19 Correlation Analysis
We again work with the father/son heights dataset collected by Pearson.
load("data/father_son.rda")
attach(father_son)19.1 Scatterplot
We saw that a scatterplot is an appropriate plot for paired numerical data.
plot(father, son, pch = 16, xlab = "father's height", ylab = "son's height", asp = 1, cex = 0.5) 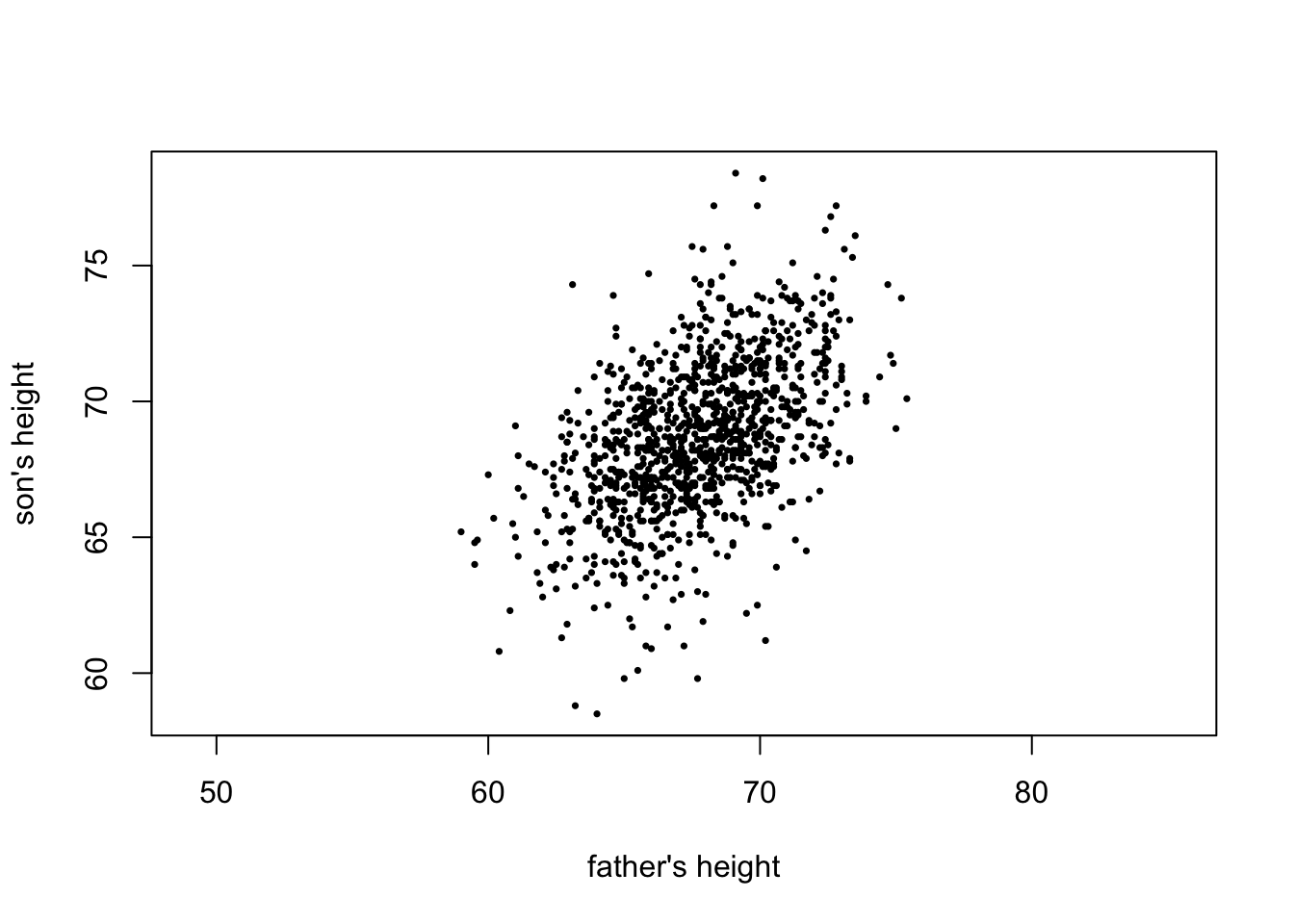
19.2 Sample correlations
We compute correlations of various types. They are all positive, in congruence with what is observed in the plot.
cor(father, son, method = "pearson")[1] 0.5012473cor(father, son, method = "spearman")[1] 0.505671cor(father, son, method = "kendall")[1] 0.352637519.3 Correlations tests
Although it is pretty clear from the scatterplot that the heights of a father and his son are positively correlated (or more generally, monotonically associated), for pedagodical reasons we perform the corresponding tests. (Refer to the manual for details on how the p-values are computed.)
cor.test(father, son, method = "pearson")
Pearson's product-moment correlation
data: father and son
t = 19.002, df = 1076, p-value < 2.2e-16
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
0.4551622 0.5446541
sample estimates:
cor
0.5012473 cor.test(father, son, method = "spearman")
Spearman's rank correlation rho
data: father and son
S = 103209762, p-value < 2.2e-16
alternative hypothesis: true rho is not equal to 0
sample estimates:
rho
0.505671 cor.test(father, son, method = "kendall")
Kendall's rank correlation tau
data: father and son
z = 17.161, p-value < 2.2e-16
alternative hypothesis: true tau is not equal to 0
sample estimates:
tau
0.3526375 19.4 Distance covariance (and test)
We also apply the distance covariance test. (The function returns the Monte Carlo permutation p-value based on R replicates.)
require(energy)
dcov.test(father, son, R = 1e3)
dCov independence test (permutation test)
data: index 1, replicates 1000
nV^2 = 742.69, p-value = 0.000999
sample estimates:
dCov
0.8300339