Chapter 2 Preparation des donnees H1
# Créer la BD finale pour les analyses de l'hypothèse 1 #
### Charger les bibliothèques ###
library(ade4)
library(ape)
library(car)
library(data.table)
library(dplyr)
library(factoextra)
library(FactoMineR)
library(gclus)
library(ggplot2)
library(ggpmisc)
library(ggpubr)
library(gridExtra)
library(janitor)
library(lme4)
library(lmodel2)
library(lubridate)
library(missMDA)
library(naniar)
library(pals)
library(performance)
library(readxl)
library(robustlmm)
library(SciViews)
library(splines)
library(tidyverse)
library(vegan)
library(visreg)
options(ggrepel.max.overlaps = Inf)2.1 Données d’ours
Je commence par charger la BD des données d’ours à partir du résultat créé par le script de Maxime, puis j’ajuste la catégorie de certaines variables et en crée quelques d’autres.
######## 1. Données d'ours ########
data <- read_excel("C:/Users/Sarah/Desktop/Maitrise/Données/Data/BD_repro_femelles_2023-05-12_Final.xlsx") # Charger les données
load("C:/Users/Sarah/Desktop/Maitrise/Données/Data/estimes.RData") # Charger les estimés
colnames(data)## [1] "Secteur" "ID_Animal"
## [3] "Age" "Annee"
## [5] "Mois" "Jour"
## [7] "Annee_Capture" "Weight_Summer"
## [9] "Date_Summer" "Total_Length_Summer"
## [11] "Neck_Circum_Summer" "Chestgirth_Summer"
## [13] "Body_Condition_Summer" "Weight_Precwinter"
## [15] "Date_Precwinter" "Total_Length_Precwinter"
## [17] "Neck_Circum_Precwinter" "Chestgirth_Precwinter"
## [19] "Body_Condition_Precwinter" "Weight_Winter"
## [21] "Date_Winter" "Total_Length_Winter"
## [23] "Neck_Circum_Winter" "Chestgirth_Winter"
## [25] "Body_Condition_Winter" "Presence_of_Youngs"
## [27] "Age_of_Youngs" "Number_of_Youngs"
## [29] "Weightx_Young_f" "Weightx_Young_m"
## [31] "Weightx_Young" "Status_Preceeding_Winter"
## [33] "Id_Young"## tibble [237 × 33] (S3: tbl_df/tbl/data.frame)
## $ Secteur : chr [1:237] "Mauricie" "Mauricie" "Mauricie" "Mauricie" ...
## $ ID_Animal : chr [1:237] "M201502" "M201503" "M201504" "M201507" ...
## $ Age : chr [1:237] "N/A" "11" "9" "11" ...
## $ Annee : chr [1:237] "2016" "2016" "2016" "2016" ...
## $ Mois : chr [1:237] "3" "3" "3" "3" ...
## $ Jour : chr [1:237] "11" "5" "5" "6" ...
## $ Annee_Capture : chr [1:237] "OUI" "OUI" "OUI" "OUI" ...
## $ Weight_Summer : chr [1:237] "108.4" "133.19999999999999" "120.8" "131.80000000000001" ...
## $ Date_Summer : chr [1:237] "2015-07-09" "2015-07-11" "2015-07-11" "2015-07-13" ...
## $ Total_Length_Summer : chr [1:237] "140" "151" "150" "150" ...
## $ Neck_Circum_Summer : chr [1:237] "42" "47" "46" "47" ...
## $ Chestgirth_Summer : chr [1:237] "67" "79" "75" "77" ...
## $ Body_Condition_Summer : chr [1:237] "BONNE" "EXCELLENTE" "EXCELLENTE" "EXCELLENTE" ...
## $ Weight_Precwinter : chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Date_Precwinter : chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Total_Length_Precwinter : chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Neck_Circum_Precwinter : chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Chestgirth_Precwinter : chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Body_Condition_Precwinter: chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Weight_Winter : chr [1:237] "108" "121.6" "118.2" "109" ...
## $ Date_Winter : chr [1:237] "2016-03-11" "2016-03-05" "2016-03-05" "2016-03-06" ...
## $ Total_Length_Winter : chr [1:237] "141" "150" "146" "146" ...
## $ Neck_Circum_Winter : chr [1:237] "42" "42" "42" "42" ...
## $ Chestgirth_Winter : chr [1:237] "69" "70" "71" "70" ...
## $ Body_Condition_Winter : chr [1:237] "EXCELLENTE" "BONNE" "BONNE" "BONNE" ...
## $ Presence_of_Youngs : chr [1:237] "NON" "OUI" "NON" "NON" ...
## $ Age_of_Youngs : chr [1:237] NA "CUB" NA NA ...
## $ Number_of_Youngs : chr [1:237] NA "2" NA NA ...
## $ Weightx_Young_f : chr [1:237] "NaN" "3" "NaN" "NaN" ...
## $ Weightx_Young_m : chr [1:237] "NaN" "1.8" "NaN" "NaN" ...
## $ Weightx_Young : chr [1:237] "NaN" "2.4" "NaN" "NaN" ...
## $ Status_Preceeding_Winter : chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Id_Young : chr [1:237] "NaN" "NA; NA" "NaN" "NaN" ...## Chr vers date ##
# Changer variables de date en format date
data <- data %>%
mutate_at(vars(Date_Precwinter, Date_Winter), as.Date, format="%Y-%m-%d")
str(data) # OK## tibble [237 × 33] (S3: tbl_df/tbl/data.frame)
## $ Secteur : chr [1:237] "Mauricie" "Mauricie" "Mauricie" "Mauricie" ...
## $ ID_Animal : chr [1:237] "M201502" "M201503" "M201504" "M201507" ...
## $ Age : chr [1:237] "N/A" "11" "9" "11" ...
## $ Annee : chr [1:237] "2016" "2016" "2016" "2016" ...
## $ Mois : chr [1:237] "3" "3" "3" "3" ...
## $ Jour : chr [1:237] "11" "5" "5" "6" ...
## $ Annee_Capture : chr [1:237] "OUI" "OUI" "OUI" "OUI" ...
## $ Weight_Summer : chr [1:237] "108.4" "133.19999999999999" "120.8" "131.80000000000001" ...
## $ Date_Summer : chr [1:237] "2015-07-09" "2015-07-11" "2015-07-11" "2015-07-13" ...
## $ Total_Length_Summer : chr [1:237] "140" "151" "150" "150" ...
## $ Neck_Circum_Summer : chr [1:237] "42" "47" "46" "47" ...
## $ Chestgirth_Summer : chr [1:237] "67" "79" "75" "77" ...
## $ Body_Condition_Summer : chr [1:237] "BONNE" "EXCELLENTE" "EXCELLENTE" "EXCELLENTE" ...
## $ Weight_Precwinter : chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Date_Precwinter : Date[1:237], format: NA NA ...
## $ Total_Length_Precwinter : chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Neck_Circum_Precwinter : chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Chestgirth_Precwinter : chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Body_Condition_Precwinter: chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Weight_Winter : chr [1:237] "108" "121.6" "118.2" "109" ...
## $ Date_Winter : Date[1:237], format: "2016-03-11" "2016-03-05" ...
## $ Total_Length_Winter : chr [1:237] "141" "150" "146" "146" ...
## $ Neck_Circum_Winter : chr [1:237] "42" "42" "42" "42" ...
## $ Chestgirth_Winter : chr [1:237] "69" "70" "71" "70" ...
## $ Body_Condition_Winter : chr [1:237] "EXCELLENTE" "BONNE" "BONNE" "BONNE" ...
## $ Presence_of_Youngs : chr [1:237] "NON" "OUI" "NON" "NON" ...
## $ Age_of_Youngs : chr [1:237] NA "CUB" NA NA ...
## $ Number_of_Youngs : chr [1:237] NA "2" NA NA ...
## $ Weightx_Young_f : chr [1:237] "NaN" "3" "NaN" "NaN" ...
## $ Weightx_Young_m : chr [1:237] "NaN" "1.8" "NaN" "NaN" ...
## $ Weightx_Young : chr [1:237] "NaN" "2.4" "NaN" "NaN" ...
## $ Status_Preceeding_Winter : chr [1:237] "Non suivie" "Non suivie" "Non suivie" "Non suivie" ...
## $ Id_Young : chr [1:237] "NaN" "NA; NA" "NaN" "NaN" ...## # A tibble: 130 × 2
## Date_Precwinter Date_Winter
## <date> <date>
## 1 2016-03-05 2017-03-01
## 2 2016-03-05 2017-02-24
## 3 2016-03-06 2017-02-27
## 4 2016-03-08 2017-02-24
## 5 2016-03-04 2017-02-28
## 6 2016-03-12 2017-02-28
## 7 2016-02-25 2017-02-26
## 8 2016-02-27 2017-02-27
## 9 2016-03-03 2017-02-26
## 10 2016-02-28 2017-02-22
## # ℹ 120 more rows# Créer variable jour cumulé
data$Day_Cumul <- yday(ymd(data$Date_Winter))
data[, c("Day_Cumul", "Date_Winter")] # OK## # A tibble: 237 × 2
## Day_Cumul Date_Winter
## <dbl> <date>
## 1 71 2016-03-11
## 2 65 2016-03-05
## 3 65 2016-03-05
## 4 66 2016-03-06
## 5 55 2016-02-24
## 6 68 2016-03-08
## 7 64 2016-03-04
## 8 72 2016-03-12
## 9 61 2016-03-01
## 10 56 2016-02-25
## # ℹ 227 more rows# Créer variables année winter et precwinter
data$Year_Precwinter <- format(as.Date(data$Date_Precwinter,format="%Y-%m-%d"), format = "%Y")
data$Year_Winter <- format(as.Date(data$Date_Winter,format="%Y-%m-%d"), format = "%Y")
data[, c("Date_Winter", "Year_Winter")] # OK## # A tibble: 237 × 2
## Date_Winter Year_Winter
## <date> <chr>
## 1 2016-03-11 2016
## 2 2016-03-05 2016
## 3 2016-03-05 2016
## 4 2016-03-06 2016
## 5 2016-02-24 2016
## 6 2016-03-08 2016
## 7 2016-03-04 2016
## 8 2016-03-12 2016
## 9 2016-03-01 2016
## 10 2016-02-25 2016
## # ℹ 227 more rows## # A tibble: 130 × 2
## Date_Precwinter Year_Precwinter
## <date> <chr>
## 1 2016-03-05 2016
## 2 2016-03-05 2016
## 3 2016-03-06 2016
## 4 2016-03-08 2016
## 5 2016-03-04 2016
## 6 2016-03-12 2016
## 7 2016-02-25 2016
## 8 2016-02-27 2016
## 9 2016-03-03 2016
## 10 2016-02-28 2016
## # ℹ 120 more rows## # A tibble: 11 × 2
## Date_Winter Year_Winter
## <date> <chr>
## 1 2019-03-08 2019
## 2 2019-03-09 2019
## 3 2019-03-08 2019
## 4 2019-03-05 2019
## 5 2019-03-10 2019
## 6 2019-03-09 2019
## 7 2019-03-12 2019
## 8 2019-03-17 2019
## 9 2019-03-17 2019
## 10 2019-03-19 2019
## 11 2019-03-18 2019## # A tibble: 6 × 2
## Date_Precwinter Year_Precwinter
## <date> <chr>
## 1 2018-03-20 2018
## 2 2018-03-21 2018
## 3 2018-03-20 2018
## 4 2018-03-21 2018
## 5 2018-03-22 2018
## 6 2018-03-23 2018## # A tibble: 6 × 2
## Date_Precwinter Year_Precwinter
## <date> <chr>
## 1 2022-03-27 2022
## 2 2022-03-17 2022
## 3 2022-03-25 2022
## 4 2022-03-28 2022
## 5 2022-03-26 2022
## 6 2022-03-20 2022## Changer variables en facteurs ##
data <- data %>%
mutate_at(vars("Secteur", "ID_Animal", "Annee", "Mois", "Jour", "Annee_Capture", "Body_Condition_Summer", "Body_Condition_Precwinter", "Body_Condition_Winter", "Presence_of_Youngs", "Age_of_Youngs", "Status_Preceeding_Winter", "Year_Precwinter", "Year_Winter"), factor)
## Changer variables en numérique ##
data[, c(3, 8, 10, 11, 12, 14, 16, 17, 18, 20, 22, 23, 24, 28, 29, 30, 31)] <- sapply(data[, c(3, 8, 10, 11, 12, 14, 16, 17, 18, 20, 22, 23, 24, 28, 29, 30, 31)], as.numeric)
str(data)## tibble [237 × 36] (S3: tbl_df/tbl/data.frame)
## $ Secteur : Factor w/ 4 levels "Gaspesie","Mauricie",..: 2 2 2 2 2 2 2 2 2 2 ...
## $ ID_Animal : Factor w/ 102 levels "G201901","G201902",..: 14 15 16 17 18 19 20 21 22 23 ...
## $ Age : num [1:237] NA 11 9 11 5 5 15 9 10 12 ...
## $ Annee : Factor w/ 8 levels "2016","2017",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ Mois : Factor w/ 3 levels "2","3","4": 2 2 2 2 1 2 2 2 2 1 ...
## $ Jour : Factor w/ 31 levels "1","10","11",..: 3 27 27 28 17 30 26 4 1 18 ...
## $ Annee_Capture : Factor w/ 2 levels "NON","OUI": 2 2 2 2 2 2 2 2 2 2 ...
## $ Weight_Summer : num [1:237] 108 133 121 132 104 ...
## $ Date_Summer : chr [1:237] "2015-07-09" "2015-07-11" "2015-07-11" "2015-07-13" ...
## $ Total_Length_Summer : num [1:237] 140 151 150 150 137 137 153 142 156 156 ...
## $ Neck_Circum_Summer : num [1:237] 42 47 46 47 41 41 47 47 52 44 ...
## $ Chestgirth_Summer : num [1:237] 67 79 75 77 67 68 75 77 85 74 ...
## $ Body_Condition_Summer : Factor w/ 5 levels "BONNE","EXCELLENTE",..: 1 2 2 2 2 2 2 2 2 2 ...
## $ Weight_Precwinter : num [1:237] NA NA NA NA NA NA NA NA NA NA ...
## $ Date_Precwinter : Date[1:237], format: NA NA ...
## $ Total_Length_Precwinter : num [1:237] NA NA NA NA NA NA NA NA NA NA ...
## $ Neck_Circum_Precwinter : num [1:237] NA NA NA NA NA NA NA NA NA NA ...
## $ Chestgirth_Precwinter : num [1:237] NA NA NA NA NA NA NA NA NA NA ...
## $ Body_Condition_Precwinter: Factor w/ 6 levels "BONNE","EXCELLENTE",..: 6 6 6 6 6 6 6 6 6 6 ...
## $ Weight_Winter : num [1:237] 108 122 118 109 97 ...
## $ Date_Winter : Date[1:237], format: "2016-03-11" "2016-03-05" ...
## $ Total_Length_Winter : num [1:237] 141 150 146 146 153 140 158 148 159 161 ...
## $ Neck_Circum_Winter : num [1:237] 42 42 42 42 40 38 46 42 49 45 ...
## $ Chestgirth_Winter : num [1:237] 69 70 71 70 69 72 75 76 85 79 ...
## $ Body_Condition_Winter : Factor w/ 5 levels "BONNE","EXCELLENTE",..: 2 1 1 1 1 2 1 2 2 2 ...
## $ Presence_of_Youngs : Factor w/ 2 levels "NON","OUI": 1 2 1 1 1 1 1 2 2 1 ...
## $ Age_of_Youngs : Factor w/ 3 levels "2 YEARS OLD",..: NA 2 NA NA NA NA NA 2 2 NA ...
## $ Number_of_Youngs : num [1:237] NA 2 NA NA NA NA NA 2 3 NA ...
## $ Weightx_Young_f : num [1:237] NaN 3 NaN NaN NaN NaN NaN 4 2.6 NaN ...
## $ Weightx_Young_m : num [1:237] NaN 1.8 NaN NaN NaN NaN NaN 4 3 NaN ...
## $ Weightx_Young : num [1:237] NaN 2.4 NaN NaN NaN ...
## $ Status_Preceeding_Winter : Factor w/ 4 levels "ALONE","CUB",..: 3 3 3 3 3 3 3 3 3 3 ...
## $ Id_Young : chr [1:237] "NaN" "NA; NA" "NaN" "NaN" ...
## $ Day_Cumul : num [1:237] 71 65 65 66 55 68 64 72 61 56 ...
## $ Year_Precwinter : Factor w/ 7 levels "2016","2017",..: NA NA NA NA NA NA NA NA NA NA ...
## $ Year_Winter : Factor w/ 8 levels "2016","2017",..: 1 1 1 1 1 1 1 1 1 1 ...## Secteur ID_Animal Age Annee Mois
## Gaspesie :13 S201604: 7 Min. : 2.00 2020 :64 2: 50
## Mauricie :72 S201622: 7 1st Qu.: 7.00 2019 :37 3:184
## Outaouais :55 O201705: 6 Median :10.50 2022 :34 4: 3
## Saguenay-Lac-St-Jean:97 S201614: 6 Mean :10.76 2018 :29
## S201617: 6 3rd Qu.:14.00 2023 :24
## S201748: 6 Max. :22.00 2021 :20
## (Other):199 NA's :3 (Other):29
## Jour Annee_Capture Weight_Summer Date_Summer
## 28 : 21 NON:123 Min. : 89.8 Length:237
## 26 : 14 OUI:114 1st Qu.:110.4 Class :character
## 27 : 14 Median :120.6 Mode :character
## 22 : 13 Mean :125.8
## 25 : 12 3rd Qu.:138.6
## 1 : 11 Max. :182.2
## (Other):152 NA's :124
## Total_Length_Summer Neck_Circum_Summer Chestgirth_Summer Body_Condition_Summer
## Min. :128.3 Min. :38.00 Min. :58.00 BONNE : 55
## 1st Qu.:142.0 1st Qu.:42.00 1st Qu.:71.75 EXCELLENTE: 40
## Median :147.0 Median :44.00 Median :75.00 MAUVAISE : 2
## Mean :147.8 Mean :44.56 Mean :74.90 MOYENNE : 17
## 3rd Qu.:154.0 3rd Qu.:46.00 3rd Qu.:78.25 Non suivie:123
## Max. :167.0 Max. :57.00 Max. :95.00
## NA's :125 NA's :125 NA's :125
## Weight_Precwinter Date_Precwinter Total_Length_Precwinter
## Min. : 82.8 Min. :2016-02-25 Min. :132.0
## 1st Qu.:118.2 1st Qu.:2018-03-21 1st Qu.:147.8
## Median :132.0 Median :2019-03-23 Median :153.0
## Mean :137.4 Mean :2019-06-25 Mean :152.7
## 3rd Qu.:157.2 3rd Qu.:2021-03-15 3rd Qu.:157.0
## Max. :225.8 Max. :2022-03-28 Max. :174.0
## NA's :110 NA's :107 NA's :121
## Neck_Circum_Precwinter Chestgirth_Precwinter Body_Condition_Precwinter
## Min. :38.00 Min. : 67.00 BONNE : 51
## 1st Qu.:42.00 1st Qu.: 75.00 EXCELLENTE: 49
## Median :45.00 Median : 82.00 MAUVAISE : 2
## Mean :45.18 Mean : 81.78 MOYENNE : 26
## 3rd Qu.:48.00 3rd Qu.: 88.00 N/A : 2
## Max. :54.00 Max. :104.00 Non suivie:107
## NA's :123 NA's :121
## Weight_Winter Date_Winter Total_Length_Winter Neck_Circum_Winter
## Min. : 42.8 Min. :2016-02-24 Min. :104.0 Min. :31.00
## 1st Qu.:118.2 1st Qu.:2019-03-08 1st Qu.:148.0 1st Qu.:43.00
## Median :137.0 Median :2020-03-07 Median :152.0 Median :45.00
## Mean :138.2 Mean :2020-01-17 Mean :152.5 Mean :45.15
## 3rd Qu.:158.3 3rd Qu.:2021-03-22 3rd Qu.:157.0 3rd Qu.:48.00
## Max. :225.8 Max. :2023-03-06 Max. :174.0 Max. :56.00
## NA's :11 NA's :24 NA's :29
## Chestgirth_Winter Body_Condition_Winter Presence_of_Youngs Age_of_Youngs
## Min. : 53.00 BONNE :100 NON: 52 2 YEARS OLD: 1
## 1st Qu.: 75.00 EXCELLENTE: 76 OUI:185 CUB :111
## Median : 82.00 MAUVAISE : 8 YEARLING : 73
## Mean : 81.45 MOYENNE : 47 NA's : 52
## 3rd Qu.: 87.00 N/A : 6
## Max. :104.00
## NA's :24
## Number_of_Youngs Weightx_Young_f Weightx_Young_m Weightx_Young
## Min. :1.000 Min. : 1.800 Min. : 1.467 Min. : 1.467
## 1st Qu.:2.000 1st Qu.: 3.117 1st Qu.: 3.587 1st Qu.: 3.500
## Median :2.000 Median : 4.600 Median : 4.600 Median : 4.833
## Mean :2.135 Mean :14.986 Mean :16.590 Mean :16.979
## 3rd Qu.:3.000 3rd Qu.:28.050 3rd Qu.:31.125 3rd Qu.:31.175
## Max. :4.000 Max. :65.200 Max. :89.000 Max. :77.100
## NA's :52 NA's :114 NA's :93 NA's :59
## Status_Preceeding_Winter Id_Young Day_Cumul Year_Precwinter
## ALONE : 27 Length:237 Min. :47.00 2019 : 28
## CUB : 64 Class :character 1st Qu.:61.00 2022 : 22
## Non suivie:107 Mode :character Median :72.00 2018 : 21
## YEARLING : 39 Mean :71.25 2020 : 20
## 3rd Qu.:80.00 2021 : 15
## Max. :94.00 (Other): 24
## NA's :107
## Year_Winter
## 2020 :64
## 2019 :37
## 2022 :34
## 2018 :29
## 2023 :24
## 2021 :20
## (Other):29## [1] "Gaspesie" "Mauricie" "Outaouais"
## [4] "Saguenay-Lac-St-Jean"## Gaspesie Mauricie Outaouais
## 13 72 55
## Saguenay-Lac-St-Jean
## 97## [1] "Gaspesie" "Mauricie" "Outaouais" "SLSJ"## Gaspesie Mauricie Outaouais SLSJ
## 13 72 55 97# Ajouter "NONE" à Age_of_Youngs quand il n'y a pas d'oursons
data$Age_of_Youngs <- as.character(data$Age_of_Youngs) %>% replace_na("NONE")
data$Age_of_Youngs <- as.factor(data$Age_of_Youngs) # remettre en facteur
summary(data$Age_of_Youngs) # OK## 2 YEARS OLD CUB NONE YEARLING
## 1 111 52 73# Ajouter 0 à Number_of_Youngs quand il n'y en a pas
data$Number_of_Youngs <- as.character(data$Number_of_Youngs) %>% replace_na("0")
data$Number_of_Youngs <- as.numeric(data$Number_of_Youngs) # remettre en num
summary(data$Number_of_Youngs) # OK## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.000 1.000 2.000 1.667 2.000 4.000# Changer la masse de lbs à kg
data <- data %>%
mutate(Weight_Precwinter_KG = Weight_Precwinter / 2.2046) %>%
mutate(Weight_Winter_KG = Weight_Winter / 2.2046) %>%
mutate(Weightx_Young_f_KG = Weightx_Young_f / 2.2046) %>%
mutate(Weightx_Young_m_KG = Weightx_Young_m / 2.2046) %>%
mutate(Weightx_Young_KG = Weightx_Young / 2.2046)
# Verif les plus utilises: Weight_Winter et Weightx_Young
na.omit(data[, c("Weight_Winter", "Weight_Winter_KG")]) # OK## # A tibble: 226 × 2
## Weight_Winter Weight_Winter_KG
## <dbl> <dbl>
## 1 108 49.0
## 2 122. 55.2
## 3 118. 53.6
## 4 109 49.4
## 5 97 44.0
## 6 93.4 42.4
## 7 131. 59.2
## 8 132. 59.8
## 9 160. 72.4
## 10 143 64.9
## # ℹ 216 more rows## # A tibble: 178 × 2
## Weightx_Young Weightx_Young_KG
## <dbl> <dbl>
## 1 2.4 1.09
## 2 4 1.81
## 3 2.73 1.24
## 4 24.7 11.2
## 5 3 1.36
## 6 2.6 1.18
## 7 1.6 0.726
## 8 2.2 0.998
## 9 3.3 1.50
## 10 2 0.907
## # ℹ 168 more rows# Toutes les valeurs = Femelles adultes en tanière et les informations correspondantes sur leurs jeunes
# Créer variable avec/sans cubs. Permettra de faire les graphiques.
data <- data %>%
mutate(Pres_Cubs =
as.factor(ifelse(Age_of_Youngs == "CUB", "AVEC", "SANS")))
# Correction body condition winter
data$Body_Condition_Winter[data$Body_Condition_Winter=="N/A"] = NA
data$Body_Condition_Winter <- droplevels(data$Body_Condition_Winter)
levels(data$Body_Condition_Winter) # ok## [1] "BONNE" "EXCELLENTE" "MAUVAISE" "MOYENNE"# Créer la variable "Reprod"
data <- data %>% mutate(Reprod = as.factor(
ifelse(Age_of_Youngs == "NONE" , "0",
ifelse(Age_of_Youngs == "CUB", "1",
NA))))
# 0: Echec, 1 = Reussite, NA = Yearling (donc compte pas)J’effectue les corrections sur les données morphométriques selon les estimés calculés précédemment (voir RMD corrections).
##### 1.1 Effectuer les corrections #####
## Mère ##
# Masse de la mère selon date en hiver
data <- data %>%
mutate(Weight_Winter_Corr_KG =
ifelse(Day_Cumul > 74 & Pres_Cubs == "AVEC" , Weight_Winter_KG + estime_masse_fem_cubs * (Day_Cumul - 74),
ifelse(Day_Cumul < 74 & Pres_Cubs == "AVEC", Weight_Winter_KG - estime_masse_fem_cubs * (74 - Day_Cumul),
ifelse(Day_Cumul > 74 & Pres_Cubs == "SANS" , Weight_Winter_KG + estime_masse_fem_nocubs * (Day_Cumul - 74),
ifelse(Day_Cumul < 74 & Pres_Cubs == "SANS", Weight_Winter_KG - estime_masse_fem_nocubs * (74 - Day_Cumul),
Weight_Winter_KG)))))
head(data[, c("Day_Cumul", "Pres_Cubs", "Weight_Winter_KG", "Weight_Winter_Corr_KG")]) #OK## # A tibble: 6 × 4
## Day_Cumul Pres_Cubs Weight_Winter_KG Weight_Winter_Corr_KG
## <dbl> <fct> <dbl> <dbl>
## 1 71 SANS 49.0 48.6
## 2 65 AVEC 55.2 52.9
## 3 65 SANS 53.6 52.4
## 4 66 SANS 49.4 48.3
## 5 55 SANS 44.0 41.3
## 6 68 SANS 42.4 41.5## # A tibble: 6 × 4
## Day_Cumul Pres_Cubs Weight_Winter_KG Weight_Winter_Corr_KG
## <dbl> <fct> <dbl> <dbl>
## 1 65 SANS 66.4 65.2
## 2 62 AVEC NA NA
## 3 52 AVEC 59.8 54.2
## 4 64 AVEC 58.0 55.4
## 5 63 SANS 65.6 64.1
## 6 55 SANS 19.4 16.8# Poitrine
data <- data %>% # Secteur
mutate(Chestgirth_Winter_Corr =
ifelse(Day_Cumul > 74 & Pres_Cubs == "AVEC" , Chestgirth_Winter + 0.25984 * (Day_Cumul - 74),
ifelse(Day_Cumul < 74 & Pres_Cubs == "AVEC", Chestgirth_Winter - 0.25984 * (74 - Day_Cumul),
ifelse(Day_Cumul > 74 & Pres_Cubs == "SANS" , Chestgirth_Winter + estime_poitrine_nocubs * (Day_Cumul - 74),
ifelse(Day_Cumul < 74 & Pres_Cubs == "SANS", Chestgirth_Winter - estime_poitrine_nocubs * (74 - Day_Cumul),
Chestgirth_Winter)))))
head(data[, c("Day_Cumul", "Pres_Cubs", "Chestgirth_Winter", "Chestgirth_Winter_Corr")]) #OK## # A tibble: 6 × 4
## Day_Cumul Pres_Cubs Chestgirth_Winter Chestgirth_Winter_Corr
## <dbl> <fct> <dbl> <dbl>
## 1 71 SANS 69 68.6
## 2 65 AVEC 70 67.7
## 3 65 SANS 71 69.7
## 4 66 SANS 70 68.9
## 5 55 SANS 69 66.3
## 6 68 SANS 72 71.2## # A tibble: 6 × 4
## Day_Cumul Pres_Cubs Chestgirth_Winter Chestgirth_Winter_Corr
## <dbl> <fct> <dbl> <dbl>
## 1 65 SANS 84 82.7
## 2 62 AVEC 82 78.9
## 3 52 AVEC 85 79.3
## 4 64 AVEC 85 82.4
## 5 63 SANS 88 86.5
## 6 55 SANS 53 50.3# Cou
data <- data %>%
mutate(Neck_Circum_Winter_Corr =
ifelse(Day_Cumul > 74 & Pres_Cubs == "AVEC" , Neck_Circum_Winter + estime_cou_cubs * (Day_Cumul - 74),
ifelse(Day_Cumul < 74 & Pres_Cubs == "AVEC", Neck_Circum_Winter - estime_cou_cubs * (74 - Day_Cumul),
ifelse(Day_Cumul > 74 & Pres_Cubs == "SANS" , Neck_Circum_Winter + estime_cou_nocubs * (Day_Cumul - 74),
ifelse(Day_Cumul < 74 & Pres_Cubs == "SANS", Neck_Circum_Winter - estime_cou_nocubs * (74 - Day_Cumul),
Neck_Circum_Winter)))))
head(data[, c("Day_Cumul", "Pres_Cubs", "Neck_Circum_Winter", "Neck_Circum_Winter_Corr")]) #OK## # A tibble: 6 × 4
## Day_Cumul Pres_Cubs Neck_Circum_Winter Neck_Circum_Winter_Corr
## <dbl> <fct> <dbl> <dbl>
## 1 71 SANS 42 42.0
## 2 65 AVEC 42 41.3
## 3 65 SANS 42 41.9
## 4 66 SANS 42 41.9
## 5 55 SANS 40 39.7
## 6 68 SANS 38 37.9## # A tibble: 6 × 4
## Day_Cumul Pres_Cubs Neck_Circum_Winter Neck_Circum_Winter_Corr
## <dbl> <fct> <dbl> <dbl>
## 1 65 SANS 45 44.9
## 2 62 AVEC 45 44.1
## 3 52 AVEC 45 43.3
## 4 64 AVEC 43 42.2
## 5 63 SANS 48 47.8
## 6 55 SANS 31 30.7# Longueur
# 2.3 Longueur selon âge
# Ajuster à 10 ans
data <- merge(x = data, y = age_length[, c(1, 3)], by = "Age", all.x = TRUE)
data$Total_Length_Winter_Age <- data$Total_Length_Winter + data$diff_to_10
# Vérif - exemple avec age 12
data %>%
drop_na(Total_Length_Winter) %>%
select(Age, Total_Length_Winter, diff_to_10, Total_Length_Winter_Age) %>%
filter(Age == 12) %>%
head()## Age Total_Length_Winter diff_to_10 Total_Length_Winter_Age
## 1 12 161 NA NA
## 2 12 152 NA NA
## 3 12 143 NA NA
## 4 12 151 NA NA
## 5 12 159 NA NA
## 6 12 153 NA NA# Selon l'analyse, il sera précisé si la longueur corrigée pour l'âge ou réelle sera utilisée.
## Cubs ##
# Il revient au même de corriger directement ici. Voir fichier estimés pour les calculs.
data <- data %>%
mutate(Weightx_Young_Corr_KG =
ifelse(Age_of_Youngs == "CUB" & Day_Cumul > 74, Weightx_Young_KG - estime_masse_cubs * (Day_Cumul - 74),
ifelse(Age_of_Youngs == "CUB" & Day_Cumul < 74, Weightx_Young_KG + estime_masse_cubs * (74 - Day_Cumul),
ifelse(Age_of_Youngs == "CUB" & Day_Cumul == 74, Weightx_Young_KG,
NA))))
head(na.omit(data[, c("Day_Cumul", "Weightx_Young_KG", "Weightx_Young_Corr_KG")]))## Day_Cumul Weightx_Young_KG Weightx_Young_Corr_KG
## 5 80 1.5573498 1.453712
## 13 56 0.9979135 1.308827
## 14 59 1.2020321 1.461127
## 18 61 1.6027095 1.827258
## 19 86 1.7539085 1.546632
## 20 82 2.2679851 2.129801## Day_Cumul Weightx_Young_KG Weightx_Young_Corr_KG
## 217 74 2.2679851 2.267985
## 220 65 1.2700717 1.425529
## 221 59 1.5724697 1.831565
## 225 60 1.6934289 1.935251
## 231 60 0.9979135 1.239735
## 234 59 0.9071940 1.166289#OK
# Calcul masse totale de la portée
data$Weight_Young_KG_Sum <- data$Weightx_Young_Corr_KG * data$Number_of_Youngs
summary(data$Weight_Young_KG_Sum)## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## 1.240 2.946 3.974 3.989 4.899 7.478 128# vérif
data %>%
drop_na(Weightx_Young_Corr_KG) %>%
select(ID_Animal, Number_of_Youngs, Weightx_Young_Corr_KG, Weight_Young_KG_Sum) %>% head() # ok## ID_Animal Number_of_Youngs Weightx_Young_Corr_KG Weight_Young_KG_Sum
## 1 S201608 3 1.453712 4.361135
## 2 S201617 2 1.308827 2.617655
## 3 S201622 2 1.461127 2.922254
## 4 O201961 3 1.827258 5.481775
## 5 G201903 3 1.546632 4.639897
## 6 G201916 2 2.129801 4.259602# Pas besoin de la masse des yearlings et de Weightx_Young_f/M, donc ils ne seront pas corrigés. À moins que l'on ne rajoute la croissance cub -> yearling comme variable.
# Les masses des femelles et oursons utilisées dans les analyses seront toujours les masse corrigées, sauf si le contraire est mentionné.Je crée l’indice de condition corporelle (ICC) avec les données morphométriques.
##### 1.2 Indice de condition corporelle ######
## PCA 1: PCA Impute Condition
data_pca1 <- data %>%
# filter(Age > 2) %>% # si jamais on veut enleve la 2 mais on fait quoi avec apres
select("ID_Animal", "Year_Winter", "Weight_Winter_Corr_KG", "Total_Length_Winter", "Neck_Circum_Winter_Corr", "Chestgirth_Winter_Corr")
dat <- data_pca1[, -c(1, 2)]
# Évaluer le nombre de components
#nb <- estim_ncpPCA(dat, scale=TRUE) # 1
#nb <- estim_ncpPCA(dat, ncp.min=0, ncp.max=5, method.cv="Kfold", scale=F) #2
#nb <- estim_ncpPCA(dat, ncp.min=0, ncp.max=5, method.cv="gcv", scale=F) # 1
#nb <- estim_ncpPCA(dat, ncp.min=0, ncp.max=5, method.cv="loo", scale=F) # 3
#nb <- estim_ncpPCA(dat, ncp.min=0, ncp.max=5, method.cv="gcv", scale=T)# 1
# Faire l'imputation
imp <- imputePCA(data_pca1[, -c(1, 2)], ncp=1, scale=T, nb.init = 5)
attributes(imp)## $names
## [1] "completeObs" "fittedX"## Weight_Winter_Corr_KG Total_Length_Winter Neck_Circum_Winter_Corr
## [1,] 16.76383 104 30.70474
## [2,] 21.43359 118 31.10878
## [3,] 52.70232 153 40.76690
## [4,] 60.21163 147 41.67366
## [5,] 73.56271 170 47.97064
## [6,] 68.38088 161 47.60878
## Chestgirth_Winter_Corr
## [1,] 50.33734
## [2,] 53.98098
## [3,] 74.89790
## [4,] 76.05706
## [5,] 93.55904
## [6,] 86.98098##
## Call:
## PCA(X = imp$completeObs, graph = FALSE)
##
##
## Eigenvalues
## Dim.1 Dim.2 Dim.3 Dim.4
## Variance 3.019 0.466 0.333 0.182
## % of var. 75.464 11.659 8.323 4.553
## Cumulative % of var. 75.464 87.123 95.447 100.000
##
## Individuals (the 10 first)
## Dist Dim.1 ctr cos2 Dim.2 ctr cos2
## 1 | 8.834 | -8.571 10.269 0.941 | -2.050 3.803 0.054
## 2 | 7.422 | -7.327 7.503 0.974 | -0.836 0.632 0.013
## 3 | 1.636 | -1.362 0.259 0.694 | 0.763 0.526 0.217
## 4 | 1.328 | -1.174 0.193 0.782 | -0.119 0.013 0.008
## 5 | 2.906 | 2.693 1.014 0.859 | 0.925 0.774 0.101
## 6 | 1.576 | 1.514 0.320 0.923 | 0.358 0.116 0.052
## 7 | 0.994 | -0.173 0.004 0.030 | -0.234 0.049 0.055
## 8 | 2.684 | -2.641 0.975 0.968 | 0.353 0.113 0.017
## 9 | 3.273 | -2.698 1.017 0.679 | -0.496 0.223 0.023
## 10 | 2.890 | -2.536 0.899 0.770 | 1.385 1.735 0.230
## Dim.3 ctr cos2
## 1 | -0.617 0.483 0.005 |
## 2 | -0.832 0.878 0.013 |
## 3 | -0.451 0.257 0.076 |
## 4 | -0.590 0.441 0.197 |
## 5 | -0.112 0.016 0.001 |
## 6 | 0.195 0.048 0.015 |
## 7 | 0.928 1.092 0.873 |
## 8 | 0.014 0.000 0.000 |
## 9 | 1.761 3.931 0.290 |
## 10 | 0.000 0.000 0.000 |
##
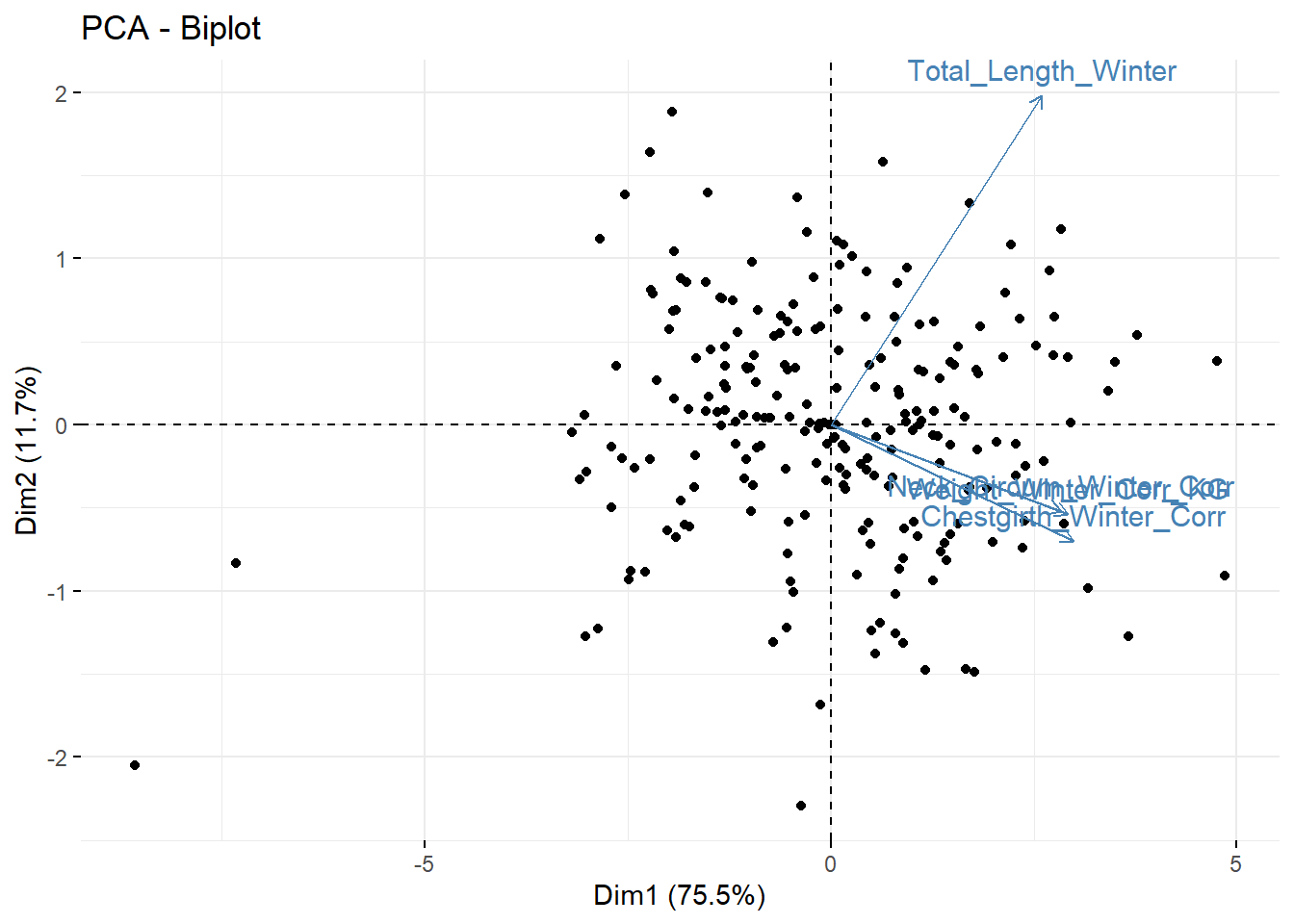
## Variables
## Dim.1 ctr cos2 Dim.2 ctr cos2 Dim.3
## Weight_Winter_Corr_KG | 0.894 26.486 0.799 | -0.165 5.834 0.027 | -0.332
## Total_Length_Winter | 0.795 20.934 0.632 | 0.606 78.833 0.368 | -0.007
## Neck_Circum_Winter_Corr | 0.867 24.911 0.752 | -0.160 5.470 0.026 | 0.460
## Chestgirth_Winter_Corr | 0.914 27.669 0.835 | -0.214 9.863 0.046 | -0.105
## ctr cos2
## Weight_Winter_Corr_KG 33.181 0.110 |
## Total_Length_Winter 0.013 0.000 |
## Neck_Circum_Winter_Corr 63.476 0.211 |
## Chestgirth_Winter_Corr 3.331 0.011 |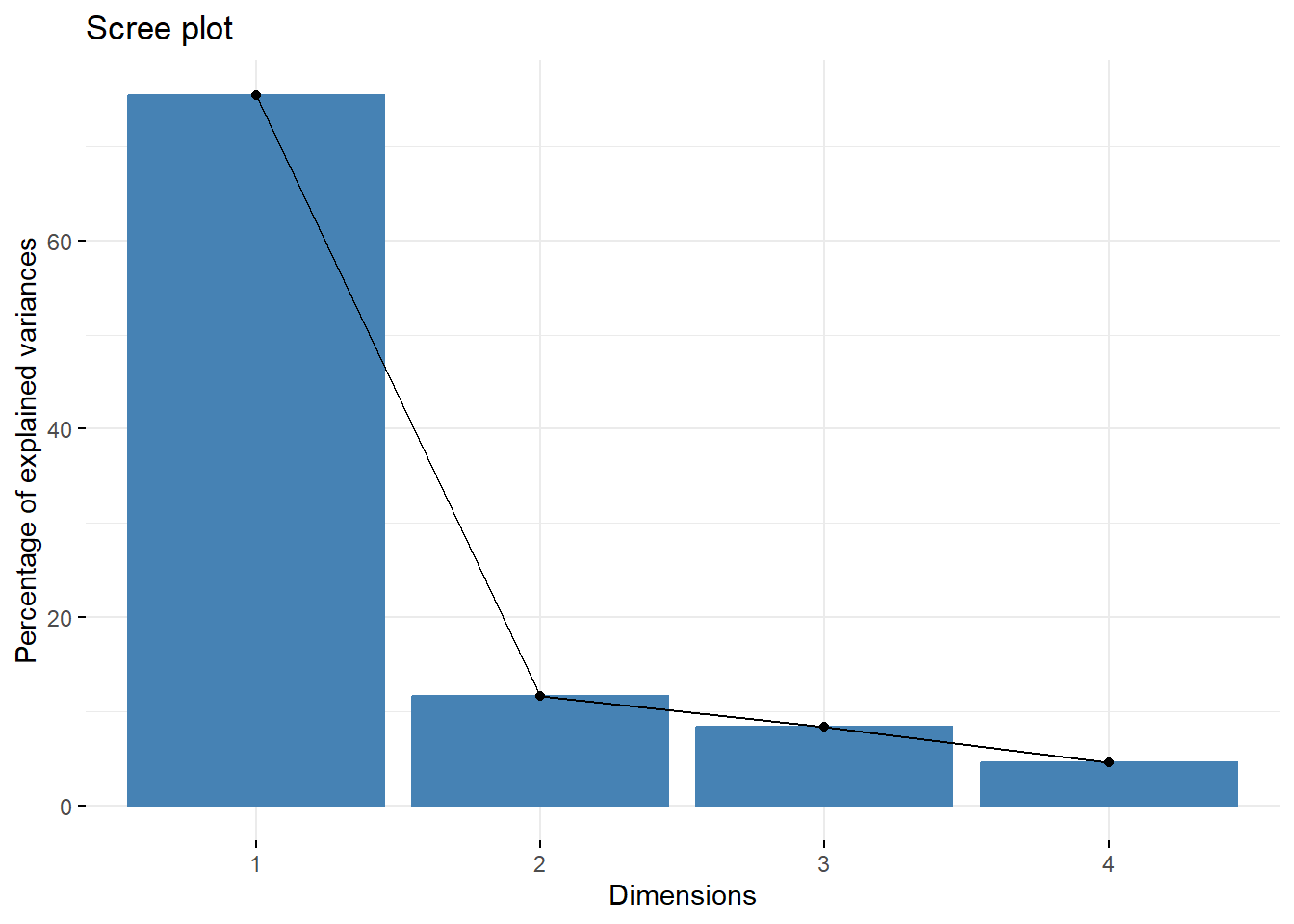
## Ajouter les données
# Ajouter scores PCA
res.ind <- as.data.frame(get_pca_ind(pca1)$coord[, 1])
res.ind$ID <- 1:nrow(res.ind)
colnames(res.ind) <- c("PCA1_Score", "ID") # 237 valeurs
# Ajouter les 237 ID
data_pca1$ID <- 1:nrow(data_pca1) # Variable ID
# Ajouter le score PCA à data_pca1
data_pca1 <- data_pca1 %>% left_join(res.ind, by = "ID")
# Ajouter les valeurs imputées à data_pca1
imp_values <- as.data.frame(imp$completeObs)
colnames(imp_values)<-paste(colnames(imp_values),"IMP",sep="_")
imp_values$ID <- 1:nrow(imp_values) # Variable ID
colnames(imp_values)## [1] "Weight_Winter_Corr_KG_IMP" "Total_Length_Winter_IMP"
## [3] "Neck_Circum_Winter_Corr_IMP" "Chestgirth_Winter_Corr_IMP"
## [5] "ID"data_pca1 <- data_pca1 %>% left_join(imp_values, by = "ID")
# Sauvegarder data_pca1
# saveRDS(data_pca1, file = "data_pca1.rds")
# Ajouter le tout à data
data <- data %>% left_join(data_pca1[, -c(3:7)], by = c("ID_Animal", "Year_Winter")) # Pas besoin d'ajouter aussi les valeurs de masse etc. originales
# Enregistrer
#saveRDS(data, "Data/data_femelles_full.rds")
# Version avec juste les colonnes pertinentes
data2 <- data %>%
select(c(1:3, 25:28, 33:34, 36, 42:43, 49, 51:52))
# Enregistrer
#saveRDS(data2, "Data/data_femelles_small.rds")2.2 Données d’oursons
Je charge maintenant les données d’oursons, encore une fois provenant de la base de données générée par le script de Maxime.
#### 2. Données d'oursons ####
data_cubs <- read_excel("C:/Users/Sarah/Desktop/Maitrise/Données/Data/BD_survie_oursons_2023-05-12_Final.xlsx")
load("C:/Users/Sarah/Desktop/Maitrise/Données/Data/estimes.RData") # Charger les estimés
colnames(data_cubs)## [1] "Secteur" "ID_Young"
## [3] "Sex" "Den_Weight_an_1"
## [5] "Den_Weight_an_2" "Annee"
## [7] "Mois" "Jour"
## [9] "ID_Mere" "Capture_Mother_an_1"
## [11] "Mother_Age" "Mother_Weight_an_1"
## [13] "Mother_Total_Length_an_1" "Mother_Neck_Circum_an_1"
## [15] "Mother_Chestgirth_an_1" "Mother_Body_Condition_an_1"
## [17] "Number_of_Youngs_an_1" "Weightx_Young_an_1"
## [19] "Capture_Mother_an_2" "Mother_Weight_an_2"
## [21] "Mother_Total_Length_an_2" "Mother_Neck_Circum_an_2"
## [23] "Mother_Chestgirth_an_2" "Mother_Body_Condition_an_2"
## [25] "Number_of_Youngs_an_2" "Weightx_Young_an_2"
## [27] "Cause_Suivi_Incomplet" "Survie"
## [29] "Duplicata"## tibble [298 × 29] (S3: tbl_df/tbl/data.frame)
## $ Secteur : chr [1:298] "Mauricie" "Mauricie" "Mauricie" "Mauricie" ...
## $ ID_Young : chr [1:298] "M201732" "M201975" "968000010358822" "M201865" ...
## $ Sex : chr [1:298] "M" "F" "M" "M" ...
## $ Den_Weight_an_1 : chr [1:298] "1.8" "4" "3.6" "3" ...
## $ Den_Weight_an_2 : chr [1:298] "25.2" "22" "NA" "28.8" ...
## $ Annee : chr [1:298] "2016" "2018" "2020" "2017" ...
## $ Mois : chr [1:298] "3" "3" "3" "2" ...
## $ Jour : chr [1:298] "5" "24" "7" "24" ...
## $ ID_Mere : chr [1:298] "M201503" "M201503" "M201503" "M201504" ...
## $ Capture_Mother_an_1 : chr [1:298] "OUI" "OUI" "OUI" "NON" ...
## $ Mother_Age : num [1:298] 11 13 15 10 12 12 6 9 16 9 ...
## $ Mother_Weight_an_1 : chr [1:298] "121.6" "125.6" "164.8" "152.19999999999999" ...
## $ Mother_Total_Length_an_1 : chr [1:298] "150" "146" "157" "156" ...
## $ Mother_Neck_Circum_an_1 : chr [1:298] "42" "42" "47" "49" ...
## $ Mother_Chestgirth_an_1 : chr [1:298] "70" "76" "85" "84" ...
## $ Mother_Body_Condition_an_1: chr [1:298] "BONNE" "BONNE" "EXCELLENTE" "BONNE" ...
## $ Number_of_Youngs_an_1 : chr [1:298] "2" "2" "1" "2" ...
## $ Weightx_Young_an_1 : chr [1:298] "2.4" "4.1" "3.6" "3" ...
## $ Capture_Mother_an_2 : chr [1:298] "NON" "NON" "NON" "NON" ...
## $ Mother_Weight_an_2 : chr [1:298] "106.2" "100.2" "NA" "94.2" ...
## $ Mother_Total_Length_an_2 : chr [1:298] "143" "140" "NA" "142" ...
## $ Mother_Neck_Circum_an_2 : chr [1:298] "45" "42" "NA" "43" ...
## $ Mother_Chestgirth_an_2 : chr [1:298] "75" "74" "NA" "74" ...
## $ Mother_Body_Condition_an_2: chr [1:298] "BONNE" "MOYENNE" "NA" "BONNE" ...
## $ Number_of_Youngs_an_2 : chr [1:298] "2" "2" "NA" "2" ...
## $ Weightx_Young_an_2 : chr [1:298] "24.7" "23.1" "NA" "29.2" ...
## $ Cause_Suivi_Incomplet : chr [1:298] "AUCUN" "AUCUN" "MORT" "AUCUN" ...
## $ Survie : chr [1:298] "OUI" "OUI" "NON" "OUI" ...
## $ Duplicata : chr [1:298] "NON" "NON" "NON" "NON" ...## Chr en Date ##
# Doit être fait en premier
# Date cub en tanière
data_cubs$Date <- as.Date(sprintf("%04d-%02d-%02d", as.numeric(data_cubs$Annee), as.numeric(data_cubs$Mois), as.numeric(data_cubs$Jour)))
data_cubs[,c("Date", "Annee", "Mois", "Jour")] #OK## # A tibble: 298 × 4
## Date Annee Mois Jour
## <date> <chr> <chr> <chr>
## 1 2016-03-05 2016 3 5
## 2 2018-03-24 2018 3 24
## 3 2020-03-07 2020 3 7
## 4 2017-02-24 2017 2 24
## 5 2019-03-24 2019 3 24
## 6 2017-02-27 2017 2 27
## 7 2017-02-24 2017 2 24
## 8 2020-03-14 2020 3 14
## 9 2017-02-28 2017 2 28
## 10 2016-03-12 2016 3 12
## # ℹ 288 more rows# Variable Jour cumulé
data_cubs$Day_Cumul <- yday(ymd(data_cubs$Date))
data_cubs[,c("Date", "Day_Cumul")] #OK## # A tibble: 298 × 2
## Date Day_Cumul
## <date> <dbl>
## 1 2016-03-05 65
## 2 2018-03-24 83
## 3 2020-03-07 67
## 4 2017-02-24 55
## 5 2019-03-24 83
## 6 2017-02-27 58
## 7 2017-02-24 55
## 8 2020-03-14 74
## 9 2017-02-28 59
## 10 2016-03-12 72
## # ℹ 288 more rows## Chr en facteurs ##
data_cubs <- data_cubs %>%
mutate_at(vars("Secteur", "Annee", "ID_Young", "Sex", "ID_Mere", "Capture_Mother_an_1", "Mother_Body_Condition_an_1", "Capture_Mother_an_2", "Mother_Body_Condition_an_2", "Cause_Suivi_Incomplet", "Survie", "Duplicata"), factor)
str(data_cubs) # OK## tibble [298 × 31] (S3: tbl_df/tbl/data.frame)
## $ Secteur : Factor w/ 4 levels "Gaspesie","Mauricie",..: 2 2 2 2 2 2 2 2 2 2 ...
## $ ID_Young : Factor w/ 228 levels "1","2","3","968000010355516",..: 112 138 74 131 52 NA NA 80 NA NA ...
## $ Sex : Factor w/ 3 levels "F","M","N/A": 2 1 2 2 1 1 2 2 1 2 ...
## $ Den_Weight_an_1 : chr [1:298] "1.8" "4" "3.6" "3" ...
## $ Den_Weight_an_2 : chr [1:298] "25.2" "22" "NA" "28.8" ...
## $ Annee : Factor w/ 8 levels "2016","2017",..: 1 3 5 2 4 2 2 5 2 1 ...
## $ Mois : chr [1:298] "3" "3" "3" "2" ...
## $ Jour : chr [1:298] "5" "24" "7" "24" ...
## $ ID_Mere : Factor w/ 83 levels "G201901","G201902",..: 12 12 12 13 13 14 15 15 16 17 ...
## $ Capture_Mother_an_1 : Factor w/ 2 levels "NON","OUI": 2 2 2 1 1 1 1 1 1 2 ...
## $ Mother_Age : num [1:298] 11 13 15 10 12 12 6 9 16 9 ...
## $ Mother_Weight_an_1 : chr [1:298] "121.6" "125.6" "164.8" "152.19999999999999" ...
## $ Mother_Total_Length_an_1 : chr [1:298] "150" "146" "157" "156" ...
## $ Mother_Neck_Circum_an_1 : chr [1:298] "42" "42" "47" "49" ...
## $ Mother_Chestgirth_an_1 : chr [1:298] "70" "76" "85" "84" ...
## $ Mother_Body_Condition_an_1: Factor w/ 5 levels "BONNE","EXCELLENTE",..: 1 1 2 1 3 1 1 3 1 2 ...
## $ Number_of_Youngs_an_1 : chr [1:298] "2" "2" "1" "2" ...
## $ Weightx_Young_an_1 : chr [1:298] "2.4" "4.1" "3.6" "3" ...
## $ Capture_Mother_an_2 : Factor w/ 2 levels "NON","OUI": 1 1 1 1 1 1 1 1 1 1 ...
## $ Mother_Weight_an_2 : chr [1:298] "106.2" "100.2" "NA" "94.2" ...
## $ Mother_Total_Length_an_2 : chr [1:298] "143" "140" "NA" "142" ...
## $ Mother_Neck_Circum_an_2 : chr [1:298] "45" "42" "NA" "43" ...
## $ Mother_Chestgirth_an_2 : chr [1:298] "75" "74" "NA" "74" ...
## $ Mother_Body_Condition_an_2: Factor w/ 6 levels "BONNE","EXCELLENTE",..: 1 4 6 1 6 2 4 6 6 5 ...
## $ Number_of_Youngs_an_2 : chr [1:298] "2" "2" "NA" "2" ...
## $ Weightx_Young_an_2 : chr [1:298] "24.7" "23.1" "NA" "29.2" ...
## $ Cause_Suivi_Incomplet : Factor w/ 9 levels "AUCUN","COLLAR_FAIL",..: 1 1 7 1 2 5 5 7 2 8 ...
## $ Survie : Factor w/ 3 levels "INDETERMINE",..: 3 3 2 3 1 3 1 2 1 3 ...
## $ Duplicata : Factor w/ 2 levels "NON","OUI": 1 1 1 1 1 1 1 1 1 1 ...
## $ Date : Date[1:298], format: "2016-03-05" "2018-03-24" ...
## $ Day_Cumul : num [1:298] 65 83 67 55 83 58 55 74 59 72 ...## Chr en numérique ##
data_cubs[, c(4, 5, 7, 8, 11, 12, 13, 14, 15, 17, 18, 20, 21, 22, 23, 25, 26)] <- sapply(data_cubs[, c(4, 5, 7, 8, 11, 12, 13, 14, 15, 17, 18, 20, 21, 22, 23, 25, 26)], as.numeric)
str(data_cubs) # OK, reste plus de chr## tibble [298 × 31] (S3: tbl_df/tbl/data.frame)
## $ Secteur : Factor w/ 4 levels "Gaspesie","Mauricie",..: 2 2 2 2 2 2 2 2 2 2 ...
## $ ID_Young : Factor w/ 228 levels "1","2","3","968000010355516",..: 112 138 74 131 52 NA NA 80 NA NA ...
## $ Sex : Factor w/ 3 levels "F","M","N/A": 2 1 2 2 1 1 2 2 1 2 ...
## $ Den_Weight_an_1 : num [1:298] 1.8 4 3.6 3 3.6 2.2 1.4 2.6 2.2 4 ...
## $ Den_Weight_an_2 : num [1:298] 25.2 22 NA 28.8 NA NA NA NA NA NA ...
## $ Annee : Factor w/ 8 levels "2016","2017",..: 1 3 5 2 4 2 2 5 2 1 ...
## $ Mois : num [1:298] 3 3 3 2 3 2 2 3 2 3 ...
## $ Jour : num [1:298] 5 24 7 24 24 27 24 14 28 12 ...
## $ ID_Mere : Factor w/ 83 levels "G201901","G201902",..: 12 12 12 13 13 14 15 15 16 17 ...
## $ Capture_Mother_an_1 : Factor w/ 2 levels "NON","OUI": 2 2 2 1 1 1 1 1 1 2 ...
## $ Mother_Age : num [1:298] 11 13 15 10 12 12 6 9 16 9 ...
## $ Mother_Weight_an_1 : num [1:298] 122 126 165 152 108 ...
## $ Mother_Total_Length_an_1 : num [1:298] 150 146 157 156 153 151 136 148 154 148 ...
## $ Mother_Neck_Circum_an_1 : num [1:298] 42 42 47 49 40 51 42 40 NA 42 ...
## $ Mother_Chestgirth_an_1 : num [1:298] 70 76 85 84 71 87 80 76 79 76 ...
## $ Mother_Body_Condition_an_1: Factor w/ 5 levels "BONNE","EXCELLENTE",..: 1 1 2 1 3 1 1 3 1 2 ...
## $ Number_of_Youngs_an_1 : num [1:298] 2 2 1 2 2 3 2 2 3 2 ...
## $ Weightx_Young_an_1 : num [1:298] 2.4 4.1 3.6 3 3.3 2.6 1.6 2.6 2.2 4 ...
## $ Capture_Mother_an_2 : Factor w/ 2 levels "NON","OUI": 1 1 1 1 1 1 1 1 1 1 ...
## $ Mother_Weight_an_2 : num [1:298] 106.2 100.2 NA 94.2 NA ...
## $ Mother_Total_Length_an_2 : num [1:298] 143 140 NA 142 NA 145 138 NA NA NA ...
## $ Mother_Neck_Circum_an_2 : num [1:298] 45 42 NA 43 NA 49 40 NA NA NA ...
## $ Mother_Chestgirth_an_2 : num [1:298] 75 74 NA 74 NA 83 67 NA NA NA ...
## $ Mother_Body_Condition_an_2: Factor w/ 6 levels "BONNE","EXCELLENTE",..: 1 4 6 1 6 2 4 6 6 5 ...
## $ Number_of_Youngs_an_2 : num [1:298] 2 2 NA 2 NA 2 1 NA NA 2 ...
## $ Weightx_Young_an_2 : num [1:298] 24.7 23.1 NA 29.2 NA 55.8 24.4 NA NA NaN ...
## $ Cause_Suivi_Incomplet : Factor w/ 9 levels "AUCUN","COLLAR_FAIL",..: 1 1 7 1 2 5 5 7 2 8 ...
## $ Survie : Factor w/ 3 levels "INDETERMINE",..: 3 3 2 3 1 3 1 2 1 3 ...
## $ Duplicata : Factor w/ 2 levels "NON","OUI": 1 1 1 1 1 1 1 1 1 1 ...
## $ Date : Date[1:298], format: "2016-03-05" "2018-03-24" ...
## $ Day_Cumul : num [1:298] 65 83 67 55 83 58 55 74 59 72 ...## Secteur ID_Young Sex Den_Weight_an_1
## Gaspesie : 26 N/A : 21 F :125 Min. :1.300
## Mauricie : 89 1 : 8 M :170 1st Qu.:3.000
## Outaouais : 73 2 : 8 N/A : 2 Median :3.600
## Saguenay-Lac-St-Jean:110 3 : 3 NA's: 1 Mean :3.678
## 968000010355516: 1 3rd Qu.:4.400
## (Other) :223 Max. :6.600
## NA's : 34 NA's :50
## Den_Weight_an_2 Annee Mois Jour ID_Mere
## Min. :12.00 2020 :115 Min. :2.000 Min. : 1.00 S201859: 10
## 1st Qu.:28.80 2022 : 41 1st Qu.:2.000 1st Qu.: 9.50 S201622: 9
## Median :34.40 2018 : 38 Median :3.000 Median :20.00 M201518: 8
## Mean :37.24 2019 : 33 Mean :2.765 Mean :17.48 S201614: 8
## 3rd Qu.:44.70 2017 : 29 3rd Qu.:3.000 3rd Qu.:26.00 S201748: 8
## Max. :89.00 2023 : 22 Max. :4.000 Max. :31.00 S201617: 7
## NA's :175 (Other): 20 (Other):248
## Capture_Mother_an_1 Mother_Age Mother_Weight_an_1
## NON:163 Min. : 4.00 Min. :104.0
## OUI:135 1st Qu.: 8.00 1st Qu.:132.6
## Median :11.00 Median :152.0
## Mean :10.82 Mean :153.0
## 3rd Qu.:13.00 3rd Qu.:166.4
## Max. :22.00 Max. :225.8
## NA's :41
## Mother_Total_Length_an_1 Mother_Neck_Circum_an_1 Mother_Chestgirth_an_1
## Min. :136.0 Min. :40.00 Min. : 69.00
## 1st Qu.:149.0 1st Qu.:44.00 1st Qu.: 80.00
## Median :153.0 Median :47.00 Median : 86.00
## Mean :154.2 Mean :46.31 Mean : 85.34
## 3rd Qu.:159.0 3rd Qu.:48.00 3rd Qu.: 90.00
## Max. :174.0 Max. :56.00 Max. :104.00
## NA's :47 NA's :54 NA's :47
## Mother_Body_Condition_an_1 Number_of_Youngs_an_1 Weightx_Young_an_1
## BONNE :118 Min. :1.000 Min. :1.467
## EXCELLENTE:117 1st Qu.:2.000 1st Qu.:2.950
## MOYENNE : 29 Median :2.000 Median :3.600
## N/A : 2 Mean :2.489 Mean :3.678
## NA : 32 3rd Qu.:3.000 3rd Qu.:4.400
## Max. :4.000 Max. :6.600
## NA's :32 NA's :50
## Capture_Mother_an_2 Mother_Weight_an_2 Mother_Total_Length_an_2
## NON:260 Min. : 87.2 Min. :131.0
## OUI: 38 1st Qu.:108.8 1st Qu.:148.0
## Median :124.2 Median :151.0
## Mean :125.6 Mean :150.2
## 3rd Qu.:142.4 3rd Qu.:154.0
## Max. :185.2 Max. :163.0
## NA's :136 NA's :147
## Mother_Neck_Circum_an_2 Mother_Chestgirth_an_2 Mother_Body_Condition_an_2
## Min. :39.00 Min. :67.00 BONNE : 70
## 1st Qu.:43.00 1st Qu.:74.00 EXCELLENTE: 44
## Median :45.00 Median :79.00 MAUVAISE : 11
## Mean :44.74 Mean :78.87 MOYENNE : 39
## 3rd Qu.:46.50 3rd Qu.:83.00 N/A : 7
## Max. :53.00 Max. :97.00 NA :127
## NA's :147 NA's :147
## Number_of_Youngs_an_2 Weightx_Young_an_2 Cause_Suivi_Incomplet
## Min. :1.000 Min. :17.67 AUCUN :77
## 1st Qu.:2.000 1st Qu.:29.25 MORT :49
## Median :2.000 Median :34.03 COLLAR_FAIL :46
## Mean :2.117 Mean :36.86 DECES_MERE :37
## 3rd Qu.:3.000 3rd Qu.:42.40 MERE_NON_SUIVIE :31
## Max. :3.000 Max. :77.10 MANQUE_INFO_OURSON:30
## NA's :127 NA's :136 (Other) :28
## Survie Duplicata Date Day_Cumul
## INDETERMINE:143 NON:287 Min. :2016-03-01 Min. :47.00
## NON : 53 OUI: 11 1st Qu.:2019-03-08 1st Qu.:60.00
## OUI :102 Median :2020-03-05 Median :71.50
## Mean :2019-12-24 Mean :70.32
## 3rd Qu.:2021-03-15 3rd Qu.:80.00
## Max. :2023-03-05 Max. :94.00
## ## [1] "Gaspesie" "Mauricie" "Outaouais"
## [4] "Saguenay-Lac-St-Jean"## Gaspesie Mauricie Outaouais
## 26 89 73
## Saguenay-Lac-St-Jean
## 110levels(data_cubs$Secteur) = c(levels(data_cubs$Secteur)[1:3],"SLSJ") # Niveau 4 = SLSJ
levels(data_cubs$Secteur)## [1] "Gaspesie" "Mauricie" "Outaouais" "SLSJ"## Gaspesie Mauricie Outaouais SLSJ
## 26 89 73 110# Changer la masse de lbs à kg
data_cubs <- data_cubs %>%
mutate(Den_Weight_an_1_KG = Den_Weight_an_1 / 2.2046) %>%
mutate(Den_Weight_an_2_KG = Den_Weight_an_2 / 2.2046) %>%
mutate(Mother_Weight_an_1_KG = Mother_Weight_an_1 / 2.2046) %>%
mutate(Mother_Weight_an_2_KG = Mother_Weight_an_2 / 2.2046) %>%
mutate(Weightx_Young_an_1_KG = Weightx_Young_an_1 / 2.2046) %>%
mutate(Weightx_Young_an_2_KG = Weightx_Young_an_2 / 2.2046)
ls(data_cubs) # Variables bien créées## [1] "Annee" "Capture_Mother_an_1"
## [3] "Capture_Mother_an_2" "Cause_Suivi_Incomplet"
## [5] "Date" "Day_Cumul"
## [7] "Den_Weight_an_1" "Den_Weight_an_1_KG"
## [9] "Den_Weight_an_2" "Den_Weight_an_2_KG"
## [11] "Duplicata" "ID_Mere"
## [13] "ID_Young" "Jour"
## [15] "Mois" "Mother_Age"
## [17] "Mother_Body_Condition_an_1" "Mother_Body_Condition_an_2"
## [19] "Mother_Chestgirth_an_1" "Mother_Chestgirth_an_2"
## [21] "Mother_Neck_Circum_an_1" "Mother_Neck_Circum_an_2"
## [23] "Mother_Total_Length_an_1" "Mother_Total_Length_an_2"
## [25] "Mother_Weight_an_1" "Mother_Weight_an_1_KG"
## [27] "Mother_Weight_an_2" "Mother_Weight_an_2_KG"
## [29] "Number_of_Youngs_an_1" "Number_of_Youngs_an_2"
## [31] "Secteur" "Sex"
## [33] "Survie" "Weightx_Young_an_1"
## [35] "Weightx_Young_an_1_KG" "Weightx_Young_an_2"
## [37] "Weightx_Young_an_2_KG"head(cbind(data_cubs$Den_Weight_an_1, data_cubs$Den_Weight_an_1_KG, data_cubs$Den_Weight_an_1_KG * 2.2046)) # OK## [,1] [,2] [,3]
## [1,] 1.8 0.8164746 1.8
## [2,] 4.0 1.8143881 4.0
## [3,] 3.6 1.6329493 3.6
## [4,] 3.0 1.3607911 3.0
## [5,] 3.6 1.6329493 3.6
## [6,] 2.2 0.9979135 2.2# Corriger les NA dans Mother_Body_Condition_an_1/AN2
data_cubs <- data_cubs %>%
replace_with_na(replace = list(Mother_Body_Condition_an_1 = c("NA", "N/A"), Mother_Body_Condition_an_2 = c("NA", "N/A")))
data_cubs$Mother_Body_Condition_an_1 <- droplevels(data_cubs$Mother_Body_Condition_an_1)
data_cubs$Mother_Body_Condition_an_2 <- droplevels(data_cubs$Mother_Body_Condition_an_2)
levels(data_cubs$Mother_Body_Condition_an_1)## [1] "BONNE" "EXCELLENTE" "MOYENNE"## [1] "BONNE" "EXCELLENTE" "MAUVAISE" "MOYENNE"## [1] "F" "M" "N/A"data_cubs <- data_cubs %>%
replace_with_na(replace = list(Sex = c("NA", "N/A")))
data_cubs$Sex <- droplevels(data_cubs$Sex)
levels(data_cubs$Sex) # OK!## [1] "F" "M"# Charger score pca
data_pca1 <- readRDS(file = "C:/Users/Sarah/Desktop/Maitrise/Données/data_pca1.rds")
# Renommer les colonnes pour fitter avec data_cubs
colnames(data_pca1) # enlever: length, ID## [1] "ID_Animal" "Year_Winter"
## [3] "Weight_Winter_Corr_KG" "Total_Length_Winter"
## [5] "Neck_Circum_Winter_Corr" "Chestgirth_Winter_Corr"
## [7] "ID" "PCA1_Score"
## [9] "Weight_Winter_Corr_KG_IMP" "Total_Length_Winter_IMP"
## [11] "Neck_Circum_Winter_Corr_IMP" "Chestgirth_Winter_Corr_IMP"colnames(data_pca1) <- c("ID_Mere", "Annee", "Mother_Weight_an_1_KG_Corr", "Mother_Total_Length_an_1", "Mother_Neck_Circum_an_1_Corr", "Mother_Chestgirth_an_1_Corr", "ID", "PCA1_Score", "Weight_Winter_Corr_KG_Imp", "Total_Length_Winter_Imp", "Neck_Circum_Winter_Corr_Imp", "Chestgirth_Winter_Corr_Imp")
colnames(data_pca1)## [1] "ID_Mere" "Annee"
## [3] "Mother_Weight_an_1_KG_Corr" "Mother_Total_Length_an_1"
## [5] "Mother_Neck_Circum_an_1_Corr" "Mother_Chestgirth_an_1_Corr"
## [7] "ID" "PCA1_Score"
## [9] "Weight_Winter_Corr_KG_Imp" "Total_Length_Winter_Imp"
## [11] "Neck_Circum_Winter_Corr_Imp" "Chestgirth_Winter_Corr_Imp"# enlever: length (5), ID (7)
data_cubs <- data_cubs %>% left_join(data_pca1[, -c(5, 7)], by = c("ID_Mere", "Annee"))
colnames(data_cubs)## [1] "Secteur" "ID_Young"
## [3] "Sex" "Den_Weight_an_1"
## [5] "Den_Weight_an_2" "Annee"
## [7] "Mois" "Jour"
## [9] "ID_Mere" "Capture_Mother_an_1"
## [11] "Mother_Age" "Mother_Weight_an_1"
## [13] "Mother_Total_Length_an_1.x" "Mother_Neck_Circum_an_1"
## [15] "Mother_Chestgirth_an_1" "Mother_Body_Condition_an_1"
## [17] "Number_of_Youngs_an_1" "Weightx_Young_an_1"
## [19] "Capture_Mother_an_2" "Mother_Weight_an_2"
## [21] "Mother_Total_Length_an_2" "Mother_Neck_Circum_an_2"
## [23] "Mother_Chestgirth_an_2" "Mother_Body_Condition_an_2"
## [25] "Number_of_Youngs_an_2" "Weightx_Young_an_2"
## [27] "Cause_Suivi_Incomplet" "Survie"
## [29] "Duplicata" "Date"
## [31] "Day_Cumul" "Den_Weight_an_1_KG"
## [33] "Den_Weight_an_2_KG" "Mother_Weight_an_1_KG"
## [35] "Mother_Weight_an_2_KG" "Weightx_Young_an_1_KG"
## [37] "Weightx_Young_an_2_KG" "Mother_Weight_an_1_KG_Corr"
## [39] "Mother_Total_Length_an_1.y" "Mother_Chestgirth_an_1_Corr"
## [41] "PCA1_Score" "Weight_Winter_Corr_KG_Imp"
## [43] "Total_Length_Winter_Imp" "Neck_Circum_Winter_Corr_Imp"
## [45] "Chestgirth_Winter_Corr_Imp"Il faut effectuer les corrections pour les oursons aussi.
##### 2.1 Corrections #####
# Mères: déjà fait avec data pca1
# Corriger masse CUB
data_cubs <- data_cubs %>%
mutate(Den_Weight_an_1_KG_Corr =
ifelse(Day_Cumul > 74, Den_Weight_an_1_KG - estime_masse_cubs * (Day_Cumul - 74),
ifelse(Day_Cumul < 74, Den_Weight_an_1_KG + estime_masse_cubs * (74 - Day_Cumul),
Den_Weight_an_1_KG)))
data_cubs[, c("Day_Cumul", "Den_Weight_an_1_KG", "Den_Weight_an_1_KG_Corr")] #OK## # A tibble: 298 × 3
## Day_Cumul Den_Weight_an_1_KG Den_Weight_an_1_KG_Corr
## <dbl> <dbl> <dbl>
## 1 65 0.816 0.972
## 2 83 1.81 1.66
## 3 67 1.63 1.75
## 4 55 1.36 1.69
## 5 83 1.63 1.48
## 6 58 0.998 1.27
## 7 55 0.635 0.963
## 8 74 1.18 1.18
## 9 59 0.998 1.26
## 10 72 1.81 1.85
## # ℹ 288 more rows# Nombre de données
# Manque-t-il des valeurs de masse
sum(!is.na(data_cubs$Den_Weight_an_1)) # 248 avec valeur masse## [1] 248## [1] 50## INDETERMINE NON OUI
## 143 53 102summary(data_cubs$Duplicata) # Si un jeune a survécu du stade ourson à yearling, mais qu'il est impossible de savoir de quel ourson il s'agissait au sein d'une portée (car pas de ID_Young, Cause_Suivi_Incomplet = MANQUE_INFO_OURSON), ce jeune apparaît deux fois dans la BD, il apparaît comme ourson et comme yearling. La variable Duplicata prend alors la valeur OUI pour ces jeunes au stade YEARLING.## NON OUI
## 287 11## [1] "AUCUN" "COLLAR_FAIL" "DECES_MERE"
## [4] "FIN_DU_PROJET" "MANQUE_INFO_OURSON" "MERE_NON_SUIVIE"
## [7] "MORT" "TANIERE_INACC" "VERIFIER"# Enregistrer
#saveRDS(data_cubs, "Data/data_cubs_full.rds")
# Garder juste les colonnes utiles
as.data.frame(colnames(data_cubs))## colnames(data_cubs)
## 1 Secteur
## 2 ID_Young
## 3 Sex
## 4 Den_Weight_an_1
## 5 Den_Weight_an_2
## 6 Annee
## 7 Mois
## 8 Jour
## 9 ID_Mere
## 10 Capture_Mother_an_1
## 11 Mother_Age
## 12 Mother_Weight_an_1
## 13 Mother_Total_Length_an_1.x
## 14 Mother_Neck_Circum_an_1
## 15 Mother_Chestgirth_an_1
## 16 Mother_Body_Condition_an_1
## 17 Number_of_Youngs_an_1
## 18 Weightx_Young_an_1
## 19 Capture_Mother_an_2
## 20 Mother_Weight_an_2
## 21 Mother_Total_Length_an_2
## 22 Mother_Neck_Circum_an_2
## 23 Mother_Chestgirth_an_2
## 24 Mother_Body_Condition_an_2
## 25 Number_of_Youngs_an_2
## 26 Weightx_Young_an_2
## 27 Cause_Suivi_Incomplet
## 28 Survie
## 29 Duplicata
## 30 Date
## 31 Day_Cumul
## 32 Den_Weight_an_1_KG
## 33 Den_Weight_an_2_KG
## 34 Mother_Weight_an_1_KG
## 35 Mother_Weight_an_2_KG
## 36 Weightx_Young_an_1_KG
## 37 Weightx_Young_an_2_KG
## 38 Mother_Weight_an_1_KG_Corr
## 39 Mother_Total_Length_an_1.y
## 40 Mother_Chestgirth_an_1_Corr
## 41 PCA1_Score
## 42 Weight_Winter_Corr_KG_Imp
## 43 Total_Length_Winter_Imp
## 44 Neck_Circum_Winter_Corr_Imp
## 45 Chestgirth_Winter_Corr_Imp
## 46 Den_Weight_an_1_KG_Corrdata_cubs2 <- data_cubs %>%
select(c(1:3, 6, 9, 27:31, 46)) %>%
filter(Duplicata == "NON")
# Enregistrer
#saveRDS(data_cubs2, "Data/data_cubs_small.rds")
#### 3. Survie des oursons ####
data_cubs = readRDS("C:/Users/Sarah/Desktop/Maitrise/Données/Data/data_cubs_full.rds")
survie_cubs <- data_cubs %>%
filter(Duplicata == "NON") %>%
filter(!(is.na(Den_Weight_an_1)))
# Enlever les duplicats et ceux qu'on a juste les infos yearling
survie_cubs %>%
group_by(Survie) %>%
summarise(n = n()) #ok## # A tibble: 3 × 2
## Survie n
## <fct> <int>
## 1 INDETERMINE 106
## 2 NON 52
## 3 OUI 90survie_cubs2 = survie_cubs %>%
group_by(ID_Mere, Year_Winter = Annee, Nb_Youngs = Number_of_Youngs_an_1, Survie) %>%
summarise(n = n()) %>%
pivot_wider(names_from = Survie,
values_from = n,
values_fill = 0,
names_prefix = "SurvieCubs_") %>%
rename(ID_Animal = ID_Mere) # Unifier le nom## `summarise()` has grouped output by 'ID_Mere',
## 'Year_Winter', 'Nb_Youngs'. You can override using the
## `.groups` argument.# Charger les femelles
data_fem = readRDS("C:/Users/Sarah/Desktop/Maitrise/Données/Data/Data_femelles_small.rds")
# Merger avec les données de survie
data_fem2 = merge(x = data_fem, y = survie_cubs2, by = c("ID_Animal", "Year_Winter"), all.x = TRUE)
# Calcul du ratio de survie
data_fem3 <- data_fem2 %>%
mutate(Ratio_Survie = (ifelse(SurvieCubs_OUI > 0 | SurvieCubs_NON > 0,
SurvieCubs_OUI/Nb_Youngs,
NA)))
# On calcule juste le ratio s'il y a des OUI ou NON. Si survie indéterminée, on laisse les NA.
# verif
data_fem3 %>% group_by(ind = as.factor(SurvieCubs_INDETERMINE)) %>% slice_sample(n = 2) %>% select("Nb_Youngs", ind = "SurvieCubs_INDETERMINE", oui = "SurvieCubs_OUI", non = "SurvieCubs_NON", "Ratio_Survie")## # A tibble: 12 × 5
## Nb_Youngs ind oui non Ratio_Survie
## <dbl> <int> <int> <int> <dbl>
## 1 2 0 2 0 1
## 2 2 0 2 0 1
## 3 1 1 0 0 NA
## 4 1 1 0 0 NA
## 5 2 2 0 0 NA
## 6 2 2 0 0 NA
## 7 4 3 1 0 0.25
## 8 3 3 0 0 NA
## 9 4 4 0 0 NA
## 10 4 4 0 0 NA
## 11 NA NA NA NA NA
## 12 NA NA NA NA NA#### 4. Remettre les masses des oursons ####
masse_cubs = data_cubs %>%
filter(!is.na(Den_Weight_an_1_KG_Corr)) %>% # Sert a rien si pas de masse
group_by(ID_Mere, Annee) %>% # Données par Mere/Annee pour fitter avec l'autre BD
mutate(nb_cub = row_number()) %>% # Nb du jeune dans la portée
select(ID_Mere, Annee, Number_of_Youngs_an_1, ID_Young, Den_Weight_an_1_KG_Corr, nb_cub) %>%
arrange(ID_Mere, Annee)
# Vérifier que ca marche
head(masse_cubs, n=10) # ok## # A tibble: 10 × 6
## # Groups: ID_Mere, Annee [5]
## ID_Mere Annee Number_of_Youngs_an_1 ID_Young Den_Weight_an_1_KG_Corr nb_cub
## <fct> <fct> <dbl> <fct> <dbl> <int>
## 1 G201902 2020 2 N/A 2.14 1
## 2 G201902 2020 2 N/A 2.05 2
## 3 G201903 2020 3 N/A 1.61 1
## 4 G201903 2020 3 N/A 1.43 2
## 5 G201903 2020 3 N/A 1.61 3
## 6 G201916 2020 2 N/A 2.04 1
## 7 G201916 2020 2 N/A 2.22 2
## 8 G201919 2020 2 N/A 0.834 1
## 9 G201919 2020 2 N/A 1.02 2
## 10 G201925 2020 3 N/A 1.94 1## # A tibble: 6 × 6
## # Groups: ID_Mere, Annee [6]
## ID_Mere Annee Number_of_Youngs_an_1 ID_Young Den_Weight_an_1_KG_C…¹ nb_cub
## <fct> <fct> <dbl> <fct> <dbl> <int>
## 1 G201938 2020 1 N/A 1.99 1
## 2 M201503 2020 1 96800001035… 1.75 1
## 3 M201738 2019 1 96800001035… 2.00 1
## 4 M201742 2018 1 <NA> 2.12 1
## 5 M201747 2019 1 96800001035… 1.84 1
## 6 M201986 2020 1 96800001035… 3.03 1
## # ℹ abbreviated name: ¹Den_Weight_an_1_KG_Corrmasse_cubs2 = masse_cubs %>%
select(-c(ID_Young, Number_of_Youngs_an_1)) %>% # Enlève les colonnes inutiles
pivot_wider(names_from = nb_cub,
values_from = Den_Weight_an_1_KG_Corr,
names_prefix = "Den_Weight_an_1_KG_Corr_Cub") %>%
rename(ID_Animal = ID_Mere,
Year_Winter = Annee) # Unifier le nom
# Merger
data_fem4 = merge(x = data_fem3, y = masse_cubs2, by = c("ID_Animal", "Year_Winter"), all.x = TRUE)
# Verif
data_fem4 %>%
filter(Age_of_Youngs == "CUB") %>%
select(ID_Animal, Year_Winter, Number_of_Youngs, cub1=Den_Weight_an_1_KG_Corr_Cub1, cub2=Den_Weight_an_1_KG_Corr_Cub2, cub3=Den_Weight_an_1_KG_Corr_Cub3, cub4=Den_Weight_an_1_KG_Corr_Cub4, moy=Weightx_Young_Corr_KG) %>%
sample_n(10)## ID_Animal Year_Winter Number_of_Youngs cub1 cub2 cub3 cub4
## 1 M201503 2018 2 1.6589311 1.749651 NA NA
## 2 G201919 2020 2 0.8338923 1.015331 NA NA
## 3 S201604 2022 2 1.5401250 1.540125 NA NA
## 4 S201617 2017 2 1.3088275 1.308827 NA NA
## 5 M201528 2020 2 1.9007531 2.082192 NA NA
## 6 S201614 2017 2 1.4729933 1.472993 NA NA
## 7 S2020109 2022 2 1.6071121 1.652472 NA NA
## 8 M201522 2019 3 2.2205205 2.039082 1.857643 NA
## 9 O201719 2020 3 2.4061691 2.496889 2.406169 NA
## 10 G201902 2020 2 2.1385099 2.047791 NA NA
## moy
## 1 1.7042908
## 2 0.9246117
## 3 1.5401250
## 4 1.3088275
## 5 1.9914725
## 6 1.4729933
## 7 1.6297919
## 8 2.0390817
## 9 2.4364089
## 10 2.0931502# Verif2
data_fem4 %>%
group_by(as.factor(Number_of_Youngs)) %>%
summarise(n = n(), cub1 = mean(Den_Weight_an_1_KG_Corr_Cub1, na.rm=T),
cub2= mean(Den_Weight_an_1_KG_Corr_Cub2, na.rm=T),
cub3 = mean(Den_Weight_an_1_KG_Corr_Cub3, na.rm=T),
cub4 = mean(Den_Weight_an_1_KG_Corr_Cub4, na.rm=T))## # A tibble: 5 × 6
## `as.factor(Number_of_Youngs)` n cub1 cub2 cub3 cub4
## <fct> <int> <dbl> <dbl> <dbl> <dbl>
## 1 0 52 NaN NaN NaN NaN
## 2 1 34 2.02 NaN NaN NaN
## 3 2 95 1.74 1.78 NaN NaN
## 4 3 53 1.80 1.73 1.75 NaN
## 5 4 3 1.42 1.45 1.30 1.21# Verif3
data_fem4 %>% filter(!is.na(Den_Weight_an_1_KG_Corr_Cub1)) %>% select(ID_Animal, Year_Winter) %>% sample_n(1) # M201529 2017## ID_Animal Year_Winter
## 1 S201871 2020## ID_Animal Year_Winter Age Secteur Body_Condition_Winter Presence_of_Youngs
## 1 M201529 2017 11 Mauricie BONNE OUI
## Age_of_Youngs Number_of_Youngs Id_Young Day_Cumul Pres_Cubs Reprod
## 1 CUB 2 NA; NA 53 AVEC 1
## Weightx_Young_Corr_KG PCA1_Score Weight_Winter_Corr_KG_IMP Nb_Youngs
## 1 1.451366 1.346687 76.83153 2
## SurvieCubs_INDETERMINE SurvieCubs_OUI SurvieCubs_NON Ratio_Survie
## 1 0 2 0 1
## Den_Weight_an_1_KG_Corr_Cub1 Den_Weight_an_1_KG_Corr_Cub2
## 1 1.542085 1.360646
## Den_Weight_an_1_KG_Corr_Cub3 Den_Weight_an_1_KG_Corr_Cub4
## 1 NA NAdata_cubs %>% filter(ID_Mere == "M201529" & Annee == "2017") %>% select(Den_Weight_an_1_KG_Corr) # 1.54 et 1.36 = ok## # A tibble: 2 × 1
## Den_Weight_an_1_KG_Corr
## <dbl>
## 1 1.54
## 2 1.36# Rajouter colonnes nb oursons survivants et morts
data_fem4$Nb_Cubs_Survivants = data_fem4$Ratio_Survie * data_fem4$Number_of_Youngs
data_fem4$Nb_Cubs_Morts = data_fem4$Number_of_Youngs - data_fem4$Nb_Cubs_Survivants
# Verif
data_fem4 %>%
filter(Age_of_Youngs != "YEARLING") %>%
select(ID_Animal, Number_of_Youngs, Nb_Cubs_Survivants, Nb_Cubs_Morts) %>%
group_by(Number_of_Youngs) %>%
slice_sample(n = 2) # OK## # A tibble: 10 × 4
## # Groups: Number_of_Youngs [5]
## ID_Animal Number_of_Youngs Nb_Cubs_Survivants Nb_Cubs_Morts
## <fct> <dbl> <dbl> <dbl>
## 1 M201522 0 NA NA
## 2 S201855 0 NA NA
## 3 M201503 1 0 1
## 4 O202196 1 NA NA
## 5 M201518 2 0 2
## 6 S201995 2 2 0
## 7 O201719 3 3 0
## 8 G201903 3 NA NA
## 9 S201995 4 1 3
## 10 O201945 4 NA NA# Verif 2 avec juste ceux avec données de survie
data_fem4 %>%
filter(Age_of_Youngs != "YEARLING" & !is.na(Ratio_Survie)) %>%
select(ID_Animal, Number_of_Youngs, Nb_Cubs_Survivants, Nb_Cubs_Morts) %>%
group_by(Number_of_Youngs) %>%
slice_sample(n = 2) # OK## # A tibble: 7 × 4
## # Groups: Number_of_Youngs [4]
## ID_Animal Number_of_Youngs Nb_Cubs_Survivants Nb_Cubs_Morts
## <fct> <dbl> <dbl> <dbl>
## 1 S201622 1 1 0
## 2 M201742 1 0 1
## 3 S201604 2 2 0
## 4 M201522 2 1 1
## 5 O201719 3 3 0
## 6 O201831 3 1 2
## 7 S201995 4 1 3Les données utilisées pour tester l’H1 sont prêtes.