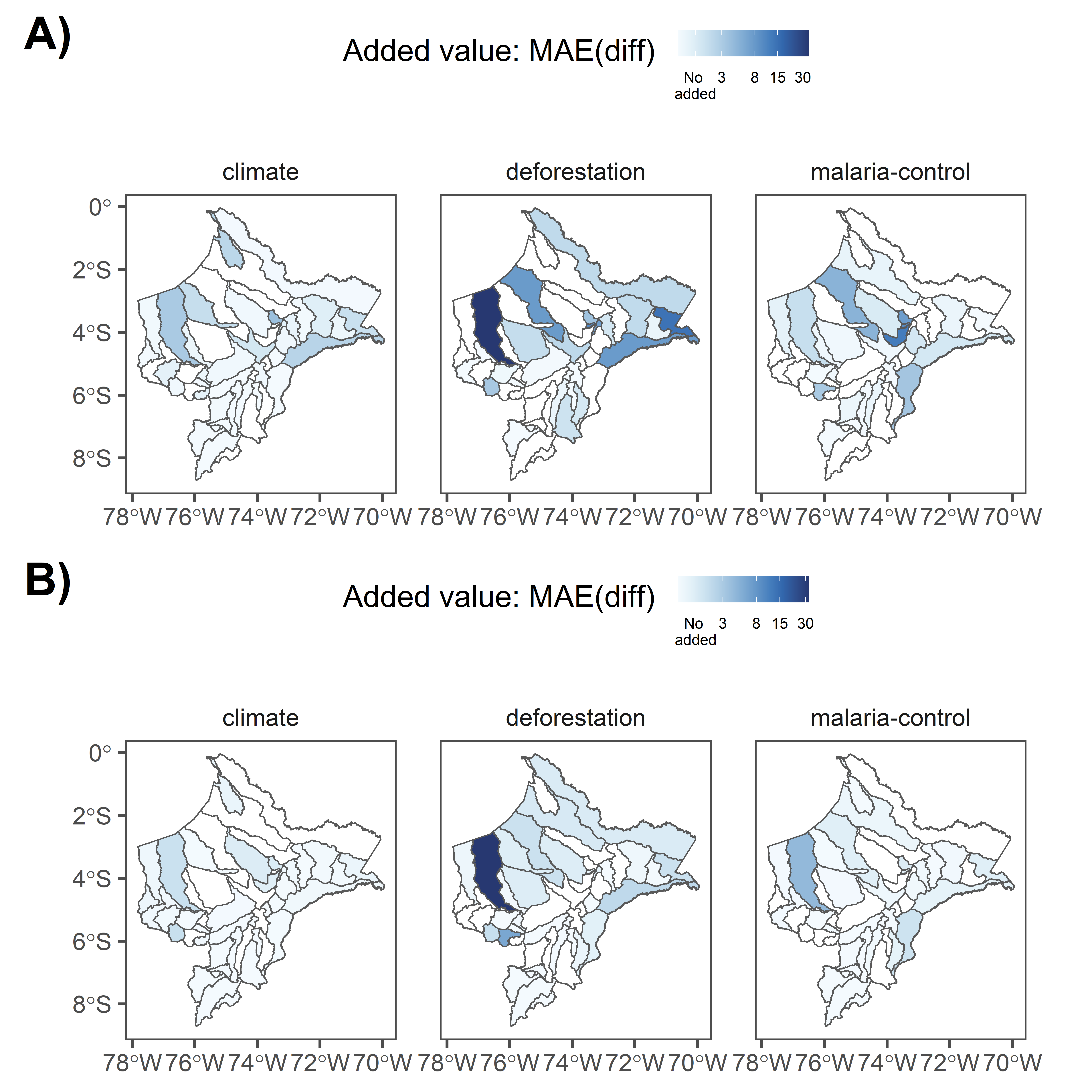
5.6 MAE: Dumbbell plot
# P. Falciparum
dat2 <-dat.rev.vf.cplt
full <-test_pf_7_mae[[1]]
model1<-test_pf_7_mae[[2]]
model2<-test_pf_7_mae[[3]]
model3<-test_pf_7_mae[[4]]
dat2$fit.full <- full$summary.fitted.values$`0.5quant`
dat2$fit.m1 <- model1$summary.fitted.values$`0.5quant`
dat2$fit.m2 <- model2$summary.fitted.values$`0.5quant`
dat2$fit.m3 <- model3$summary.fitted.values$`0.5quant`
length(dat2$fit.full)
#variables extra que al final no se usaron (se preservan para ver su equivalente en la f(x))
id2 <- dat2$NOMBDIST
var1<-dat2$falciparum
rmse <- dat2 %>%
mutate(r_full = abs(falciparum-fit.full),
r_m1 = abs(falciparum-fit.m1),
r_m2 = abs(falciparum-fit.m2),
r_m3 = abs(falciparum-fit.m3)) %>%
group_by(NOMBDIST) %>%
summarise(rmse_full = mean(r_full, na.rm = T),
rmse_m1 = mean(r_m1, na.rm = T),
rmse_m2 = mean(r_m2, na.rm = T),
rmse_m3 = mean(r_m3, na.rm = T))
colnames(rmse)<-c("District","Full","climate","Malaria-Control","Deforestation")
falciparum.dumbble<- ggplot() +
#grafico de puntos primera geometria
#el primer geoma determina el orden de los distritos
geom_segment(
data = gather(rmse, measure, value, -District) %>%
group_by(District) %>%
summarise(start = range(value)[1], end = range(value)[2]) %>%
ungroup(),
aes(x = start, xend = end, y = reorder(District, rmse[[2]], mean), # a ordenar/valor por el cual ordenar/extra. rmse[[2]] es el Full
yend = reorder(District, end, mean)),color = "grey20", size = 0.5)+
#puntos
geom_point(data = gather(rmse, measure, value, -District),
aes(value,reorder(District, value, mean) , color = measure), #District
size = 4,alpha=0.45)+
scale_color_npg(palette = c("nrc"), alpha = 0.2, name="Variable set",
limits=c("Full","climate","Deforestation","Malaria-Control"))+
theme(legend.position = "top",
panel.background = element_blank(),
legend.key = element_rect(fill = "white"),
panel.grid.major.x = element_blank() ,
# explicitly set the horizontal lines (or they will disappear too)
panel.grid.major.y = element_line( size=.05, color="darkgray" ) )+
scale_x_continuous(limits = c(0, 250)) +
xlab("Mean Absolute Error") +
ylab("District")
falciparum.dumbble
dat2 <-dat.rev.vf.cplt
fullv <-test_pv_8_mae[[4]]
model1v<-test_pv_8_mae[[1]] # clima
model2v<-test_pv_8_mae[[2]] # Malaria-Control
model3v<-test_pv_8_mae[[3]] # Deforestation
dat2$fit.fullv <- fullv$summary.fitted.values$`0.5quant`
dat2$fit.m1v <- model1v$summary.fitted.values$`0.5quant`
dat2$fit.m2v <- model2v$summary.fitted.values$`0.5quant`
dat2$fit.m3v <- model3v$summary.fitted.values$`0.5quant`
length( dat2$fit.fullv)
#variables extra que al final no se usaron (se preservan para ver su equivalente en la f(x))
id2 <- dat2$NOMBDIST
rmsev <- dat2 %>%
mutate(r_fullv = abs(vivax-fit.fullv),
r_m1v = abs(vivax-fit.m1v),
r_m2v = abs(vivax-fit.m2v),
r_m3v = abs(vivax-fit.m3v)) %>%
group_by(NOMBDIST) %>%
summarise(rmse_fullv = mean(r_fullv, na.rm = T),
rmse_m1v = mean(r_m1v, na.rm = T),
rmse_m2v = mean(r_m2v, na.rm = T),
rmse_m3v = mean(r_m3v, na.rm = T))
colnames(rmsev)<-c("District","Full","climate","Malaria-Control","Deforestation")
vivax.dumbble<- ggplot() +
#grafico de puntos primera geometria
#el primer geoma determina el orden de los distritos
geom_segment(
data = gather(rmsev, measure, value, -District) %>%
group_by(District) %>%
summarise(start = range(value)[1], end = range(value)[2]) %>%
ungroup(),
aes(x = start, xend = end, y = reorder(District, rmsev[[2]], mean), # a ordenar/valor por el cual ordenar/extra. rmse[[2]] es el Full
yend = reorder(District, end, mean)),color = "grey20", size = 0.5)+
#puntos
geom_point(data = gather(rmsev, measure, value, -District),
aes(value,reorder(District, value, mean) , color = measure), #District
size = 4,alpha=0.45)+
scale_color_npg(palette = c("nrc"), alpha = 0.2, name="Variable set",
limits=c("Full","climate","Deforestation","Malaria-Control"))+
theme(legend.position = "top",
panel.background = element_blank(),
legend.key = element_rect(fill = "white"),
panel.grid.major.x = element_blank() ,
# explicitly set the horizontal lines (or they will disappear too)
panel.grid.major.y = element_line( size=.05, color="darkgrey" ) )+
scale_x_continuous(limits = c(0, 250)) +
xlab("Mean Absolute Error") +
ylab("District")
vivax.dumbble
library(cowplot)
(rm_seasonal_d<-plot_grid(vivax.dumbble,falciparum.dumbble, ncol=2, labels = c("A) P.vivax ", "B) P. falciparum"), hjust = -0.05))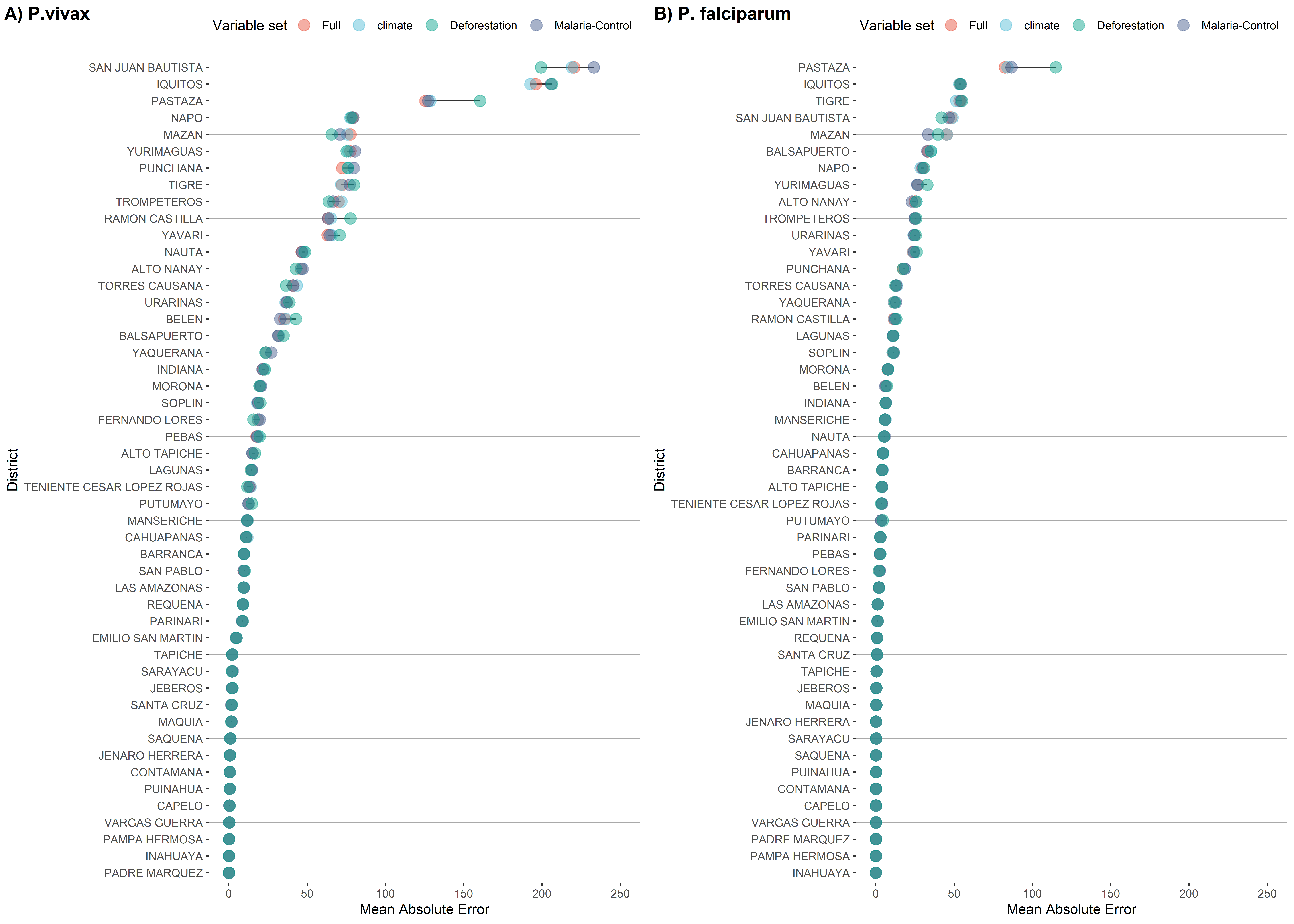
5.7 Dumbbell plot and Spatial heterogeneity of covariates effects in terms of mean absolute error
library(colorspace)
library(sf)
library(ggthemes)
library(Hmisc)
library(cowplot)
##############################################################################
# Dumbbell plot
##############################################################################
# P. Falciparum
dat2 <-dat.rev.vf.cplt
full <-test_pf_7_mae[[1]]
model1<-test_pf_7_mae[[2]]
model2<-test_pf_7_mae[[3]]
model3<-test_pf_7_mae[[4]]
dat2$fit.full <- full$summary.fitted.values$`0.5quant`
dat2$fit.m1 <- model1$summary.fitted.values$`0.5quant`
dat2$fit.m2 <- model2$summary.fitted.values$`0.5quant`
dat2$fit.m3 <- model3$summary.fitted.values$`0.5quant`
length(dat2$fit.full)
#variables extra que al final no se usaron (se preservan para ver su equivalente en la f(x))
id2 <- dat2$NOMBDIST
var1<-dat2$falciparum
rmse <- dat2 %>%
mutate(r_full = abs(falciparum-fit.full),
r_m1 = abs(falciparum-fit.m1),
r_m2 = abs(falciparum-fit.m2),
r_m3 = abs(falciparum-fit.m3)) %>%
group_by(NOMBDIST) %>%
summarise(rmse_full = mean(r_full, na.rm = T),
rmse_m1 = mean(r_m1, na.rm = T),
rmse_m2 = mean(r_m2, na.rm = T),
rmse_m3 = mean(r_m3, na.rm = T))
colnames(rmse)<-c("District","Full","climate","Malaria-Control","Deforestation")
colfunc <- c("#E64B35B2","#4DBBD5B2","#00A087B2","#3C5488B2")
rmse2 <- rmse %>%
arrange(desc(Full)) %>%
head(n = 10)
datapoint = gather(rmse2, measure, value, -District)
falciparum.dumbble<- ggplot() +
#grafico de puntos primera geometria
#el primer geoma determina el orden de los distritos
geom_segment(
data = gather(rmse2, measure, value, -District) %>%
group_by(District) %>%
summarise(start = range(value)[1], end = range(value)[2]) %>%
ungroup(),
aes(x = start,
xend = end,
y = reorder(District, end, mean),
yend = reorder(District, end, mean)),
color = "grey20", size = 0.5)+ #puntos
geom_point(data = gather(rmse2, measure, value, -District),
aes(value,reorder(District, value, mean),
shape = measure,
color = measure), #District
size = 4,alpha=0.45)+
theme(legend.position = "none")+
theme(plot.margin = unit(c(3,1,3,1), "lines"), legend.position="none")+
scale_shape_manual(values=c(15,16,17,18),
name="Variable set",
limits=c("Full","climate","Deforestation","Malaria-Control"))+
scale_color_manual(name="Variable set",
limits=c("Full","climate","Deforestation","Malaria-Control"),
values = c("Full" = colfunc[1],
"climate" = colfunc[2],
"Deforestation" = colfunc[3],
"Malaria-Control" = colfunc[4])) +
theme(legend.position = "top",
panel.background = element_blank(),
legend.key = element_rect(fill = "white"),
panel.grid.major.x = element_blank(),
# explicitly set the horizontal lines (or they will disappear too)
panel.grid.major.y = element_line( size=.05, color="darkgrey"))+
theme(panel.border = element_rect(fill = NA, color = "black", size = 1))+
scale_x_continuous(limits = c(0, 250)) +
xlab("Mean Absolute Error") +
ylab("District")
falciparum.dumbble
dat2 <-dat.rev.vf.cplt
fullv <-test_pv_8_mae[[4]]
model1v<-test_pv_8_mae[[1]] # clima
model2v<-test_pv_8_mae[[2]] # Malaria-Control
model3v<-test_pv_8_mae[[3]] # Deforestation
dat2$fit.fullv <- fullv$summary.fitted.values$`0.5quant`
dat2$fit.m1v <- model1v$summary.fitted.values$`0.5quant`
dat2$fit.m2v <- model2v$summary.fitted.values$`0.5quant`
dat2$fit.m3v <- model3v$summary.fitted.values$`0.5quant`
length( dat2$fit.fullv)
#variables extra que al final no se usaron (se preservan para ver su equivalente en la f(x))
id2 <- dat2$NOMBDIST
rmsev <- dat2 %>%
mutate(r_fullv = abs(vivax-fit.fullv),
r_m1v = abs(vivax-fit.m1v),
r_m2v = abs(vivax-fit.m2v),
r_m3v = abs(vivax-fit.m3v)) %>%
group_by(NOMBDIST) %>%
summarise(rmse_fullv = mean(r_fullv, na.rm = T),
rmse_m1v = mean(r_m1v, na.rm = T),
rmse_m2v = mean(r_m2v, na.rm = T),
rmse_m3v = mean(r_m3v, na.rm = T))
colnames(rmsev)<-c("District","Full","climate","Malaria-Control","Deforestation")
rmsev2 <- rmsev %>%
arrange(desc(Full)) %>%
head(n = 10)
vivax.dumbble<- ggplot() +
#grafico de puntos primera geometria
#el primer geoma determina el orden de los distritos
geom_segment(
data = gather(rmsev2, measure, value, -District) %>%
group_by(District) %>%
summarise(start = range(value)[1], end = range(value)[2]) %>%
ungroup(),
aes(x = start, xend = end, y = reorder(District, end, mean),
yend = reorder(District, end, mean)),color = "grey20", size = 0.5)+ #puntos
geom_point(data = gather(rmsev2, measure, value, -District),
aes(value,reorder(District, value, mean),
shape = measure,
color = measure), #District
size = 4,alpha=0.45)+
theme(legend.position = "none")+#geom_text(show.legend = FALSE)+
theme(plot.margin = unit(c(3,1,3,1), "lines"), legend.position="none")+
scale_shape_manual(values=c(15,16,17,18),
name="Variable set",
limits=c("Full","climate","Deforestation","Malaria-Control"))+
scale_color_manual(name="Variable set",
limits=c("Full","climate","Deforestation","Malaria-Control"),
values = c("Full" = colfunc[1],
"climate" = colfunc[2],
"Deforestation" = colfunc[3],
"Malaria-Control" = colfunc[4])) +
theme(legend.position = "top",
panel.background = element_blank(),
legend.key = element_rect(fill = "white"),
panel.grid.major.x = element_blank(),
# explicitly set the horizontal lines (or they will disappear too)
panel.grid.major.y = element_line( size=.05, color="darkgrey"))+
theme(panel.border = element_rect(fill = NA, color = "black", size = 1))+
scale_x_continuous(limits = c(0, 250)) +
xlab("Mean Absolute Error") +
ylab("District")
vivax.dumbble
##############################################################################
# Spatial heterogeneity of covariates effects in terms of mean absolute error
##############################################################################
dat<-dat %>%
mutate(q_std =((q - mean(q,na.rm=T)) / sd(q,na.rm=T)),
aet_std =((aet - mean(aet,na.rm=T)) / sd(aet,na.rm=T)),
soilm_std =((soilm - mean(soilm,na.rm=T)) / sd(soilm,na.rm=T)),
prcp_day_std =(((prcp/30) - mean((prcp/30),na.rm=T)) / sd((prcp/30),na.rm=T)),
tmax_std =((tmax - mean(tmax,na.rm=T)) / sd(tmax,na.rm=T)),
tmin_std =((tmin - mean(tmin,na.rm=T)) / sd(tmin,na.rm=T)),
cum_loss_km2_std =((cum_loss_km2 - mean(cum_loss_km2,na.rm=T)) / sd(cum_loss_km2,na.rm=T)),
water_deficit_std =((water_deficit - mean(water_deficit,na.rm=T)) / sd(water_deficit,na.rm=T)))
dat<- dat %>%
group_by(NOMBDIST) %>%
mutate(workers_lag_12=Lag(workers,+12),
nets_lag_12=Lag(nets,+12),
n_eess_lag_12=Lag(n_eess,+12),
enviro_lag_12=Lag(enviro,+12),
cum_loss_km2_std_lag_12=Lag(cum_loss_km2_std,+12),
soilm_std_lag1=Lag(soilm_std,+1),
q_std_lag1=Lag(q_std,+1),
aet_std_lag1=Lag(aet_std,+1),
tmax_std_lag1=Lag(tmax_std,+1),
tmin_std_lag1=Lag(tmin_std,+1),
prcp_day_std_lag1=Lag(prcp_day_std,+1),
water_deficit_std_lag1=Lag(water_deficit_std,+1))
dat.rev.vf<-dat %>%
dplyr::select(NOMBDIST,vivax,falciparum,index,year,month2,
prcp_day_std,prcp_day_std_lag1,tmax_std,tmax_std_lag1,
cum_loss_km2_std,
n_eess,workers,enviro,nets,
n_eess_lag_12, workers_lag_12, enviro_lag_12,
nets_lag_12,pop2015,water_deficit_std,water_deficit_std_lag1,
soilm_std,aet_std,q_std,tmin_std,
soilm_std_lag1,aet_std_lag1,q_std_lag1,tmin_std_lag1)
dat.rev.vf.cplt<-dat.rev.vf[complete.cases(dat.rev.vf),]
###########################################
# P. Vivax
###########################################
pv_8.ma<- c(
# clima
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12,
# control-malaria
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# deforestacion
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1"),
# full
vivax ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw2") +
f(inla.group(prcp_day_std_lag1), model = "rw1") + f(inla.group(tmax_std),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(aet_std_lag1), model = "rw1") + f(inla.group(q_std),
model = "rw1")
)
###########################################
# P. Falciparum
###########################################
pf_7.ma<-c(
#Full model
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
n_eess_lag_12 + workers_lag_12 + enviro_lag_12 + nets_lag_12 +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
#clima
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(cum_loss_km2_std), model = "rw1") + n_eess_lag_12 +
workers_lag_12 + enviro_lag_12 + nets_lag_12,
#controles malaria
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + f(inla.group(cum_loss_km2_std), model = "rw1") +
f(inla.group(q_std_lag1), model = "rw1") + f(inla.group(aet_std_lag1),
model = "rw1"),
#deforestacion
formula = falciparum ~ 1 + f(index, model = "bym", graph = "map.graph") +
f(year, model = "iid") + f(month2, model = "rw1") +
f(inla.group(prcp_day_std), model = "rw1") + f(inla.group(tmax_std_lag1),
model = "rw1") + n_eess_lag_12 + workers_lag_12 + enviro_lag_12 +
nets_lag_12 + f(inla.group(q_std_lag1), model = "rw1") +
f(inla.group(aet_std_lag1), model = "rw1")
)
# P. Vivax ##############################
f3_a<-plot_mae2(dat1 = dat.rev.vf.cplt, var = vivax, full = test_pv_8_mae[[4]],
model1 = test_pv_8_mae[[1]], model2 = test_pv_8_mae[[2]], model3 = test_pv_8_mae[[3]],
map = area.sf, id = NOMBDIST)
dat.f3a <- f3_a[[3]] %>%
dplyr::select(NOMBDIST, m1:m3, i1:i3) %>%
dplyr::rename(climate_val=m1, `malaria-control_val`=m2, deforestation_val=m3,
climate_i=i1, `malaria-control_i`=i2, deforestation_i=i3) %>%
gather(type, val, climate_val:deforestation_i) %>%
separate(type, into = c("type","i"), sep = "_") %>%
spread(i,val) %>%
mutate(i = as.numeric(i),
# i = ifelse(i<0,NA,i),
i = ifelse(i>100,NA,i)
) %>%
mutate(val = ifelse(val=="No Added", val, "Added"))
f3a <- dat.f3a %>%
ggplot() +
geom_sf(aes(fill=i)) +
# scale_fill_continuous_sequential(palette="blues", na="white",
# trans="pseudo_log",
# breaks = c(1,3,8,15,30),
# labels = c("No \nadded","3","8","15","30")) +
scale_fill_gradient2(low = "red3", mid = "white", high = "#18517C", #na="#18517C",
midpoint = 0, aesthetics = "fill", trans="pseudo_log",
breaks = c(-11,0,32))+
# scale_colour_brewer(palette="RdBu", na="white",
# trans="pseudo_log",
# breaks = c(1,3,8,15,30),
# labels = c("No \nadded","3","8","15","30")) +
labs(fill = "MAE(diff) \n ") +
theme_few(base_size = 20) +
theme(legend.position = "right",
panel.spacing = unit(2, "lines"),
legend.text = element_text(size = 10)) +
theme(panel.border = element_rect(fill = NA, color = "black", size = 1))+
facet_wrap(.~type)
# f3a
# P. Falciparum ##############################
f3_b<-plot_mae2(dat1 = dat.rev.vf.cplt, var = falciparum, full = test_pf_7_mae[[1]],
model1 = test_pf_7_mae[[2]], model2 = test_pf_7_mae[[3]], model3 = test_pf_7_mae[[4]],
map = area.sf, id = NOMBDIST)
dat.f3b <- f3_b[[3]] %>%
dplyr::select(NOMBDIST, m1:m3, i1:i3) %>%
dplyr::rename(climate_val=m1, `malaria-control_val`=m2, deforestation_val=m3,
climate_i=i1, `malaria-control_i`=i2, deforestation_i=i3) %>%
gather(type, val, climate_val:deforestation_i) %>%
separate(type, into = c("type","i"), sep = "_") %>%
spread(i,val) %>%
mutate(i = as.numeric(i),
# i = ifelse(i<0,NA,i),
i = ifelse(i>100,NA,i)
) %>%
mutate(val = ifelse(val=="No Added", val, "Added"))
library(RColorBrewer)
f3b <- dat.f3b %>%
ggplot() +
geom_sf(aes(fill=i)) +
# scale_fill_continuous_sequential(palette="blues", na="white",
# trans="pseudo_log",
# breaks = c(1,3,8,15,30),
# labels = c("No \nadded","3","8","15","30")) +
scale_fill_gradient2(low = "red3", mid = "white", high = "#18517C", #na="#18517C",
midpoint = 0, aesthetics = "fill", trans="pseudo_log",
breaks = c(-11,0,32))+
# scale_colour_brewer(palette="RdBu", na="white",
# trans="pseudo_log",
# breaks = c(1,3,8,15,30),
# labels = c("No \nadded","3","8","15","30")) +
labs(fill = "MAE(diff) \n ") +
theme_few(base_size = 20) +
theme(legend.position = "right",
panel.spacing = unit(2, "lines"),
legend.text = element_text(size = 10)) +
theme(panel.border = element_rect(fill = NA, color = "black", size = 1))+
facet_wrap(.~type)
# f3b
##############################################################################
# Final Figure
(dfigure<-plot_grid(vivax.dumbble,f3a,falciparum.dumbble,f3b, ncol=2, nrow=2, labels = c("A) P. Vivax ", "", "B) P. Falciparum", ""), hjust = -0.05, label_size=17, rel_widths = c(1,2)))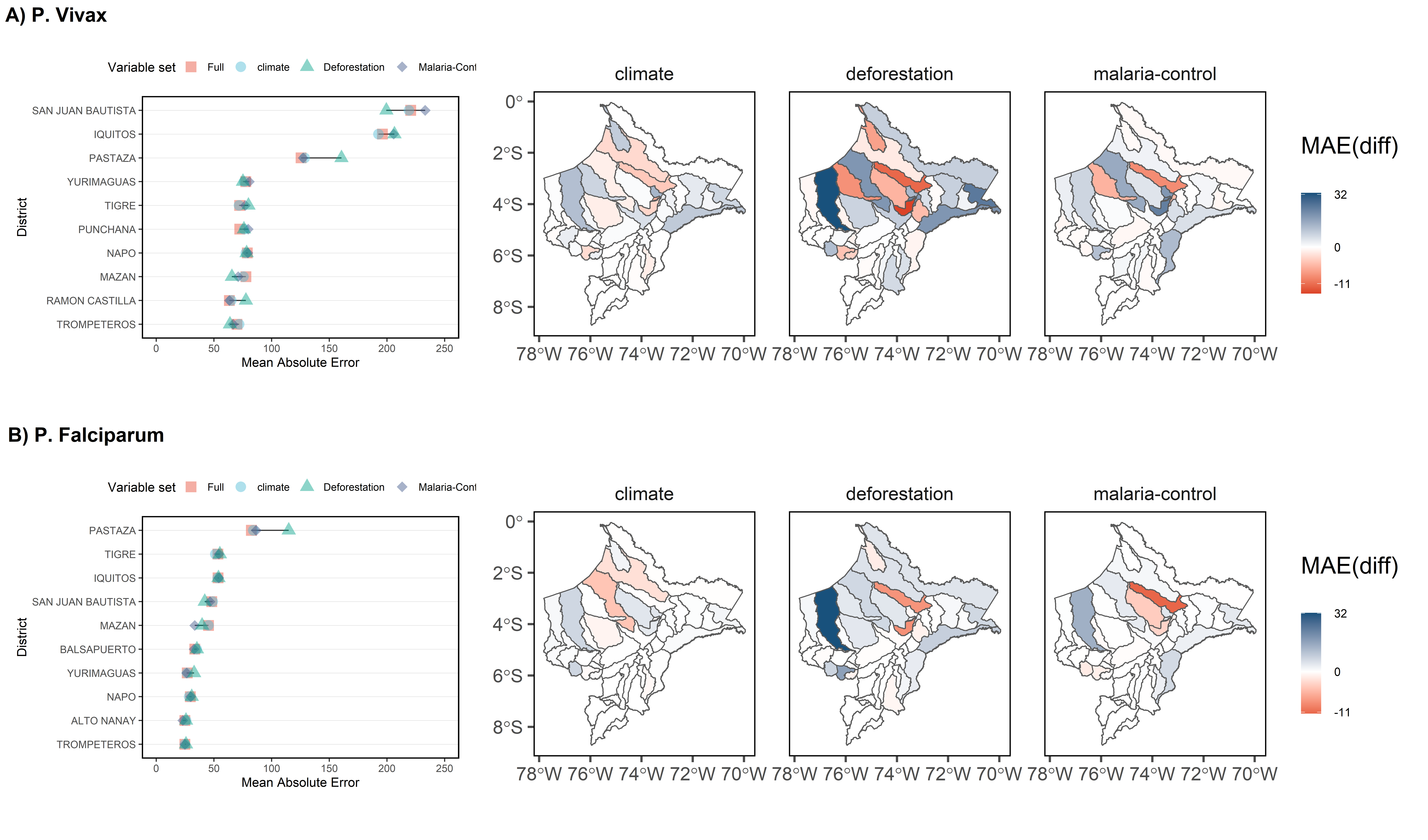
5.8 Supplementary Figure 7: Nonlinear effects
5.8.1 Figure 7.1
plot_random2_ver3 <- function(model, lab1 = NULL, scaled = NULL,
scaled_vars = 1,
analysis_vars = unique(mydata$var),
scaled_means = NULL,
scaled_sds = NULL,
y_lab = "Incidence Rate Ratio (IRR)",
name="", vars = NULL) {
print("Deprecated, update to plot_random3")
summ <- summary(model)
a <- scaled_vars
b <- scaled_means
c <- scaled_sds
{if(is.null(scaled)) {
mydata <- model$summary.random %>% bind_rows( .id = "var") %>% filter(var!="index")}
else{
mydata <- model$summary.random %>% bind_rows( .id = "var") %>%
filter(var %in% scaled_vars)
names(scaled_sds)<-scaled_vars
names(scaled_means)<-scaled_vars
mydata$ID<-(mydata$ID * scaled_sds[mydata$var]) + scaled_means[mydata$var]
mydata <- mydata %>% # no estaba enviando esta operacion a ningun lado
bind_rows(model$summary.random %>% bind_rows( .id = "var") %>%
filter(var!="index", !var %in% scaled_vars)) %>% filter(var %in% analysis_vars)
}
}
library(ggthemes)
lab2 <- lab1
if(!is.null(vars)) {mydata <- mydata %>% dplyr::filter(var %in% vars)}
# !is.null(vars) no se activará por defecto pues vars es NULL
if(!is.null(lab1)) {names(lab2) <- mydata %>% arrange(var) %>% distinct(var) %>% unlist()}
new_order <- scaled_vars
mydata2 <- arrange(transform(mydata, var = factor(var, levels = new_order)), var)
mydata_names <- colnames(mydata)
colnames(mydata2) <- mydata_names
levels(mydata2$var) <- lab2
plot<- mydata2 %>% ggplot(aes(x = ID, y = exp(`0.5quant`))) +
geom_ribbon(aes(ymin = exp(`0.025quant`), ymax = exp(`0.975quant`)), fill="grey90", size=.3, alpha = .6) +
geom_line(color = "firebrick") +
scale_x_continuous(expand=c(0,0)) +
geom_hline(aes(yintercept = 1), col="black", linetype="dashed", size=.4) +
labs(title=name, x="", y=y_lab) +
theme_base() +
theme(plot.background = element_blank(),
strip.placement = "outside") +
{if(!is.null(lab1)) {facet_wrap(.~var, scales = "free", strip.position = "bottom")}
else{facet_wrap(.~var, scales = "free", strip.position = "bottom")}
}
return(plot)
}
# P. Vivax
sv8a <- test_pv_8.3_back_2[[5]] %>%
plot_random2_ver3(lab1 = c("Avg. Precipitation\n (mm) lag", "Tmax (ºC)","Cum deforest x 100 km2",
"Evapotranspiration\n (mm) lag", "Runoff (mm)"),
scaled = T,
analysis_vars = c("inla.group(prcp_day_std_lag1)","inla.group(tmax_std)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std)"),
scaled_vars = c("inla.group(prcp_day_std_lag1)","inla.group(tmax_std)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std)"),
scaled_means = c(mean(dat$prcp/30, na.rm=T),mean(dat$tmax, na.rm=T),
mean(dat$cum_loss_km2, na.rm=T), mean(dat$aet, na.rm=T), mean(dat$q, na.rm=T)),
scaled_sds = c(sd(dat$prcp/30, na.rm=T),sd(dat$tmax, na.rm=T),sd(dat$cum_loss_km2, na.rm=T),
sd(dat$aet, na.rm=T), sd(dat$q, na.rm=T))
)
#lab1,scaled_vars, deben tener el mismo orden
# P. Falciparum
sf8b <- test_pf_7_back_3[[5]] %>%
plot_random2_ver3(lab1 = c("Avg. Precipitation (mm)", "Tmax (ºC) lag","Cum deforest x 100 km2",
"Evapotranspiration\n (mm) lag", "Runoff (mm) lag"),
scaled = T,
analysis_vars = c("inla.group(prcp_day_std)","inla.group(tmax_std_lag1)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std_lag1)"),
scaled_vars = c("inla.group(prcp_day_std)","inla.group(tmax_std_lag1)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std_lag1)"),
scaled_means = c(mean(dat$prcp/30, na.rm=T),mean(dat$tmax, na.rm=T),
mean(dat$cum_loss_km2, na.rm=T), mean(dat$aet, na.rm=T), mean(dat$q, na.rm=T)),
scaled_sds = c(sd(dat$prcp/30, na.rm=T),sd(dat$tmax, na.rm=T),sd(dat$cum_loss_km2, na.rm=T),
sd(dat$aet, na.rm=T), sd(dat$q, na.rm=T))
)
library(cowplot)
(sf8 <- plot_grid(sv8a, sf8b, labels = c("A) P. Vivax","B) P. Falciparum"), hjust = -0.05, ncol = 1))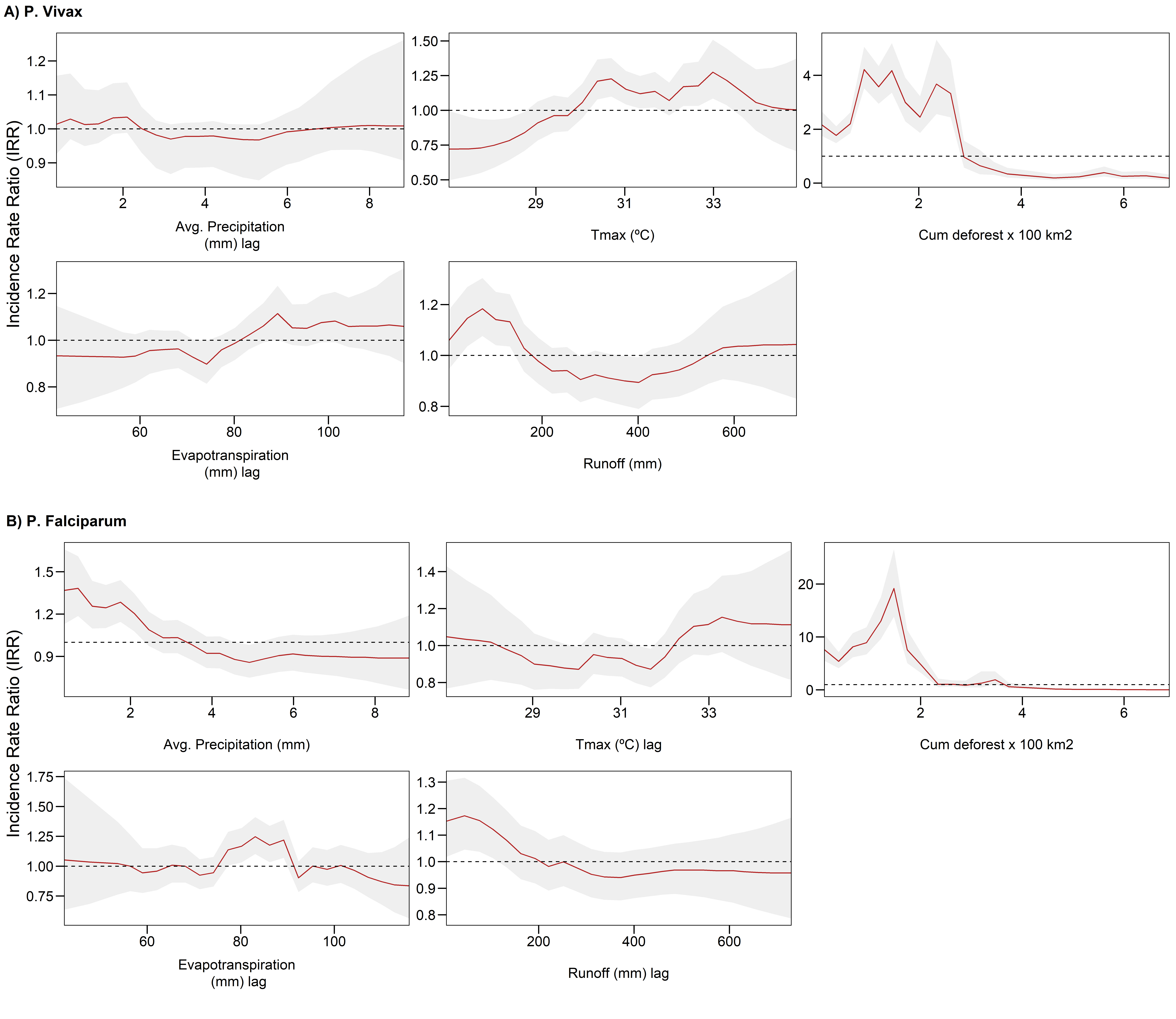
5.8.2 Figure 7.2
plot_random2_ver4 <- function(model, lab1 = NULL, scaled = NULL,
scaled_vars = 1,
analysis_vars = unique(mydata$var),
scaled_means = NULL,
scaled_sds = NULL,
y_lab = "Log (IR)",
name="", vars = NULL) {
print("Deprecated, update to plot_random3")
summ <- summary(model)
a <- scaled_vars
b <- scaled_means
c <- scaled_sds
{if(is.null(scaled)) {
mydata <- model$summary.random %>% bind_rows( .id = "var") %>% filter(var!="index")}
else{
mydata <- model$summary.random %>% bind_rows( .id = "var") %>%
filter(var %in% scaled_vars)
names(scaled_sds)<-scaled_vars
names(scaled_means)<-scaled_vars
mydata$ID<-(mydata$ID * scaled_sds[mydata$var]) + scaled_means[mydata$var]
mydata <- mydata %>% # no estaba enviando esta operacion a ningun lado
bind_rows(model$summary.random %>% bind_rows( .id = "var") %>%
filter(var!="index", !var %in% scaled_vars)) %>% filter(var %in% analysis_vars)
}
}
library(ggthemes)
lab2 <- lab1
if(!is.null(vars)) {mydata <- mydata %>% dplyr::filter(var %in% vars)}
# !is.null(vars) no se activará por defecto pues vars es NULL
if(!is.null(lab1)) {names(lab2) <- mydata %>% arrange(var) %>% distinct(var) %>% unlist()}
new_order <- scaled_vars
mydata2 <- arrange(transform(mydata, var = factor(var, levels = new_order)), var)
mydata_names <- colnames(mydata)
colnames(mydata2) <- mydata_names
levels(mydata2$var) <- lab2
plot<- mydata2 %>% ggplot(aes(x = ID, y = `0.5quant`)) +
geom_ribbon(aes(ymin = `0.025quant`, ymax = `0.975quant`), fill="grey90", size=.3, alpha = .6) +
geom_line(color = "firebrick") +
scale_x_continuous(expand=c(0,0)) +
geom_hline(aes(yintercept = 0), col="black", linetype="dashed", size=.4) +
labs(title=name, x="", y=y_lab) +
theme_base() +
theme(plot.background = element_blank(),
strip.placement = "outside") +
{if(!is.null(lab1)) {facet_wrap(.~var, scales = "free", strip.position = "bottom")}
else{facet_wrap(.~var, scales = "free", strip.position = "bottom")}
}
return(plot)
}
# P. Vivax
sv8a <- test_pv_8.3_back_2[[5]] %>%
plot_random2_ver4(lab1 = c("Avg. Precipitation\n (mm) lag", "Tmax (ºC)","Cum deforest x 100 km2",
"Evapotranspiration\n (mm) lag", "Runoff (mm)"),
scaled = T,
analysis_vars = c("inla.group(prcp_day_std_lag1)","inla.group(tmax_std)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std)"),
scaled_vars = c("inla.group(prcp_day_std_lag1)","inla.group(tmax_std)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std)"),
scaled_means = c(mean(dat$prcp/30, na.rm=T),mean(dat$tmax, na.rm=T),
mean(dat$cum_loss_km2, na.rm=T), mean(dat$aet, na.rm=T), mean(dat$q, na.rm=T)),
scaled_sds = c(sd(dat$prcp/30, na.rm=T),sd(dat$tmax, na.rm=T),sd(dat$cum_loss_km2, na.rm=T),
sd(dat$aet, na.rm=T), sd(dat$q, na.rm=T))
)
#lab1,scaled_vars, deben tener el mismo orden
# P. Falciparum
sf8b <- test_pf_7_back_3[[5]] %>%
plot_random2_ver4(lab1 = c("Avg. Precipitation (mm)", "Tmax (ºC) lag","Cum deforest x 100 km2",
"Evapotranspiration\n (mm) lag", "Runoff (mm) lag"),
scaled = T,
analysis_vars = c("inla.group(prcp_day_std)","inla.group(tmax_std_lag1)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std_lag1)"),
scaled_vars = c("inla.group(prcp_day_std)","inla.group(tmax_std_lag1)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std_lag1)"),
scaled_means = c(mean(dat$prcp/30, na.rm=T),mean(dat$tmax, na.rm=T),
mean(dat$cum_loss_km2, na.rm=T), mean(dat$aet, na.rm=T), mean(dat$q, na.rm=T)),
scaled_sds = c(sd(dat$prcp/30, na.rm=T),sd(dat$tmax, na.rm=T),sd(dat$cum_loss_km2, na.rm=T),
sd(dat$aet, na.rm=T), sd(dat$q, na.rm=T))
)
library(cowplot)
(sf8 <- plot_grid(sv8a, sf8b, labels = c("A) P. Vivax","B) P. Falciparum"), hjust = -0.05, ncol = 1))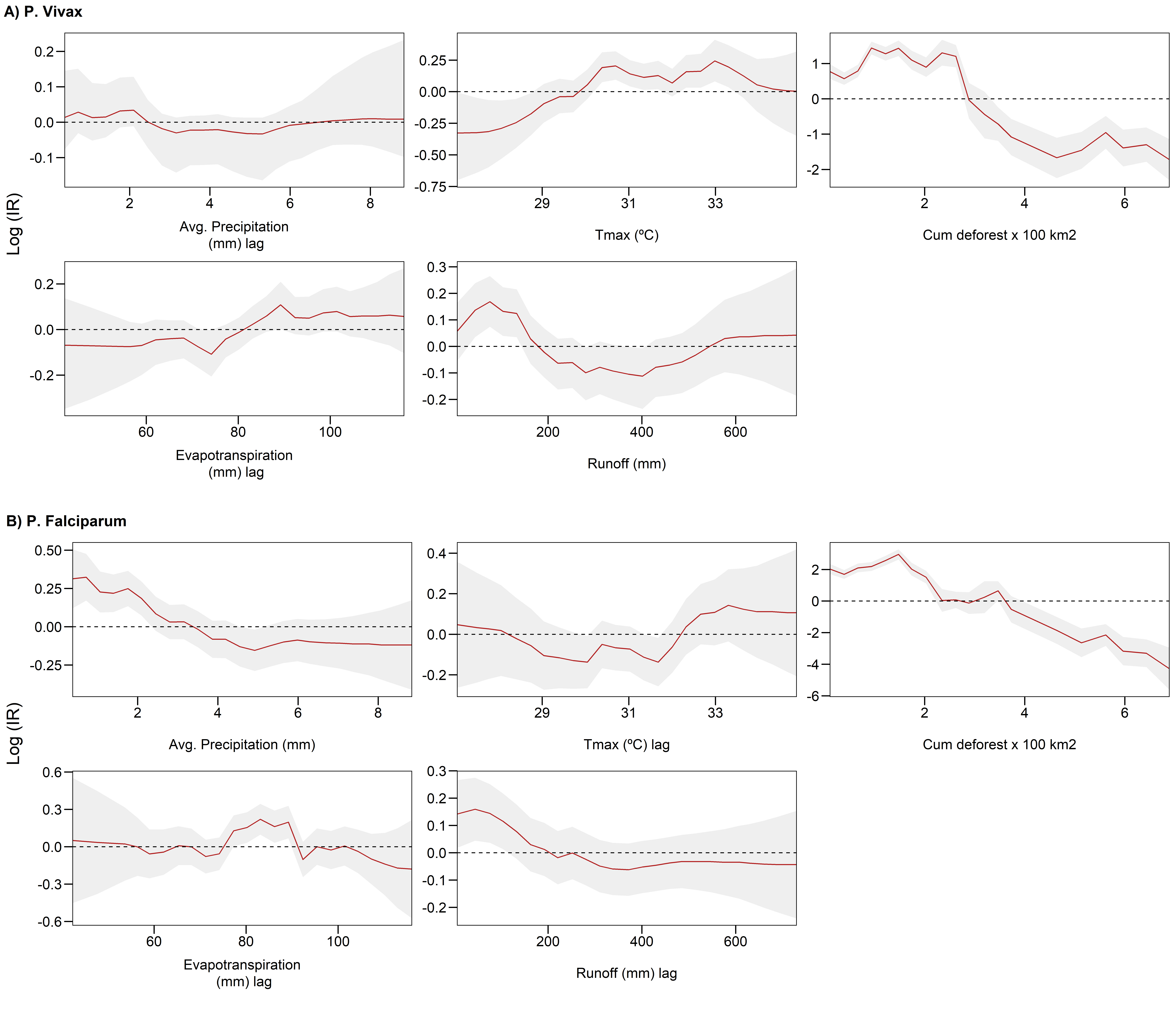
5.9 Combine plot
5.9.1 Combine plot 1
plot_fixed_ver2<-function(model, label = id, col1="black", dire=-1, breaks, lab_mod, lim, title, legend=NULL, filter, ylab = "Incidence Rate Ratio (IRR)", ylim = 10, angle=45, hjust = 1, xlabs=lim, b.size=20, flip=F) {
library(ggsci)
label <- enquo(label)
extract_dat <- function(model) {
mydata <- model$summary.fixed %>%
rownames_to_column(var = "var")
return(mydata)
}
dat_na <- data.frame(var=NA)
extract_dat.safe <- possibly(extract_dat, otherwise = dat_na)
qqq <- model %>% purrr::map(~extract_dat.safe(model=.)) %>%
bind_rows(.id = "formula") %>%
mutate(id = match(formula, unique(formula))) %>%
filter(!is.na(`0.025quant`), `0.5quant`<filter) %>%
ggplot(aes(x = var, y = exp(`0.5quant`), col=factor(id))) +
geom_point(position = position_dodge(0.3)) +
geom_errorbar(aes(ymin = exp(`0.025quant`), ymax = exp(`0.975quant`)), width=.1, position = position_dodge(0.3)) +
geom_hline(aes(yintercept = 1), col=col1, linetype="dashed", size=.4) +
ggsci::scale_color_npg(palette = c("nrc"),name="Models", breaks=breaks, labels=lab_mod) +
scale_x_discrete(limits=lim, labels = xlabs) +
ylim(0, ylim) +
{if(!is.null(legend)) {guides(color = legend)}} +
labs(title=title, x = "variables") +
theme_bw(base_size = b.size) +
labs(y=ylab) +
{if(isTRUE(flip)) {coord_flip()}} +
theme(axis.text.x = element_text(angle = angle, hjust = hjust)
,legend.position = "bottom",
legend.box = "vertical")+
guides(col=guide_legend(nrow=2,byrow=TRUE))
return(qqq)
}
plot_fixed_ver2.safe <- possibly(plot_fixed_ver2, otherwise = NA_real_)
#personalize plot ##########################################################
plot_random2_ver3 <- function(model, lab1 = NULL, scaled = NULL,
scaled_vars = 1,
analysis_vars = unique(mydata$var),
scaled_means = NULL,
scaled_sds = NULL,
y_lab = "Incidence Rate Ratio (IRR)",
name="", vars = NULL) {
print("Deprecated, update to plot_random3")
summ <- summary(model)
a <- scaled_vars
b <- scaled_means
c <- scaled_sds
{if(is.null(scaled)) {
mydata <- model$summary.random %>% bind_rows( .id = "var") %>% filter(var!="index")}
else{
mydata <- model$summary.random %>% bind_rows( .id = "var") %>%
filter(var %in% scaled_vars)
names(scaled_sds)<-scaled_vars
names(scaled_means)<-scaled_vars
mydata$ID<-(mydata$ID * scaled_sds[mydata$var]) + scaled_means[mydata$var]
mydata <- mydata %>% # no estaba enviando esta operacion a ningun lado
bind_rows(model$summary.random %>% bind_rows( .id = "var") %>%
filter(var!="index", !var %in% scaled_vars)) %>% filter(var %in% analysis_vars)
}
}
library(ggthemes)
lab2 <- lab1
if(!is.null(vars)) {mydata <- mydata %>% dplyr::filter(var %in% vars)}
# !is.null(vars) no se activará por defecto pues vars es NULL
if(!is.null(lab1)) {names(lab2) <- mydata %>% arrange(var) %>% distinct(var) %>% unlist()}
new_order <- scaled_vars
mydata2 <- arrange(transform(mydata, var = factor(var, levels = new_order)), var)
mydata_names <- colnames(mydata)
colnames(mydata2) <- mydata_names
levels(mydata2$var) <- lab2
plot<- mydata2 %>% ggplot(aes(x = ID, y = exp(`0.5quant`))) +
geom_ribbon(aes(ymin = exp(`0.025quant`), ymax = exp(`0.975quant`)), fill="grey90", size=.3, alpha = .6) +
geom_line(color = "firebrick") +
scale_x_continuous(expand=c(0,0)) +
geom_hline(aes(yintercept = 1), col="black", linetype="dashed", size=.4) +
labs(title=name, x="", y=y_lab) +
theme_base() +
theme(plot.background = element_blank(),
strip.placement = "outside") +
{if(!is.null(lab1)) {facet_wrap(.~var, scales = "free", strip.position = "bottom")}
else{facet_wrap(.~var, scales = "free", strip.position = "bottom")}
}
return(plot)
}
#############################################################################
#cp:combine plot
sv_7_pv_cp <- test_pv_8.3_back_2[c(5)] %>%
plot_fixed_ver2.safe(title = " ",
filter=10,
legend = F,
ylim = 6,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12"),
xlab = c("Lag of workers","Lag of nets","Lag of hfd","Lag of enviro"),
breaks=c("1","2"),
lab_mod=c("Full model",
""),
flip=T) +
theme(plot.margin = unit(c(1,1,1,1), "lines"), legend.position="none")
sv8a_cp <- test_pv_8.3_back_2[[5]] %>%
plot_random2_ver3(lab1 = c("Avg. Precipitation\n (mm) lag", "Tmax (ºC)","Cum deforest x 100 km2",
"Evapotranspiration\n (mm) lag", "Runoff (mm)"),
scaled = T,
analysis_vars = c("inla.group(prcp_day_std_lag1)","inla.group(tmax_std)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std)"),
scaled_vars = c("inla.group(prcp_day_std_lag1)","inla.group(tmax_std)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std)"),
scaled_means = c(mean(dat$prcp/30, na.rm=T),mean(dat$tmax, na.rm=T),
mean(dat$cum_loss_km2, na.rm=T), mean(dat$aet, na.rm=T), mean(dat$q, na.rm=T)),
scaled_sds = c(sd(dat$prcp/30, na.rm=T),sd(dat$tmax, na.rm=T),sd(dat$cum_loss_km2, na.rm=T),
sd(dat$aet, na.rm=T), sd(dat$q, na.rm=T))
)
sfcp_1 <- plot_grid(sv_7_pv_cp,sv8a_cp, labels = c("A) P. Vivax Fixed effects and Random effects"), hjust = -0.05, ncol = 2,nrow=1)
sv_7_pf_cp <- test_pf_7_back_3[c(5)] %>%
plot_fixed_ver2.safe(title = " ",
filter=10,
legend = F,
ylim = 6,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12"),
xlab = c("Lag of workers","Lag of nets","Lag of hfd","Lag of enviro"),
breaks=c("1","2"),
lab_mod=c("Full model",
""),
flip=T) +
theme(plot.margin = unit(c(1,1,1,1), "lines"), legend.position="none")
sv8bf_cp <- test_pf_7_back_3[[5]] %>%
plot_random2_ver3(lab1 = c("Avg. Precipitation (mm)", "Tmax (ºC) lag","Cum deforest x 100 km2",
"Evapotranspiration\n (mm) lag", "Runoff (mm) lag"),
scaled = T,
analysis_vars = c("inla.group(prcp_day_std)","inla.group(tmax_std_lag1)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std_lag1)"),
scaled_vars = c("inla.group(prcp_day_std)","inla.group(tmax_std_lag1)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std_lag1)"),
scaled_means = c(mean(dat$prcp/30, na.rm=T),mean(dat$tmax, na.rm=T),
mean(dat$cum_loss_km2, na.rm=T), mean(dat$aet, na.rm=T), mean(dat$q, na.rm=T)),
scaled_sds = c(sd(dat$prcp/30, na.rm=T),sd(dat$tmax, na.rm=T),sd(dat$cum_loss_km2, na.rm=T),
sd(dat$aet, na.rm=T), sd(dat$q, na.rm=T))
)
sfcp_2 <- plot_grid(sv_7_pf_cp,sv8bf_cp, labels = c("B) P. Falciparum Fixed effects and Random effects"), hjust = -0.05, ncol = 2,nrow=1)
(sfcp <- plot_grid(sfcp_1,sfcp_2, ncol = 1,nrow=2))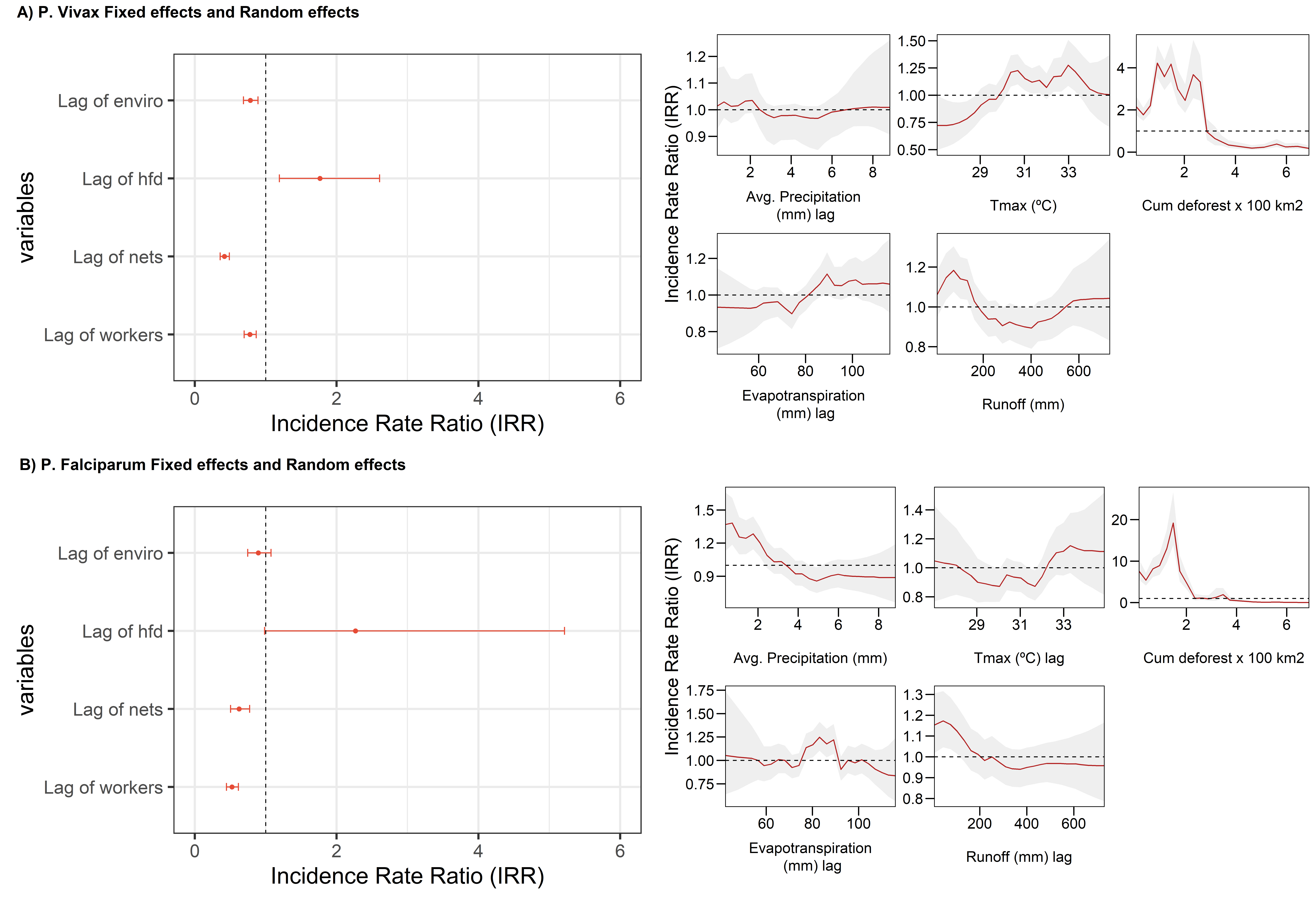
5.9.2 Combine plot 2
plot_fixed_ver2<-function(model, label = id, col1="black", dire=-1, breaks, lab_mod, lim, title, legend=NULL, filter, ylab = "Log (IR)", ylim = 10, angle=45, hjust = 1, xlabs=lim, b.size=20, flip=F) {
library(ggsci)
label <- enquo(label)
extract_dat <- function(model) {
mydata <- model$summary.fixed %>%
rownames_to_column(var = "var")
return(mydata)
}
dat_na <- data.frame(var=NA)
extract_dat.safe <- possibly(extract_dat, otherwise = dat_na)
qqq <- model %>% purrr::map(~extract_dat.safe(model=.)) %>%
bind_rows(.id = "formula") %>%
mutate(id = match(formula, unique(formula))) %>%
filter(!is.na(`0.025quant`), `0.5quant`<filter) %>%
ggplot(aes(x = var, y = exp(`0.5quant`), col=factor(id))) +
geom_point(position = position_dodge(0.3)) +
geom_errorbar(aes(ymin = exp(`0.025quant`), ymax = exp(`0.975quant`)), width=.1, position = position_dodge(0.3)) +
geom_hline(aes(yintercept = 1), col=col1, linetype="dashed", size=.4) +
ggsci::scale_color_npg(palette = c("nrc"),name="Models", breaks=breaks, labels=lab_mod) +
scale_x_discrete(limits=lim, labels = xlabs) +
ylim(0, ylim) +
{if(!is.null(legend)) {guides(color = legend)}} +
labs(title=title, x = "variables") +
theme_bw(base_size = b.size) +
labs(y=ylab) +
{if(isTRUE(flip)) {coord_flip()}} +
theme(axis.text.x = element_text(angle = angle, hjust = hjust)
,legend.position = "bottom",
legend.box = "vertical")+
guides(col=guide_legend(nrow=2,byrow=TRUE))
return(qqq)
}
plot_fixed_ver2.safe <- possibly(plot_fixed_ver2, otherwise = NA_real_)
#personalize plot ##########################################################
plot_random2_ver4 <- function(model, lab1 = NULL, scaled = NULL,
scaled_vars = 1,
analysis_vars = unique(mydata$var),
scaled_means = NULL,
scaled_sds = NULL,
y_lab = "Log (IR)",
name="", vars = NULL) {
print("Deprecated, update to plot_random3")
summ <- summary(model)
a <- scaled_vars
b <- scaled_means
c <- scaled_sds
{if(is.null(scaled)) {
mydata <- model$summary.random %>% bind_rows( .id = "var") %>% filter(var!="index")}
else{
mydata <- model$summary.random %>% bind_rows( .id = "var") %>%
filter(var %in% scaled_vars)
names(scaled_sds)<-scaled_vars
names(scaled_means)<-scaled_vars
mydata$ID<-(mydata$ID * scaled_sds[mydata$var]) + scaled_means[mydata$var]
mydata <- mydata %>% # no estaba enviando esta operacion a ningun lado
bind_rows(model$summary.random %>% bind_rows( .id = "var") %>%
filter(var!="index", !var %in% scaled_vars)) %>% filter(var %in% analysis_vars)
}
}
library(ggthemes)
lab2 <- lab1
if(!is.null(vars)) {mydata <- mydata %>% dplyr::filter(var %in% vars)}
# !is.null(vars) no se activará por defecto pues vars es NULL
if(!is.null(lab1)) {names(lab2) <- mydata %>% arrange(var) %>% distinct(var) %>% unlist()}
new_order <- scaled_vars
mydata2 <- arrange(transform(mydata, var = factor(var, levels = new_order)), var)
mydata_names <- colnames(mydata)
colnames(mydata2) <- mydata_names
levels(mydata2$var) <- lab2
plot<- mydata2 %>% ggplot(aes(x = ID, y = `0.5quant`)) +
geom_ribbon(aes(ymin = `0.025quant`, ymax = `0.975quant`), fill="grey90", size=.3, alpha = .6) +
geom_line(color = "firebrick") +
scale_x_continuous(expand=c(0,0)) +
geom_hline(aes(yintercept = 0), col="black", linetype="dashed", size=.4) +
labs(title=name, x="", y=y_lab) +
theme_base() +
theme(plot.background = element_blank(),
strip.placement = "outside") +
{if(!is.null(lab1)) {facet_wrap(.~var, scales = "free", strip.position = "bottom")}
else{facet_wrap(.~var, scales = "free", strip.position = "bottom")}
}
return(plot)
}
#############################################################################
#cp:combine plot
sv_log_pv <- test_pv_8.3_back_2[c(5)] %>%
plot.log.fixed.safe(title = "",
filter=8,
legend = F,
ylim = 3,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12"),
xlab = c("Lag of workers","Lag of nets","Lag of hfd","Lag of enviro"),
breaks = c("1","2"),
lab_mod = c("Full model", ""),
flip=T)+
ylab("Log (IR)")+
theme(plot.margin = unit(c(1,1,1,1), "lines"))
sv8a_cp <- test_pv_8.3_back_2[[5]] %>%
plot_random2_ver4(lab1 = c("Avg. Precipitation\n (mm) lag", "Tmax (ºC)","Cum deforest x 100 km2",
"Evapotranspiration\n (mm) lag", "Runoff (mm)"),
scaled = T,
analysis_vars = c("inla.group(prcp_day_std_lag1)","inla.group(tmax_std)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std)"),
scaled_vars = c("inla.group(prcp_day_std_lag1)","inla.group(tmax_std)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std)"),
scaled_means = c(mean(dat$prcp/30, na.rm=T),mean(dat$tmax, na.rm=T),
mean(dat$cum_loss_km2, na.rm=T), mean(dat$aet, na.rm=T), mean(dat$q, na.rm=T)),
scaled_sds = c(sd(dat$prcp/30, na.rm=T),sd(dat$tmax, na.rm=T),sd(dat$cum_loss_km2, na.rm=T),
sd(dat$aet, na.rm=T), sd(dat$q, na.rm=T))
)
sfcp_1 <- plot_grid(sv_log_pv,sv8a_cp, labels = c("A) P. Vivax Fixed effects and Random effects"), hjust = -0.05, ncol = 2,nrow=1)
sf_log_pf <- test_pf_7_back_3[c(5)] %>%
plot.log.fixed.safe(title = "",
filter=8,
legend = F,
ylim = 3,
angle = 0,
hjust = .5,
lim = c("workers_lag_12","nets_lag_12",
"n_eess_lag_12","enviro_lag_12"),
xlab = c("Lag of workers","Lag of nets","Lag of hfd","Lag of enviro"),
breaks = c("1","2"),
lab_mod = c("Full model", ""),
flip=T)+
ylab("Log (IR)")+
theme(plot.margin = unit(c(1,1,1,1), "lines"))
sv8bf_cp <- test_pf_7_back_3[[5]] %>%
plot_random2_ver4(lab1 = c("Avg. Precipitation (mm)", "Tmax (ºC) lag","Cum deforest x 100 km2",
"Evapotranspiration\n (mm) lag", "Runoff (mm) lag"),
scaled = T,
analysis_vars = c("inla.group(prcp_day_std)","inla.group(tmax_std_lag1)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std_lag1)"),
scaled_vars = c("inla.group(prcp_day_std)","inla.group(tmax_std_lag1)",
"inla.group(cum_loss_km2_std)", "inla.group(aet_std_lag1)", "inla.group(q_std_lag1)"),
scaled_means = c(mean(dat$prcp/30, na.rm=T),mean(dat$tmax, na.rm=T),
mean(dat$cum_loss_km2, na.rm=T), mean(dat$aet, na.rm=T), mean(dat$q, na.rm=T)),
scaled_sds = c(sd(dat$prcp/30, na.rm=T),sd(dat$tmax, na.rm=T),sd(dat$cum_loss_km2, na.rm=T),
sd(dat$aet, na.rm=T), sd(dat$q, na.rm=T))
)
sfcp_2 <- plot_grid(sf_log_pf,sv8bf_cp, labels = c("B) P. Falciparum Fixed effects and Random effects"), hjust = -0.05, ncol = 2,nrow=1)
(sfcp <- plot_grid(sfcp_1,sfcp_2, ncol = 1,nrow=2))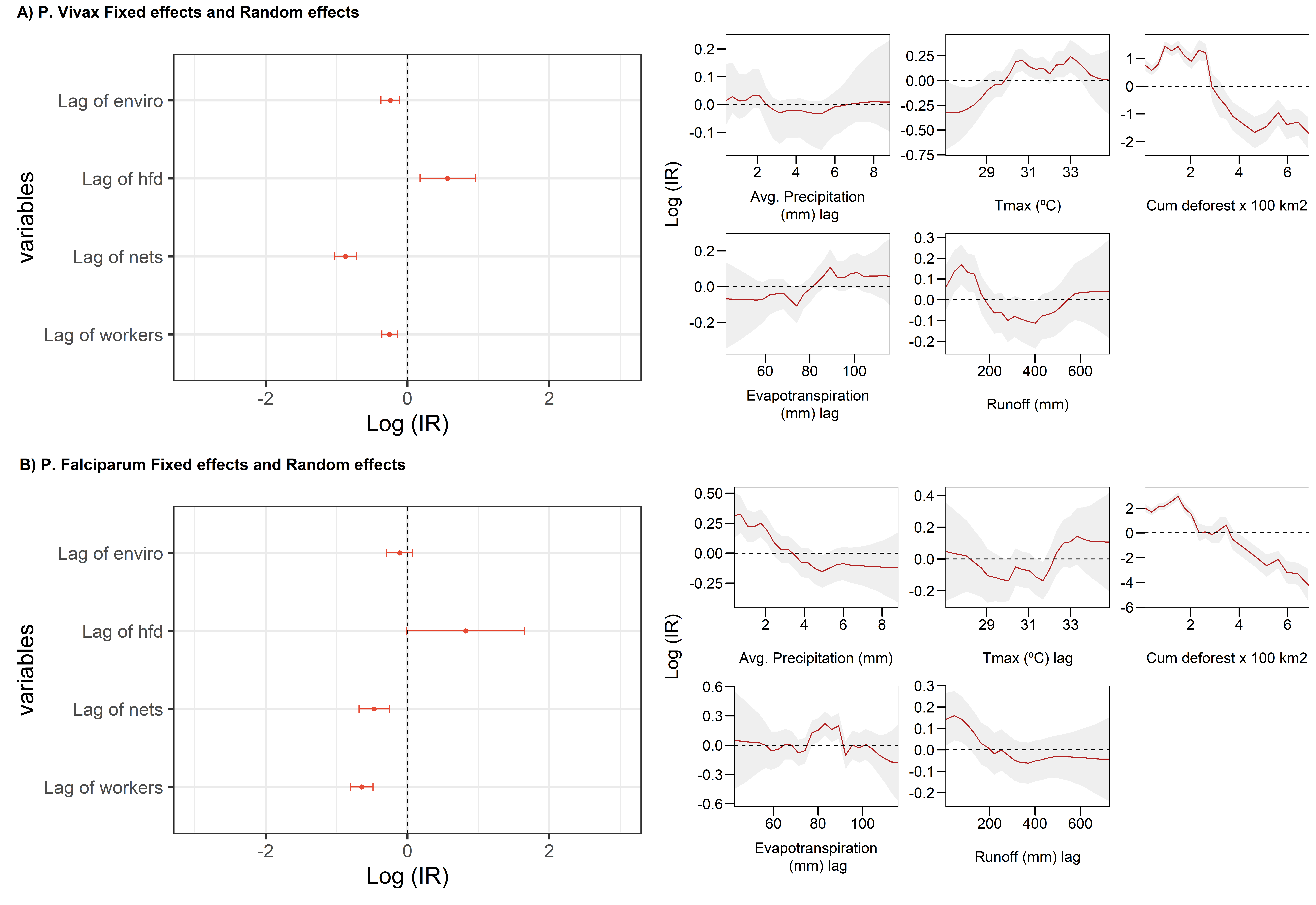
5.10 Supplementary Figure 8: Contributions of spatial random effects to P.vivax and P.falciparum risk
library(scales)
library(colorspace)
plot_random_map2_ver2 <- function(model, map, palette="Red-Green", dire=-1, trans="identity", name="", id, col1="black") {
library(sf)
n1=nrow(map)
id2 <- enquo(id)
by <- as_label(id2)
a <- model$summary.random %>% bind_rows( .id = "var") %>% filter(var=="index")
a1 <- a %>% slice(1:n1) %>%
dplyr::select(ID,mean) %>%
mutate(ID=1:n1) %>%
dplyr::rename(unstructured = mean)
a2 <- a %>% slice(n1+1:nrow(a)) %>%
dplyr::select(ID,mean) %>%
mutate(ID=1:n1) %>%
dplyr::rename(structured = mean)
library(RColorBrewer)
a3 <- map %>%
mutate(ID = 1:n1) %>%
left_join(a1, by="ID") %>%
left_join(a2, by="ID") %>%
mutate(posterior = structured+unstructured) %>%
ggplot() +
geom_sf(aes(fill=posterior), size = 0.3, col = col1) +
scale_fill_continuous_diverging(palette = "Green-Brown", rev = TRUE,
breaks=c(-12,0,5.7))+
labs(title=name, fill="Linear \nPredictor") +
theme_void() +
theme(panel.grid.major = element_line(color = "white"),
strip.text = element_text(size=15))
return(a3)
}
rm_pv_7 <-test_pv_8.3_back_2[[5]] %>%
plot_random_map2_ver2(map = area.sf, name= "")
###########################################################################
plot_random_map2_ver2f <- function(model, map, palette="RdBu", dire=-1, trans="identity", name="", id, col1="black") {
library(sf)
n1=nrow(map)
id2 <- enquo(id)
by <- as_label(id2)
a <- model$summary.random %>% bind_rows( .id = "var") %>% filter(var=="index")
a1 <- a %>% slice(1:n1) %>%
dplyr::select(ID,mean) %>%
mutate(ID=1:n1) %>%
dplyr::rename(unstructured = mean)
a2 <- a %>% slice(n1+1:nrow(a)) %>%
dplyr::select(ID,mean) %>%
mutate(ID=1:n1) %>%
dplyr::rename(structured = mean)
library(RColorBrewer)
pal1 = brewer.pal(11, palette)
a3 <- map %>%
mutate(ID = 1:n1) %>%
left_join(a1, by="ID") %>%
left_join(a2, by="ID") %>%
mutate(posterior = structured+unstructured) %>%
ggplot() +
geom_sf(aes(fill=posterior), size = 0.3, col = col1) +
scale_fill_gradient2(low = "#18517C", mid = "white", high = "red3",
midpoint = 0, aesthetics = "fill",
breaks = c(-5.3,0,3.8))+
labs(title=name, fill="Linear \nPredictor") +
theme_void() +
theme(panel.grid.major = element_line(color = "white"),
strip.text = element_text(size=15))
return(a3)
}
rm_pf_7 <-test_pf_7_back_3[[5]] %>%
plot_random_map2_ver2f(map = area.sf, name= "")
library(cowplot)
(rm_seasonal<-plot_grid(rm_pv_7,rm_pf_7, ncol=2, labels = c("A) P. vivax", "B) P. falciparum")))