# Variables indépendantes (x)
# Secteur: Région d'étude
# *NEW*: US_Graminees, US_Fruits et US_Noix
# Score_Pond: Score de l'habitat
# PCA1_Score: Indice de condition corporelle (ICC)
# Variables dépendantes (y)
# Age_of_Youngs: Yearling, Cub ou None (none = échec reproduction, cub = succès, yearling = NA)
# Number_of_Youngs: Nombre de jeunes
# Weightx_Young_Corr_KG: Masse moyenne des jeunes
# Den_Weight_an_1_KG_Corr_Cub1-4: Masse par ourson
# Ratio_Survie: Ratio de survie des cubs vers yearling
# Variables aléatoires
# ID_Animal: Identifiant de l'animal
# Year_Winter: Année de la prise de données en tanière (pas hyp1)
# Secteur: Secteur d'étude (pas hyp1)
# Covariable potentielle
# Age: Âge en années. Gardé au cas où.
# Faire un fichier avec juste ces variables
data = data0 %>%
dplyr::select("ID_Animal", "Site", "Year_Winter", "PCA1_Score", "Age", "Age_of_Youngs", "Number_of_Youngs", "Weightx_Young_Corr_KG", "Den_Weight_an_1_KG_Corr_Cub1", "Den_Weight_an_1_KG_Corr_Cub2", "Den_Weight_an_1_KG_Corr_Cub3", "Den_Weight_an_1_KG_Corr_Cub4", "Ratio_Survie", "Nb_Cubs_Survivants", "Nb_Cubs_Morts", "Score_Graminees_Pond_Tot", "Score_Fruits_Pond_Tot", "Score_Fourmis_Pond_Tot", "Score_Noix_Pond_Tot", "US_Graminees_Tot", "US_Fruits_Tot", "US_Noix_Tot") %>%
mutate(Age_of_Youngs = fct_relevel(Age_of_Youngs, "NONE", "CUB", "YEARLING", "2 YEARS OLD")) # relevel
str(data) # ok
Étape 6
#### 6.1 - Données aberrantes en x et y ####
# On doit regarder s’il y a des données aberrantes dans les X ou le Y.
### Les X ###
# Graminées
data %>%
ggplot(aes(x = US_Graminees_Tot)) +
geom_histogram(colour="black", fill="steelblue3") +
labs(title = "Score graminées")
## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.
## Warning: Removed 169 rows containing non-finite outside the
## scale range (`stat_bin()`).
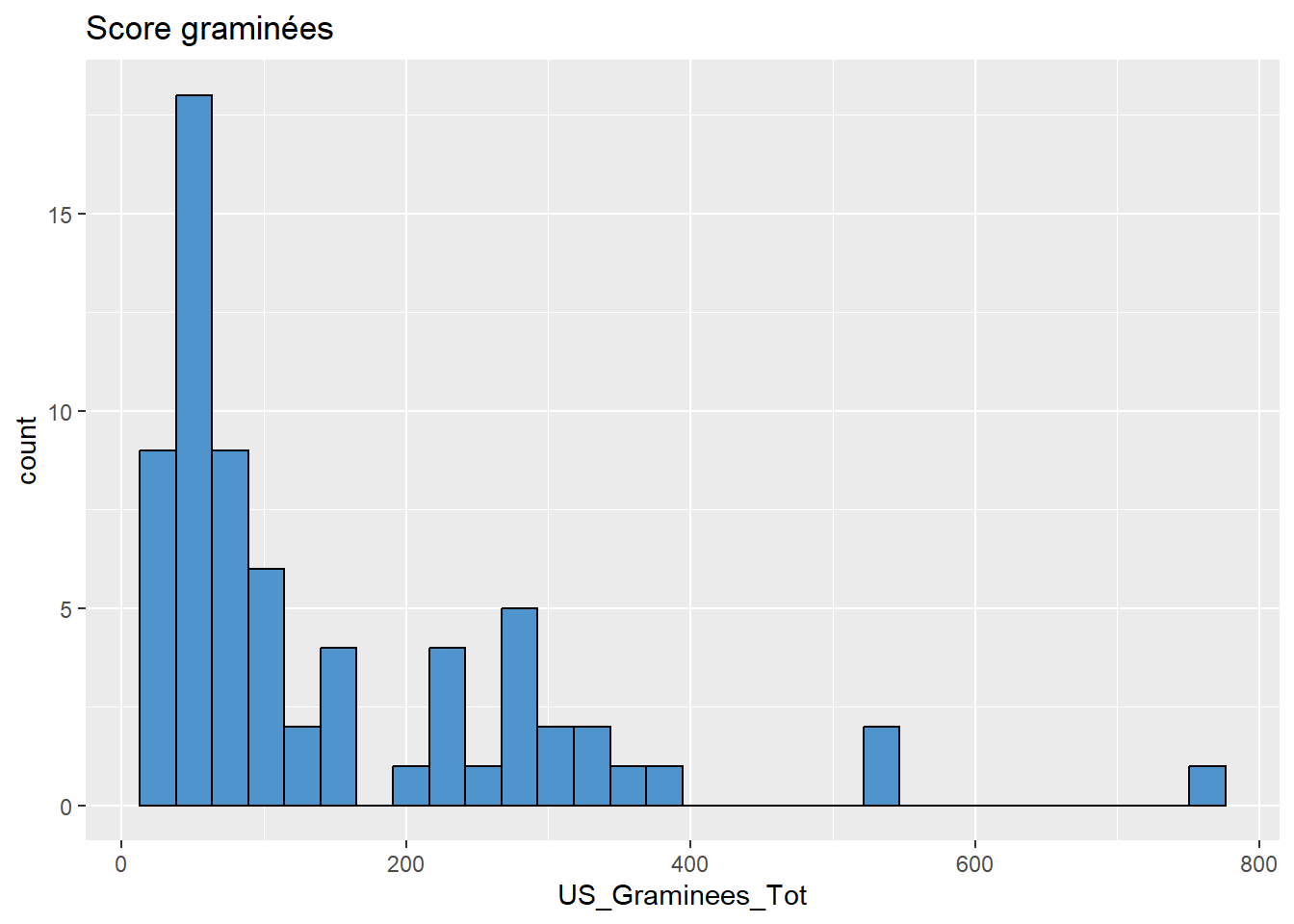
data %>%
ggplot(aes(x = log(US_Graminees_Tot+1))) +
geom_histogram(colour="black", fill="steelblue3") +
labs(title = "Score graminées log+1")
## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.
## Warning: Removed 169 rows containing non-finite outside the
## scale range (`stat_bin()`).
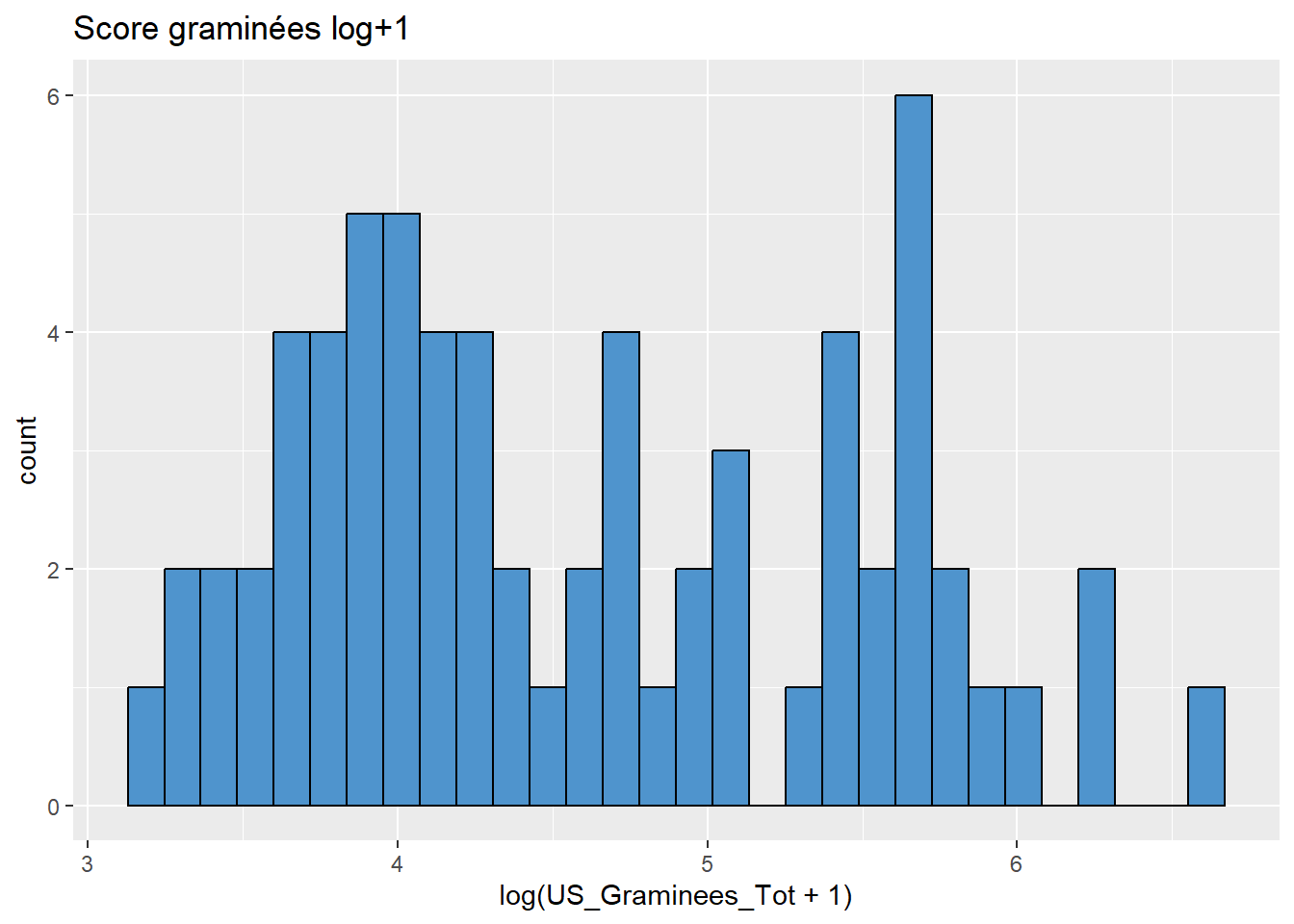
# mieux!
# Fruits
data %>%
ggplot(aes(x = US_Fruits_Tot)) +
geom_histogram(colour="black", fill="steelblue3") +
labs(title = "Score fruits")
## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.
## Warning: Removed 99 rows containing non-finite outside the scale
## range (`stat_bin()`).
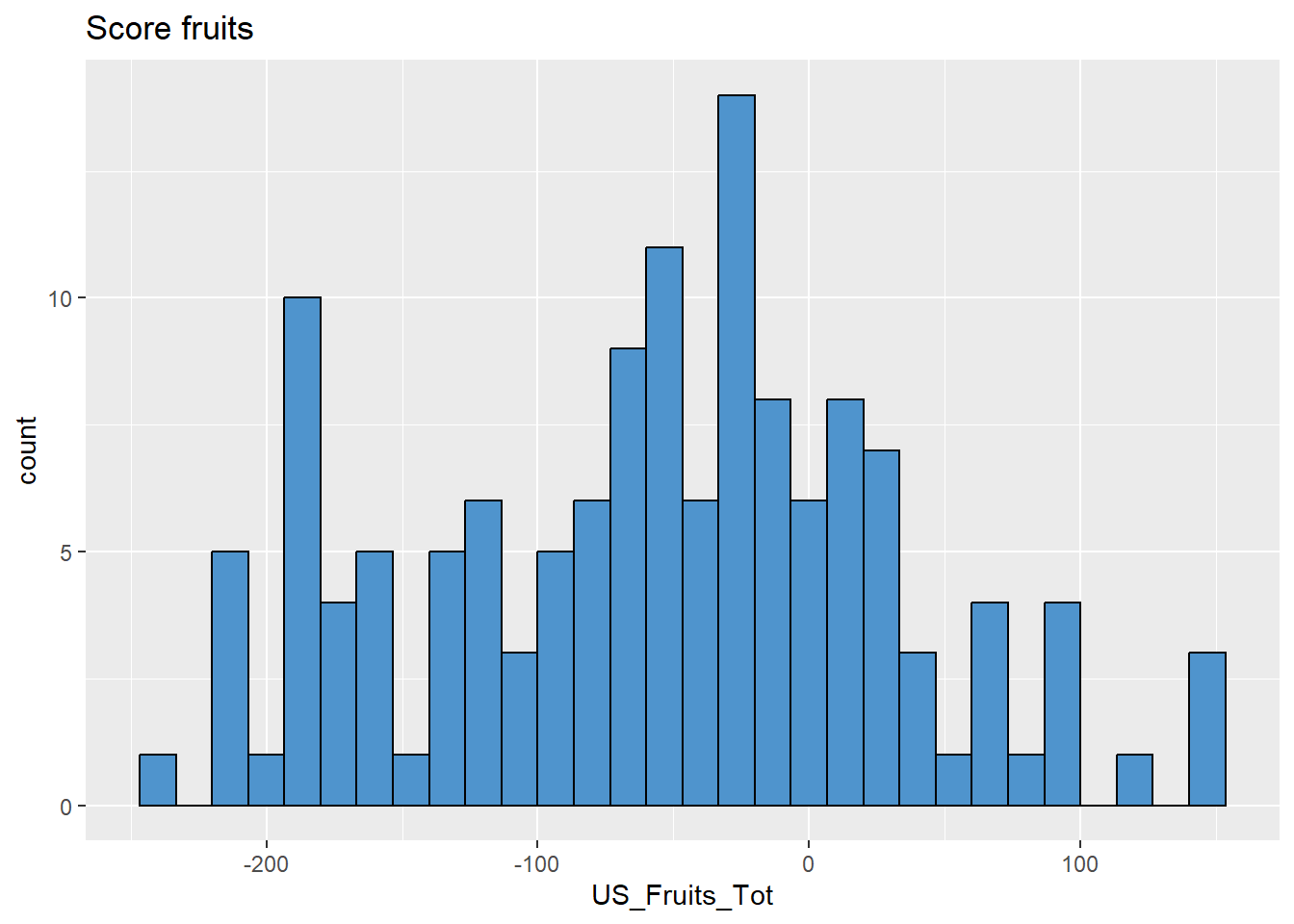
# surprenamment pas si pire?
# Noix
data %>%
ggplot(aes(x = US_Noix_Tot)) +
geom_histogram(colour="black", fill="steelblue3") +
labs(title = "Score fruits")
## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.
## Warning: Removed 100 rows containing non-finite outside the
## scale range (`stat_bin()`).
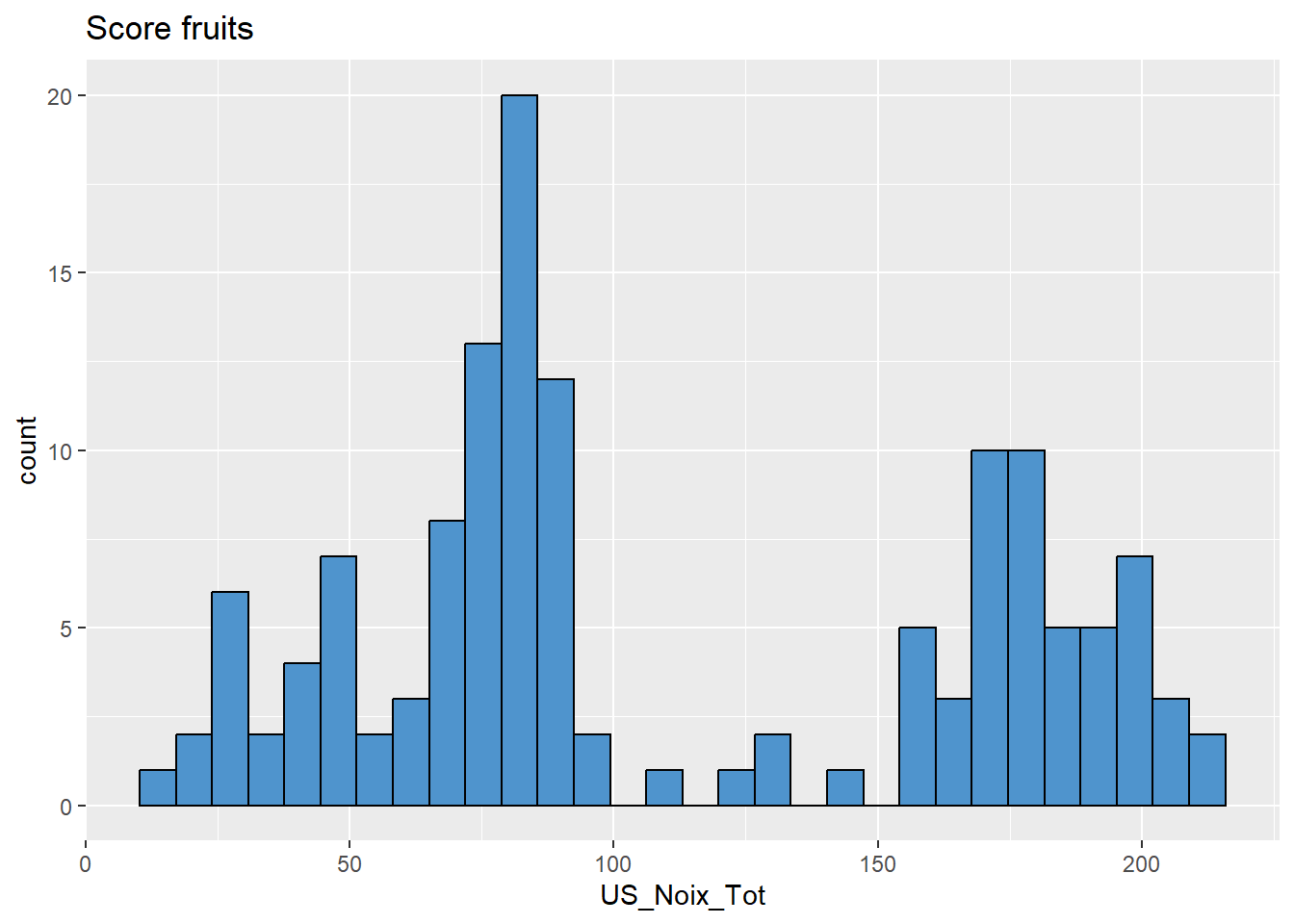
data %>%
ggplot(aes(x = log(US_Graminees_Tot+1))) +
geom_histogram(colour="black", fill="steelblue3") +
labs(title = "Score noix log +1")
## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.
## Warning: Removed 169 rows containing non-finite outside the
## scale range (`stat_bin()`).
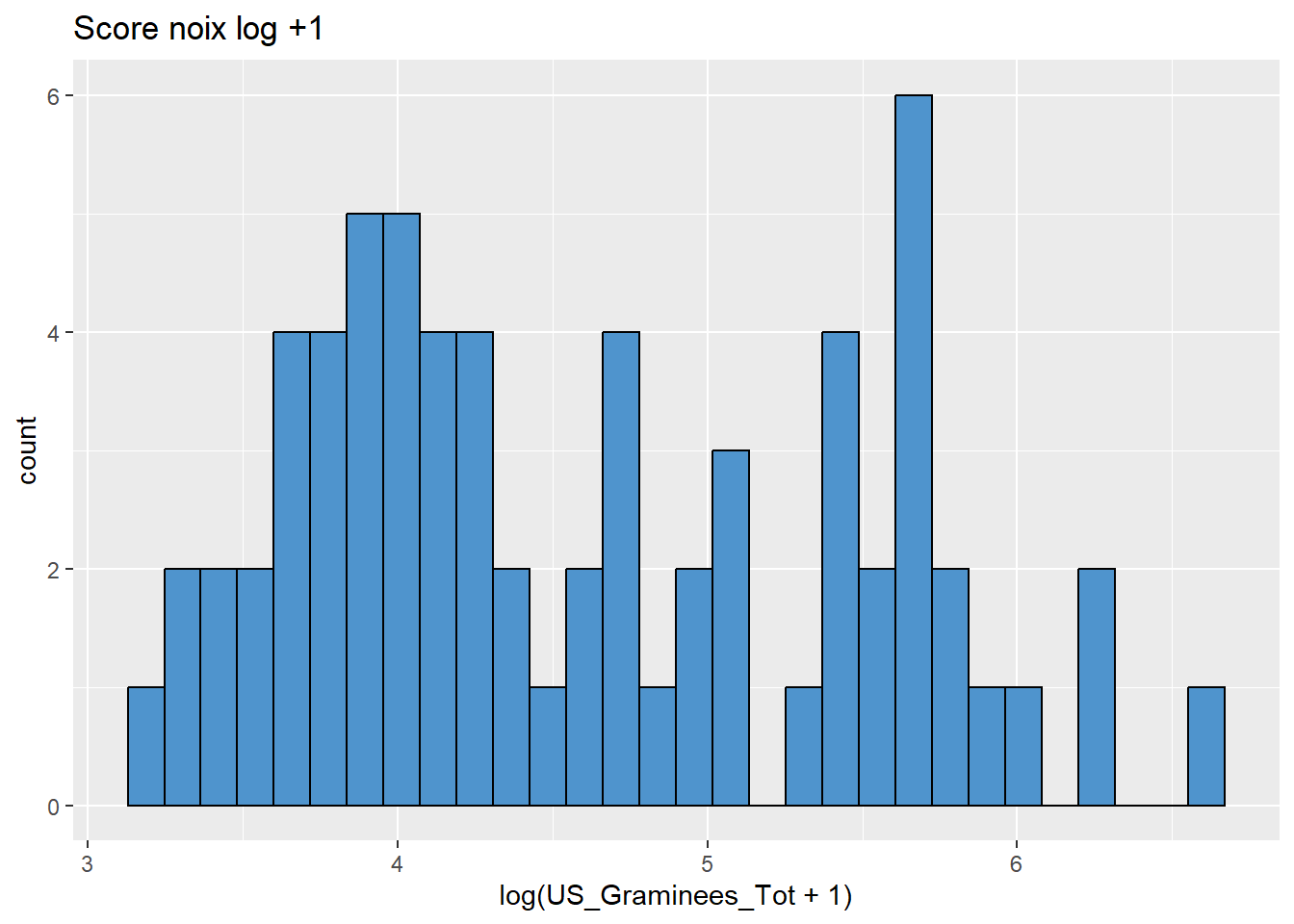
# mieux!
### Les Y ###
# Pas de nouveau Y.
#### 6.2 - Homogénéité de la variance des Y####
# Pas de nouvelle catégorie
#### 6.3 - Normalité des Y ####
# Pas de nouveau Y
#### 6.4 - Problèmes de 0 dans les Y ####
# Pas de nouveau Y
#### 6.5 - Colinéarité des X ####
as.data.frame(colnames(data))
## colnames(data)
## 1 ID_Animal
## 2 Site
## 3 Year_Winter
## 4 PCA1_Score
## 5 Age
## 6 Age_of_Youngs
## 7 Number_of_Youngs
## 8 Weightx_Young_Corr_KG
## 9 Den_Weight_an_1_KG_Corr_Cub1
## 10 Den_Weight_an_1_KG_Corr_Cub2
## 11 Den_Weight_an_1_KG_Corr_Cub3
## 12 Den_Weight_an_1_KG_Corr_Cub4
## 13 Ratio_Survie
## 14 Nb_Cubs_Survivants
## 15 Nb_Cubs_Morts
## 16 Score_Graminees_Pond_Tot
## 17 Score_Fruits_Pond_Tot
## 18 Score_Fourmis_Pond_Tot
## 19 Score_Noix_Pond_Tot
## 20 US_Graminees_Tot
## 21 US_Fruits_Tot
## 22 US_Noix_Tot
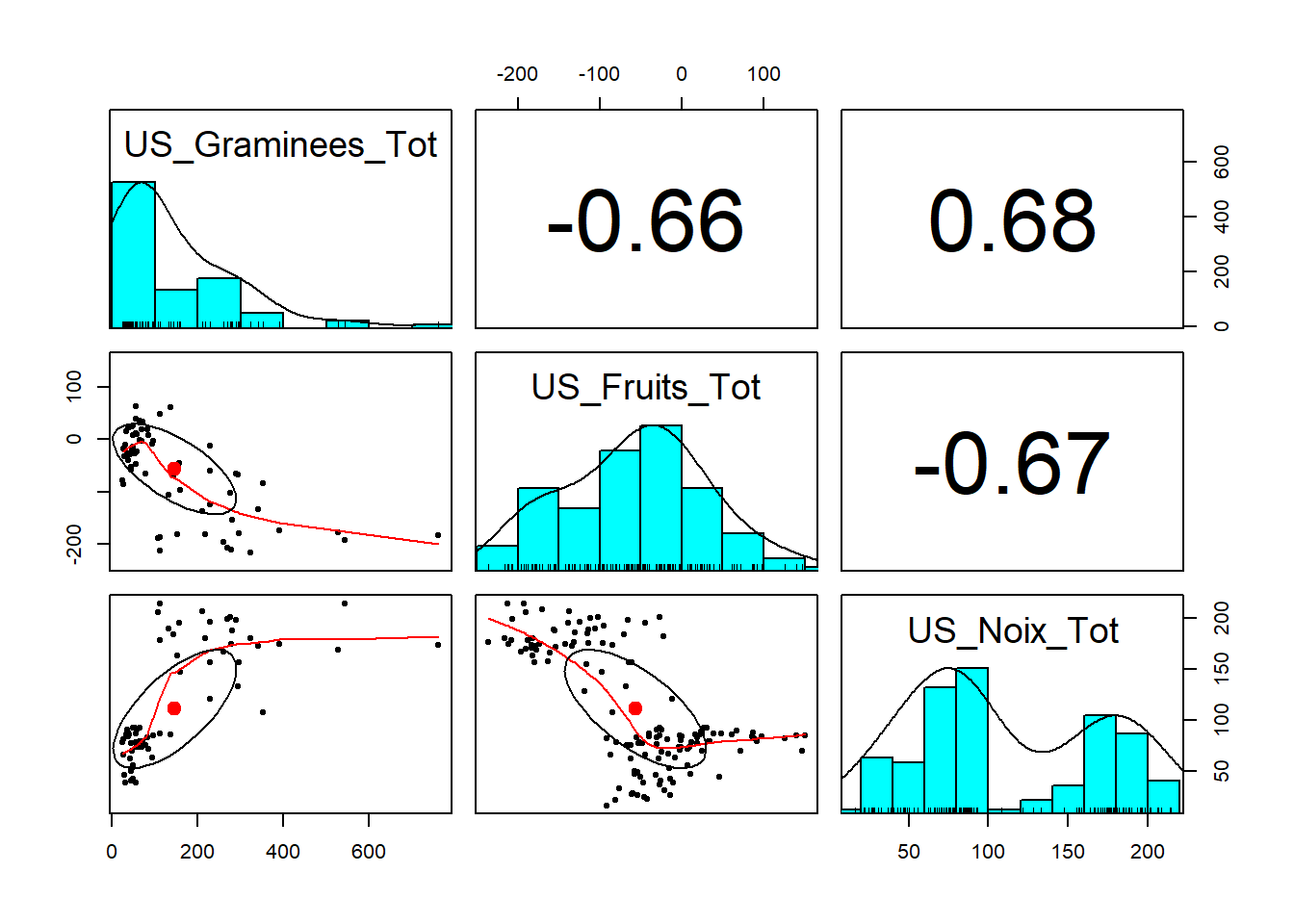
etape6_5 = unique(data[,c(20:22)])
# Pearson
pairs.panels(etape6_5, method = "pearson")
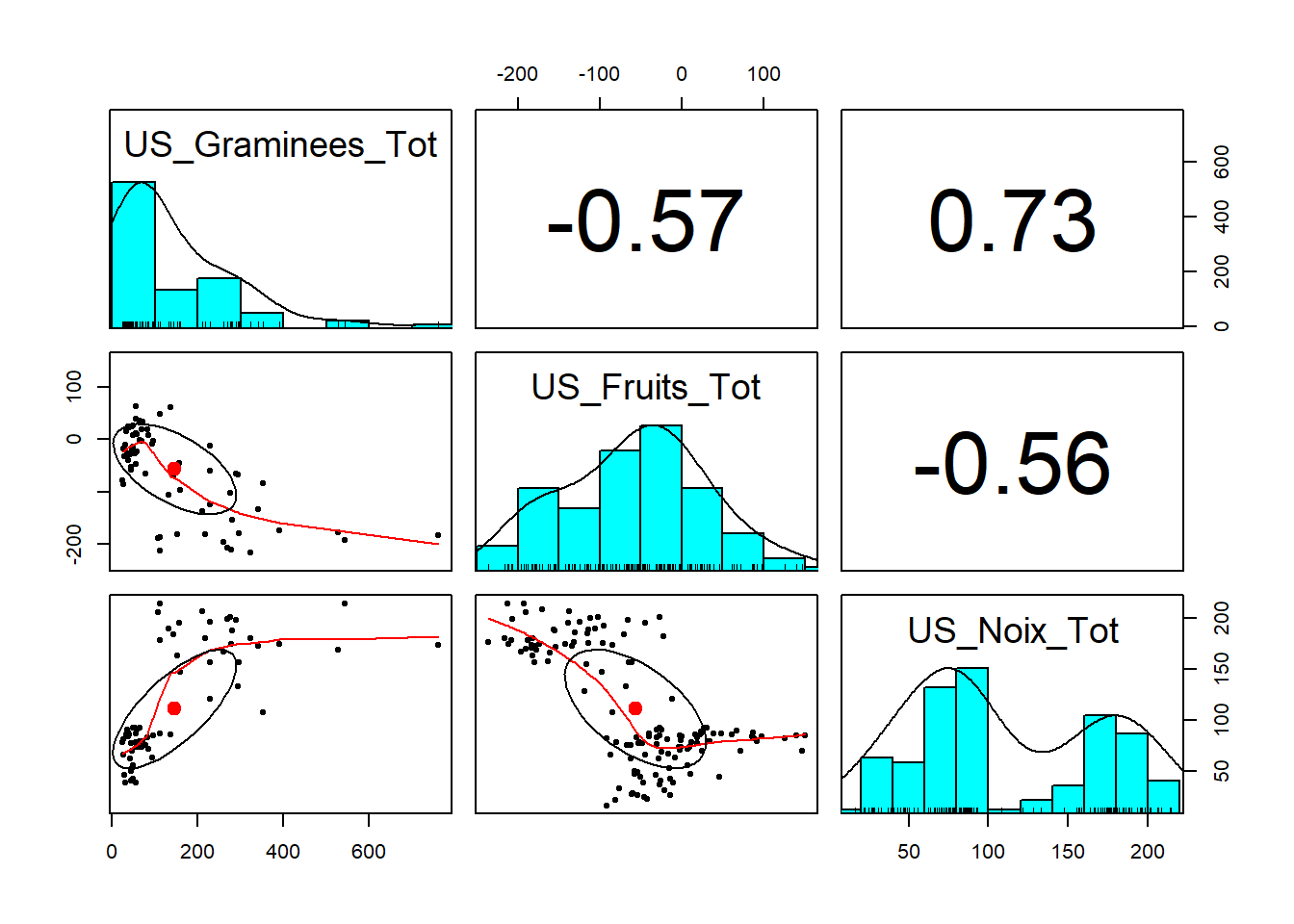
# Spearman
pairs.panels(etape6_5, method = "spearman")
#### 6.6 - Relations entre les X et Y ####
# Nombre de jeunes
Z <- as.vector(as.matrix(data[, c(20:22)]))
Y10 <- rep(data$Number_of_Youngs, 3)
MyNames <- colnames(data)[20:22]
ID10 <- rep(MyNames, each = length(data$Number_of_Youngs))
ID11 <- factor(ID10, labels = MyNames,
levels = MyNames)
xyplot(Y10 ~ Z | ID11, col = 1,
strip = function(bg='white',...) strip.default(bg='white',...),
scales = list(alternating = T,
x = list(relation = "free"),
y = list(relation = "same")),
xlab = "Habitats",
par.strip.text = list(cex = 0.8),
ylab = "Nb jeunes",
panel=function(x, y, subscripts,...){
panel.grid(h =- 1, v = 2)
panel.points(x, y, col = 1, pch = 16)
panel.loess(x,y,col=1,lwd=2)
})
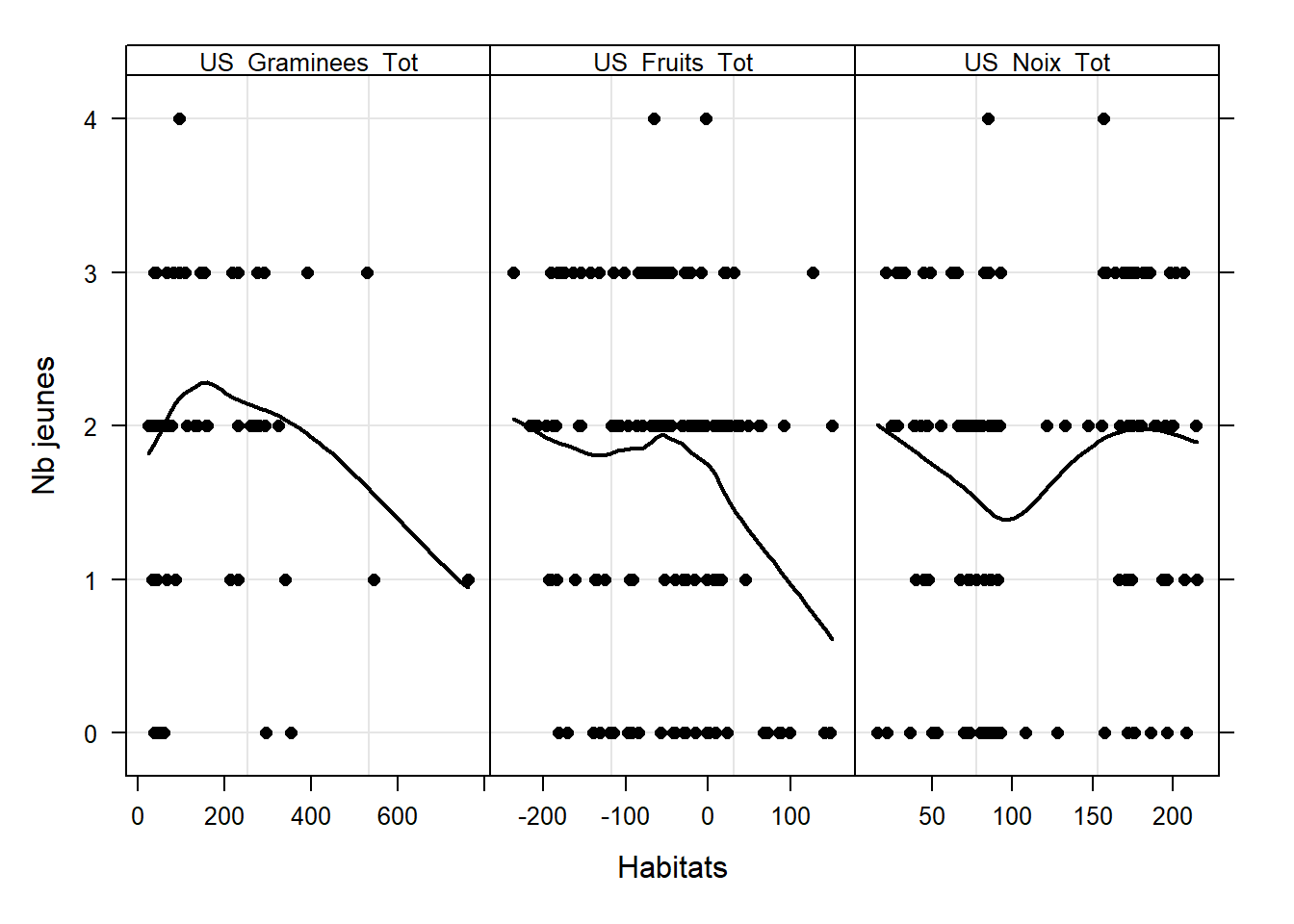
# Masse des jeunes
Z <- as.vector(as.matrix(data[, c(20:22)]))
Y10 <- rep(data$Weightx_Young_Corr_KG, 3)
MyNames <- colnames(data)[20:22]
ID10 <- rep(MyNames, each = length(data$Weightx_Young_Corr_KG))
ID11 <- factor(ID10, labels = MyNames,
levels = MyNames)
xyplot(Y10 ~ Z | ID11, col = 1,
strip = function(bg='white',...) strip.default(bg='white',...),
scales = list(alternating = T,
x = list(relation = "free"),
y = list(relation = "same")),
xlab = "Habitats",
par.strip.text = list(cex = 0.8),
ylab = "Masse moyenne de la portée (kg)",
panel=function(x, y, subscripts,...){
panel.grid(h =- 1, v = 2)
panel.points(x, y, col = 1, pch = 16)
panel.loess(x,y,col=1,lwd=2)
})
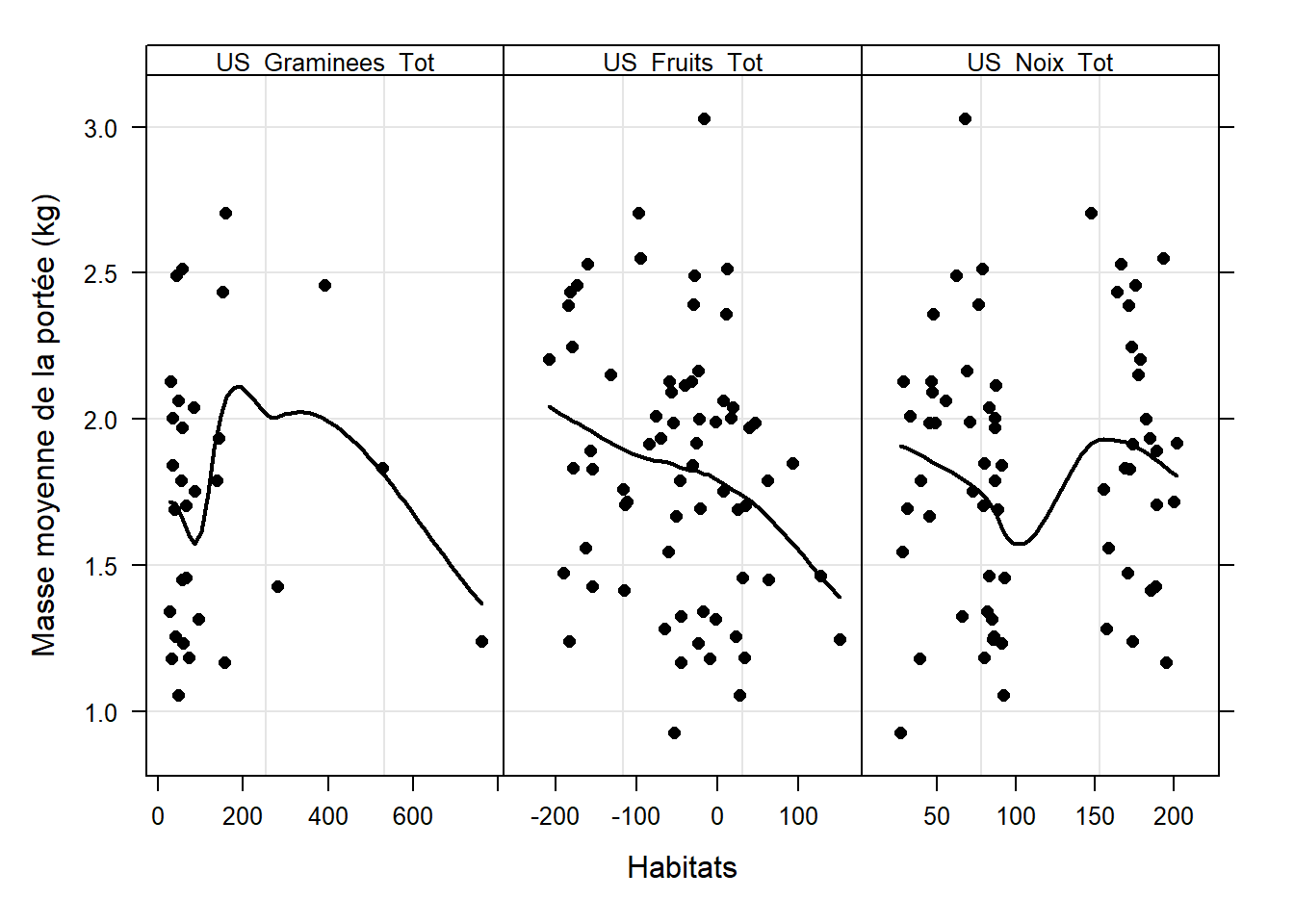
# Nb jeunes survivants
Z <- as.vector(as.matrix(data[, c(20:22)]))
Y10 <- rep(data$Nb_Cubs_Survivants, 3)
MyNames <- colnames(data)[20:22]
ID10 <- rep(MyNames, each = length(data$Nb_Cubs_Survivants))
ID11 <- factor(ID10, labels = MyNames,
levels = MyNames)
xyplot(Y10 ~ Z | ID11, col = 1,
strip = function(bg='white',...) strip.default(bg='white',...),
scales = list(alternating = T,
x = list(relation = "free"),
y = list(relation = "same")),
xlab = "Habitats",
par.strip.text = list(cex = 0.8),
ylab = "Nb cubs survivants",
panel=function(x, y, subscripts,...){
panel.grid(h =- 1, v = 2)
panel.points(x, y, col = 1, pch = 16)
panel.loess(x,y,col=1,lwd=2)
})
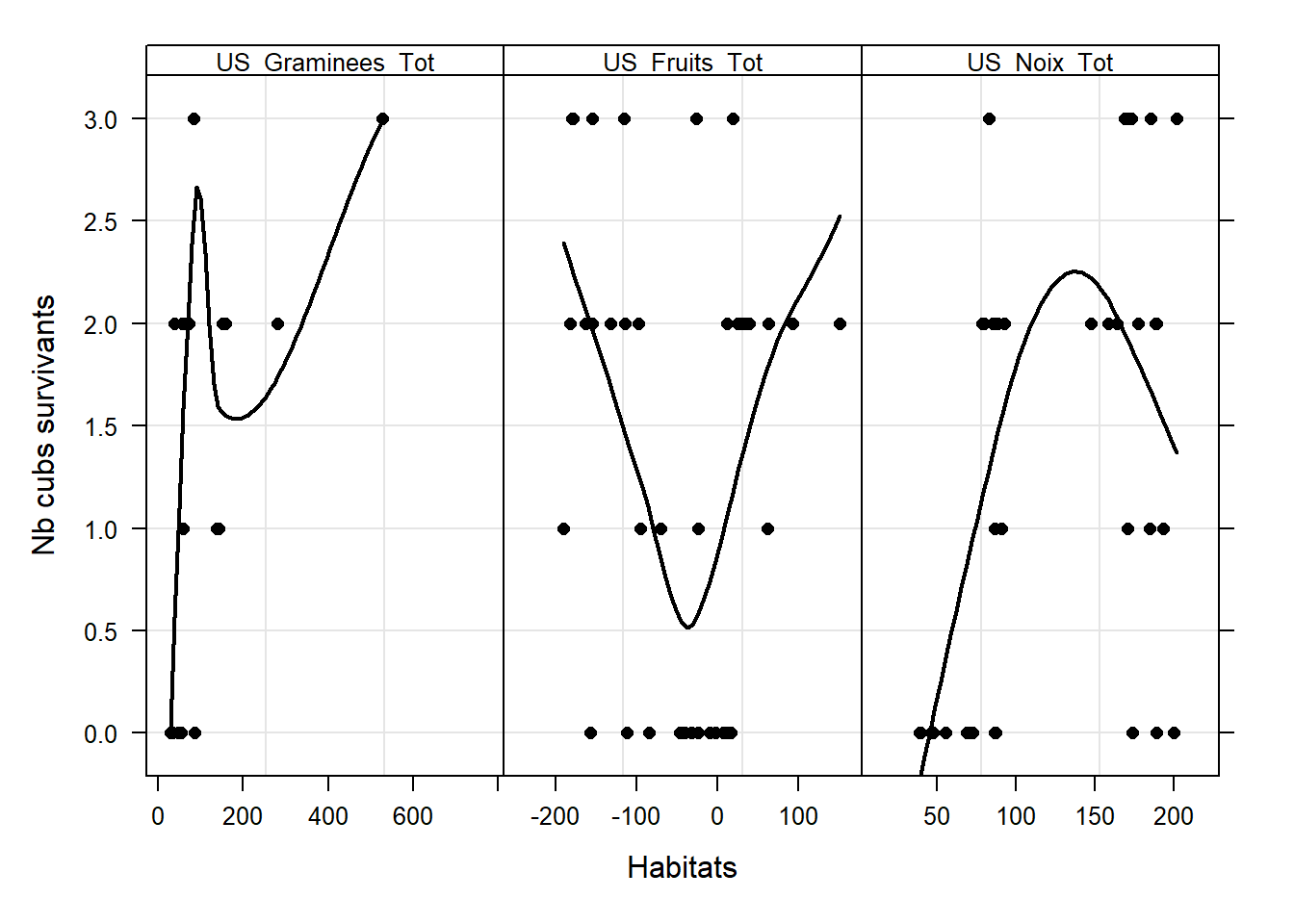
#### 6.7 - Interactions ####
# Pas d'interactions à regarder
#### 6.8 - Indépendance des Y ####
# Pas de nouveau Y









