Practical 5 Databases
5.1 Introduction
5.1.1 1. Overview
This practical is based on Chapter 4 in Comber and Brunsdon (2021). You will need space on your M drive to undertake this practical - one of data files we create is 1GB in size.
Data can be so big that they are difficult to work with on a standard computer: they may not fit on hard drive or disk if they are terabytes in size, even if they are smaller, the computer may struggle to hold them in working memory and in the memory allocated to the program you are using (including R).
For these reasons, we increasingly work with data that is held elsewhere in some kind of database system that we access remotely. Queries are sent to the database and the results are returned.
The advantage of querying databases is that it is only the data we are interested in that is returned to our computer’s working memory. This is usually a (much) smaller subset of the data, which has been manipulated and summarised in some way.
Queries to databases can be used group, summarise, link and manipulate data. Hopefully this statement has made you think that this is similar to piping in dplyr. You are correct: much of the syntax we use in piping can be used to connect to databases and extract data from them.
The key thing is that the data, in its raw unqueried form, is not in the working memory of our computers until we want it to be.
For this practical you will need to install and load the following packages.
packages <- c("sf", "tidyverse", "RSQLite")
# check which packages are not installed
not_installed <- packages[!packages %in% installed.packages()[, "Package"]]
# install missing packages
if (length(not_installed) > 0) {
install.packages(not_installed, repos = "https://cran.rstudio.com/", dep = T)
}
library(sf) # for spatial data
library(tidyverse) # for data wrangling, visualisation and mapping
library(RSQLite) # for databases
# redefine dplyr select
select <- dplyr::select
# remove unwanted variables
rm(list = c("packages", "not_installed"))5.1.2 Introduction to Databases
Up until now all of the code snippets in this module have worked with in memory data: the data were loaded into R in some way, either read from an external file or from one of R’s internal datasets. These data have been small enough in size for R to manage them to its internal memory.
However as data get larger, their access times get longer. Additionally, any kind of selection or filtering etc of data rows or columns, requires linear searches of the data, and although these can be quicker if the data are structured in some way (for example by sorting some or all of the columns), there is a trade off between storage and search efficiency. The problem is that sorting columns is expensive as sorted columns (or indexed fields) require additional storage. Thus because of the slowness of reading, writing and searching, much data are held in indexed databases, that are remotely stored (i.e. they are not on your computer or not in your computer’s memory), and are accessed remotely.
The basic idea behind databases (as opposed a data.frame or tibble and other in session data table formats) is that you connect to a database (a local one, one in working memory or a remote one), compile queries that are passed to the database and only the query results are returned.
Databases are a collection of data tables that are accessed through some kind of Database Management Systems or DBMS providing a structure that supports database queries. Databases frequently hold multiple data tables which have some field (attribute) in common supporting relational queries. In these records (observations) in different data tables are related to each other using the field they have in common (e.g. postcode, national insurance number, etc) - see Practical 2 and the section on Two table manipulations in dplyr. This allows data in different tables to be combined, before data are extracted and / or summarised in some way. All of this is done through queries. Queries in this context are specific combinations of instructions to the DBMS in order to retrieve data from the server.
Because we often link or relate data tables in databases they are often referred to as Relational DBMS. An example is shown in Figure \(\ref{fig:f1}\) of the data tables used a in much longer and deeper version of this practical in Chapter 4 of Comber and Brunsdon (2021). Notice how the different tables are linked (or related) by the attributes they have in common.
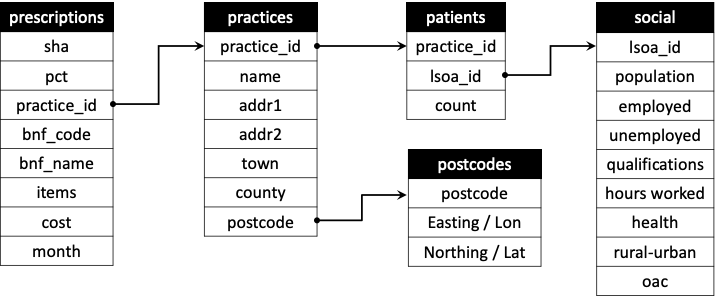
Figure 5.1: An example of a relational database, from Chapter 4 in Comber and Brunsdon (2021).
5.1.3 Databases in R
Relational databases and DBMSs, classically use SQL (Structured Query Language) to retrieve data through queries. SQL has a relatively simple syntax (supported by ISO standards) but complex queries can be difficult to code correctly. For these reasons the team behind dplyr constructed tools and functions that translate to SQL when applied to databases. Workflows combine the various dplyr verbs used for single table and two-table manipulations (Practical 2) and translates them into SQL (Structured Query Language) which is then passed to the DBMS.
You have already done something very similar in the 2nd practical when you wrote some code to join oa_sf and oa2. Remember: do not run this code as we have not loaded any data - it is simply repeated from Practical 2.
In the above code, the join was done by linking variables in the two data tables (oa_sf and oa2) that were joined by the Output Area code attribute that they had in common, although named differently in each data table.
This practical uses SQLite because it is embedded inside an R package (RSQlite) which is automatically loaded with dplyr. This provides a convenient tool for understanding how to manage large datasets because it is completely embedded inside an R package and you do not need to connect to a separate database server. A good introduction to this topic can be found in Horton, Baumer, and Wickham (2015) and the DBI package provides an interface to many different database packages. This allows you to use the same R code, for example with dplyr verbs, to access and connect to a number of back-end database formats including MySQL with the RMySQL package and Postgres with `RPostgreSQL.
In summary:
- Data are frequently held in databases as big data can need a lot of storage and we may not be able to load it locally in our computers;
- DBMSs / RDBMSs are used to hold, link and mange collections of databases;
- We can compile queries using
dplyrsyntax; - Relationships and cardinality (the direction of the relationships) are really important considerations.
Some resources that you may find useful outside of the practical:
- A good overview of cardinality etc is here: https://www.slideshare.net/IngePowell/erd-cardinality
- A good overview of using R with databases is here: https://www.ibm.com/developerworks/data/library/techarticle/dm-1402db2andr/
- A good introduction to the topic can be found in: Horton et al (2015) also a shorter version is here http://chance.amstat.org/2015/04/ setting-the-stage/
- An excellent summary of operations in R can be found in Wickham and Grolemund (2016) Chapter 13 (Relational Data with dplyr) here http://r4ds.had.co.nz/relational-data.html
5.2 Creating relation databases in R
The basic idea for creating databases is that you define or open a connection from your R / RStudio session to a named database. The database is then populated with data and then you can work with it. The connection should be closed when you finish working with the database.
There a number of different types of databases that can be created:
- local, in memory databases: these are useful for prototyping and testing. They are temporary and are automatically deleted you disconnect from them.
- local, on file databases: these are permanent databases but are still held locally (i.e. on your computer).
- remote, on file databases: these are permanent and held elsewhere (i.e. on another computer!), and require an internet connection to access them. The creation of these is not covered here, but once you are connected to the remote server, the same operations can be undertaken.
5.2.1 Data
You should have downloaded the data file from the VLE for this week. Load the prac5_data.RData file:
## [1] "ccg" "ccg_patients" "prescriptions" "select"You should see that 3 files are loaded (as well as the redefined select function):
- a data table called
prescriptions. This is month of prescribing data for January 2016 downloaded from here https://data.gov.uk/dataset/prescribing-by-gp-practice-presentation-level and converted to an.RDatafile; - a spatial object in
sfformat calledccg. This has the Care Commissioning Group (CCG) areas in England - area public health administration areas. These used to be called Primary Care Trusts (PCTs); - a data table called
ccg_patientswith the the number of patients in each CCG/PCT area.
We will use these to populate databases of different kinds
5.2.2 Creating a local, in memory database
Local, in memory databases are useful for illustrating the principles of database creation and of constructing queries. In reality, if the data fit into working memory and can be manipulated and accessed with ease, then it is likely that you do not need to use databases and queries: standard R data formats such the data.frame and tibble and related operations with dplyr will probably suffice.
To work with a database in dplyr, the connection to it needs to be specified. This is done with the DBI::dbConnect() function. This can be interpreted as saying use the Connect() function from the DBI package - i.e. the :: indicates to R in which package the function is (recall that the DBI package is loaded with dbdplyr, which in turn is loaded by dplyr, which is in turn loaded by tidyverse).
The code snippet below defines an in memory database:
The arguments to DBI::dbConnect() varies from database to database, but the first argument is always the database backend. It is RSQLite::SQLite() for RSQLite, RPostgreSQL::PostgreSQL() for RPostgreSQL, etc. The SQLite implementation in R (RSQLite) only needs one other argument: the path to the database. Here we use the special string ":memory:" which causes SQLite to make a temporary in memory database (i.e. in the working memory of R / RStudio).
The database, db has no data in it. To populate the database, the dplyr function copy_to can be used. This uploads data to the database. The code below does this for the prescriptions data table.
In fact copy_to is a wrapper for dbWriteTable in the DBI package and the same operation can be undertaken with the code below (you do not need to run this!):
# do not run!
dbWriteTable(conn = db, name = "prescripts_db",
value = prescriptions,
row.names = FALSE, header = TRUE, overwrite = T)The tbl function can then be used to make the connection to the data table which can be examined:
## # Source: table<`prescripts_db`> [?? x 10]
## # Database: sqlite 3.47.1 []
## SHA PCT PRACTICE BNF.CODE BNF.NAME ITEMS NIC ACT.COST QUANTITY PERIOD
## <chr> <chr> <chr> <chr> <chr> <int> <dbl> <dbl> <int> <int>
## 1 Q44 RTV Y04937 0401010Y0… "Zolpid… 1 2.56 2.38 56 201601
## 2 Q44 RTV Y04937 0401010Z0… "Zopicl… 1 0.67 0.73 14 201601
## 3 Q44 RTV Y04937 0401010Z0… "Zopicl… 1 1.41 1.31 28 201601
## 4 Q44 RTV Y04937 0401020K0… "Diazep… 2 1.72 1.61 56 201601
## 5 Q44 RTV Y04937 0401020K0… "Diazep… 1 0.68 0.74 21 201601
## 6 Q44 RTV Y04937 0401020K0… "Diazep… 1 1.52 1.52 42 201601
## 7 Q44 RTV Y04937 0402010AB… "Quetia… 1 1.74 1.72 84 201601
## 8 Q44 RTV Y04937 0402010AB… "Quetia… 1 39.6 36.7 21 201601
## 9 Q44 RTV Y04937 0402010AD… "Aripip… 1 20.2 18.7 28 201601
## 10 Q44 RTV Y04937 0402010D0… "Chlorp… 1 1.08 1.11 14 201601
## # ℹ more rowsIn a similar way the CCG patients data can be added to the database:
And this too can be examined:
## # Source: table<`ccg_pat_db`> [?? x 3]
## # Database: sqlite 3.47.1 []
## CCGcode CCGnm_s ccg.reg.pa
## <chr> <chr> <chr>
## 1 00C Darlington 107191
## 2 00D Durham Dales, Easington & Sedgefield 289529
## 3 00J North Durham 254568
## 4 00K Hartlepool & Stockton-On-Tees 294897
## 5 00L Northumberland 322154
## 6 00M South Tees 293801
## 7 00N South Tyneside 155426
## 8 00P Sunderland 283594
## 9 00Q Blackburn with Darwen 171623
## 10 00R Blackpool 171758
## # ℹ more rowsThe tbl function is very useful here as it allows us to extract data from the database and to pipe it around (i.e. do things with it). The way that tbl works is to create a reference to the data table in the database (e.g. prescripts_db).
The dplyr piping syntax can be applied in conjunction with tbl to select, filter manipulate summarise and extract data from the database. The example below select records from the prescripts_db data table in db that are in CCG / PCT 00D (the PCT field in the data) and cost more than £100:
tbl(db, "prescripts_db") |>
filter(PCT == "00D" & ACT.COST > 100) |>
select(PCT, BNF.NAME, ACT.COST) ## # Source: SQL [?? x 3]
## # Database: sqlite 3.47.1 []
## PCT BNF.NAME ACT.COST
## <chr> <chr> <dbl>
## 1 00D "Sod Algin/Pot Bicarb_Susp S/F " 288.
## 2 00D "Gaviscon Infant_Sach 2g (Dual Pack) S/F " 235.
## 3 00D "Mebeverine HCl_Tab 135mg " 253.
## 4 00D "Lansoprazole_Cap 30mg (E/C Gran) " 667.
## 5 00D "Lansoprazole_Cap 15mg (E/C Gran) " 141.
## 6 00D "Omeprazole_Cap E/C 20mg " 842.
## 7 00D "Omeprazole_Cap E/C 40mg " 114.
## 8 00D "Mesalazine_Tab E/C 400mg " 287.
## 9 00D "Mesalazine_Tab G/R 1.2g M/R " 173.
## 10 00D "Asacol_MR Tab E/C 400mg " 289.
## # ℹ more rowsA similar syntax can be used work out average prescription costs for different Strategic Health Authorities in sha:
## # Source: SQL [?? x 2]
## # Database: sqlite 3.47.1 []
## PCT mean_cost
## <chr> <dbl>
## 1 00C 74.7
## 2 00D 78.4
## 3 00J 77.7
## 4 00K 76.4
## 5 00L 77.4
## 6 00M 68.5
## 7 00N 70.8
## 8 00P 66.3
## 9 00Q 62.8
## 10 00R 77.6
## # ℹ more rowsThis starts to suggest how queries can be constructed.
When we have finished with any database connection we have to close it using the dbDisconnect function:
5.2.3 Creating a local, on file database
Any in memory database ceases to exists when the connection is closed (as above).
By contrast, an on file database is permanent and, after being populated with data, can be connected to in later R sessions. Here the data tables are used create a RSQlite database called prescribing.sqlite. This will contain the same 2 data tables for CCGs and prescriptions. The process is similar to creating an in memory database, but this time a database file is created in your local directory.
In the code below, the first line checks for the existence of prescribing_lite.sqlite and if it exists removes it. The second then creates a connection to the database:
if (file.exists("prescribing.sqlite") == TRUE)
file.remove("prescribing.sqlite")
db = DBI::dbConnect(RSQLite::SQLite(), dbname="prescribing.sqlite")Now if you look in your working folder using Windows Explorer (PC) or Finder (Mac), you will see that an object called prescribing.sqlite has been created. It is empty and has no size because it has not been populated with data yet.
The code below populates the db object with the data tables, using the dbWriteTable function and then closes the connection to the database:
dbWriteTable(conn = db, name = "prescriptions", value = prescriptions,
row.names = FALSE, header = TRUE)
dbWriteTable(conn = db, name = "ccg_patients", value = ccg_patients,
row.names = FALSE, header = TRUE)If you check again in your working folder you will see that prescribing_lite.sqlite now has been populated and has a size (~1GB).
You can check what the database contains using different commands form the DBI package:
## [1] "ccg_patients" "prescriptions"## [1] "SHA" "PCT" "PRACTICE" "BNF.CODE" "BNF.NAME" "ITEMS"
## [7] "NIC" "ACT.COST" "QUANTITY" "PERIOD"Again, the dbDisconnect function is used to close the connection to the database:
If you enter db at the console you will see that it is disconnected.
To access the data, we need simply to connect to the database, having created it:
It is possible to query the data tables in the database. For example, the code below summarises the mean cost of prescriptions in each of the CCGs/PCTs, arranges them in descending mean cost order, filters them for at least 100 items and prints the top 10 to the console:
tbl(db, "prescriptions") |>
group_by(PCT) |>
summarise(
mean_cost = mean(ACT.COST, na.rm = T),
n = n()
) |>
ungroup() |>
arrange(desc(mean_cost)) |>
filter(n > 100) |> print(n=10)However, this is working with only one of the data tables in the db database. It is not taking advantage of the layered analysis that is possible by linking data tables, as shown in Figure \(\ref{fig:f1}\).
To link the data tables in the db database we need to specify the links between them, just as we did using the join functions in Practical 2. However, to join in the right way, we need to think about how we wish to construct our queries and what any queries to database tables will return (and therefore the analysis and inference they will support). The DBI package has a number of functions that list data tables and their fields as above. Similarly, tbl in the dplyr package can be used to do these summaries:
The results are similar to the str, head and summary functions used earlier. However, it is important to restate what the tbl function is doing: it is sending a tbl query to db about the table (e.g. the prescriptions table) in db. As tbl returns the first 10 records this is what is returned from db: i.e. the data returned to the R session is only the result of this query and not the whole of the data table. To emphasise this, the code below compares the size of the query and the size of the prescriptions data table that is loaded into the R session.
## [1] "6 Kb"## [1] "463.4 Mb"Again the database can be closed as follows:
5.2.4 Summary
The basic process of creating a database is to first define a connection to a database, then populate it with data, and, when it is populated, to close the connection.
Two kinds of database were illustrated: an in memory one - good for development - and a local, on file database. The procedures for constructing these were essentially the same:
- define a connection to a database
- populate the database with data
- close the database
This sequence can be used to construct much larger databases.
5.3 Database queries
In this section we will explore a number of core database operations using dplyr to construct queries and apply them to the prescribing.sqlite database created above. There are 3 mains groups of operations that are commonly used in database queries, either singly or in combination. Queries specify operations that:
- Extract (specifying criteria, logical pattern matching, etc.)
- Join (linking different tables)
- Summarise (grouping, using summary functions, maintaining fields, creating new fields)
There are many overlaps with dplyr operations, but here these are applied in a database context. In many cases the syntax is exactly the same as those applied to data.frame and tibble formats as in previous practicals. On occasion, however, they need to be adapted for working with databases.
5.3.1 Extracting data from a database
It is possible to extract whole records (rows) and fields (columns), individual rows and columns that match some criteria and individual elements (cells in a data table). The two most commonly used approaches for extracting data are:
- By specifying some kind of logical test or conditions that have to be satisfied for the data to be extracted.
- By specifying location of the data you wish to extract, for example by using the \(i^th\) row or \(j^th\) column, or variable names.
Logical queries have a TRUE or FALSE answers and use logical operators (e.g. greater than, less than, equals, not equals etc). These have been covered in previous practicals full set of logical operators can be found in the R help (enter ?base::Logic). The other main way of selecting is through some kind of text pattern matching. Both may be used to subset database fields (columns) by filtering and / or database records (rows) by selecting.
A connection has to be made for a database to be queried. Connect to the prescribing.sqlite database you created earlier as before, but this time using a different syntax:
The code below uses filter to extract the prescriptions for a specific flu vaccine via the BNF code (for example see https://openprescribing.net/bnf/) and orders the extracted records by volume (items):
tbl(db, "prescriptions") |>
filter(BNF.CODE == "1404000H0AAAFAF") |>
select(BNF.CODE, BNF.NAME, ITEMS) |>
distinct()Here the filter operation was based on a single logical statement using == that returned single exact matches. For multiple exact matches, the %in% function can be used with a vector of names:
tbl(db, "prescriptions") |>
filter(BNF.CODE %in% c("1404000H0AAAFAF", "1404000H0AAAJAJ")) |>
select(BNF.CODE, BNF.NAME) |>
distinct() Now we might want to filter for all the flu vaccine prescriptions, which start with the BNF code 1404000H0. For standard dplyr operations, the str_detect function can be used to filter. However, the stringr pattern matching functions within dplyr, all in the form str_<action>, do not work with databases. Instead %like% can be applied, with carefully positioned wildcards (\%):
# one pattern
tbl(db, "prescriptions") |>
filter(BNF.CODE %like% '%1404000H0%')
# multiple patterns
tbl(db, "prescriptions") |>
filter(BNF.NAME %like% 'Dermol%' | BNF.NAME %like% 'Influenza_Vac%') |>
select(BNF.CODE, BNF.NAME) |>
distinct() However, pattern matching in this way needs to be done with extreme care because it undertakes partial matching, whereas filtering using logical approaches with == or %in% will only return exact matches.
Additionally, the wildcards (\%) need to be carefully used. For example, they can be placed at the start and end of the pattern passed to %like%. This means that the filter operation will return any records that have BNF.CODE values containing 1404000H0, not just those that start with 1404000H0.
Notice how the select function was used in the above to return specific fields from a database in a similar way to working data tables loaded into the R session:
Fields can also be selected using pattern matching, but here a set of tidy matching functions can be applied:
tbl(db, "prescriptions") |>
select(starts_with("BNF"))
tbl(db, "prescriptions") |>
select(contains("BNF")) You should examine the help for these:
Thus far, all of the results of the code snippets applied to the database have been printed out to the console. No data has been returned to the R session, meaning that all of the analysis has taken place away from the working memory of your computer. This is one of the key advantages of using databases: the queries are all undertaken by outside of your computer’s working memory (as well as the database being separate).
However, if you want to use the results (for example to link to a map, or other data) then the query results need to be collected. The collect() function returns the results. It creates an object (even if it is just printed out). To show this, run the code below which uses the object.size function to evaluate the memory cost to R of running the code snippets. This is a key advantage of working with databases.
# size of the call
tbl(db, "prescriptions") |> object.size()
# size of a longer call
tbl(db, "prescriptions") |>
filter(BNF.CODE %like% '%1404%') |>
arrange(desc(ACT.COST)) |> object.size()
# size of what is returned with collect
tbl(db, "prescriptions") |>
filter(BNF.CODE %like% '%1404%') |>
arrange(desc(ACT.COST)) |> collect() |> object.size()5.3.2 Joining (linking) database tables
Practical 2 introduced the idea of joins between different tables. Some (but not all) of the two-table dplyr joins can also be applied to tables held in databases.
These can be applied to the data tables in the database in a similar way to working with data tables loaded into an ordinary R session. The code below filters for anti-depressants, summarises the costs over CCGs / PCTs and then joins the result to the patients attribute in the ccg_patients data table, before summarising the costs per 1000 patients in each CCG / PCT area:
## [1] "CCGcode" "CCGnm_s" "ccg.reg.pa"# the query
tbl(db, "prescriptions") |>
filter(BNF.CODE %like% '%1404000H0%') |>
group_by(PCT) |>
summarise(PCT_COST = sum(ACT.COST, na.rm = T)) |>
left_join(tbl(db, "ccg_patients"), by = c("PCT" = "CCGcode")) |>
mutate(costs_pp = PCT_COST/(ccg.reg.pa/1000))## # Source: SQL [?? x 5]
## # Database: sqlite 3.47.1 [/Users/geoaco/Library/CloudStorage/OneDrive-UniversityofLeeds/leeds_work/teaching_24/geog5917/bookdowngeog5917/prescribing.sqlite]
## PCT PCT_COST CCGnm_s ccg.reg.pa costs_pp
## <chr> <dbl> <chr> <chr> <dbl>
## 1 00C 2724. Darlington 107191 25.4
## 2 00D 11229. Durham Dales, Easington & Sedgefield 289529 38.8
## 3 00J 17615. North Durham 254568 69.2
## 4 00K 10410. Hartlepool & Stockton-On-Tees 294897 35.3
## 5 00L 15926. Northumberland 322154 49.4
## 6 00M 20728. South Tees 293801 70.6
## 7 00N 6069 South Tyneside 155426 39.0
## 8 00P 19527. Sunderland 283594 68.9
## 9 00Q 22255. Blackburn with Darwen 171623 130.
## 10 00R 5143. Blackpool 171758 29.9
## # ℹ more rowsAs well as joining the data the code snippet above shows how dyplr single table verbs can be applied to joined data, after it has been joined! In this case summarising results over a grouping, the PCT / CCG groups.
Note again that without the collect function in the code above, the joined data remains in the database (i.e. is not returned to the console). To map the rates, the output of the query can be collected and linked to the ccg sf spatial layer and then mapped as in Figure \(\ref{fig:f3}\).
# as above but this time collecting the result from the database
tbl(db, "prescriptions") |>
filter(BNF.CODE %like% '%1404000H0%') |>
group_by(PCT) |>
summarise(PCT_COST = sum(ACT.COST, na.rm = T)) |>
left_join(tbl(db, "ccg_patients"), by = c("PCT" = "CCGcode")) |>
mutate(costs_pp = PCT_COST/(ccg.reg.pa/1000)) |> collect() -> tmp
# pass tmp to ggplot
ccg |> left_join(tmp, by = c("CCGcode"="PCT")) |>
ggplot() +
geom_sf(aes(fill = costs_pp)) +
scale_fill_continuous(type = "viridis")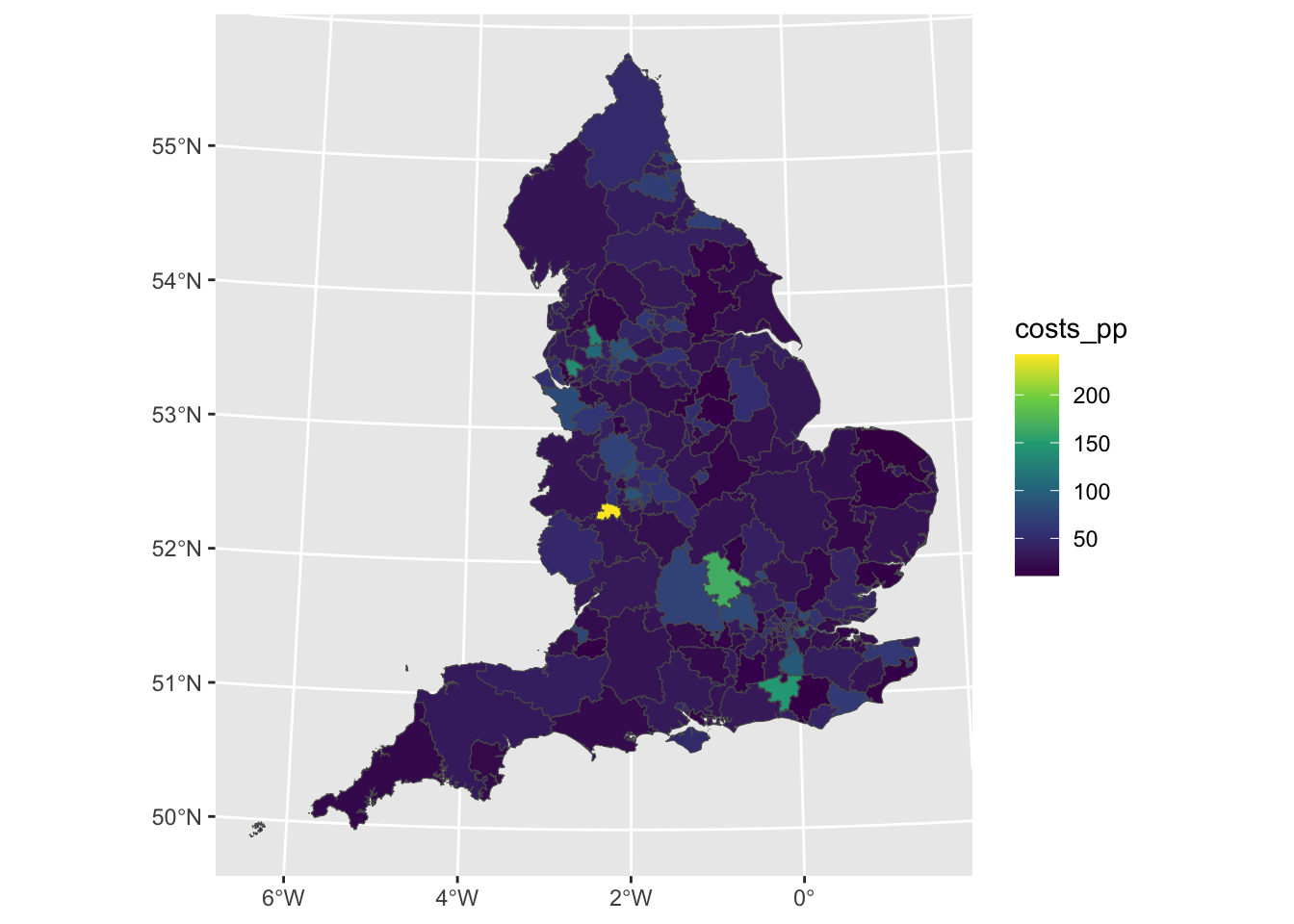
Figure 5.2: The prescribing of antidepressants for each CCG / PCT.
The summarise() function summarises existing variables by the function that is passed to it. If the data are grouped it will return a summary for each group, otherwise it will summarise over the whole dataset. To illustrate these the code below summarises costs in the prescription table over the whole dataset and then groups these by PCT, and by PCT and month:
# entire dataset
tbl(db, "prescriptions") |>
summarise(total = sum(ACT.COST, na.rm = T))
# grouped by PCT
tbl(db, "prescriptions") |>
group_by(PCT) |>
summarise(total = sum(ACT.COST, na.rm = T)) |>
arrange(desc(total))
# grouped by PCT and month
tbl(db, "prescriptions") |>
mutate(month = substr(PERIOD, 5,6)) |>
group_by(PCT, month) |>
summarise(total = sum(ACT.COST, na.rm = T)) |>
arrange(desc(total))Note a number of things in the code above:
- the use of
substrto extract the month from thePERIODvariable - that
summariseonly returns variables created by its summary operations along with any grouping; - that the
mutate()function adds new variables for all the rows in the input data table and returns (selects) all variables present when it is applied.
The code below summarises the costs of all prescriptions per practice and uses mutate to calculates the cost per item.
tbl(db, "prescriptions") |>
group_by(PRACTICE) |>
summarise(
cost = sum(ACT.COST, na.rm = T),
n = n()) |>
mutate(mean_cost = cost/n) |>
arrange(desc(mean_cost))## # Source: SQL [?? x 4]
## # Database: sqlite 3.47.1 [/Users/geoaco/Library/CloudStorage/OneDrive-UniversityofLeeds/leeds_work/teaching_24/geog5917/bookdowngeog5917/prescribing.sqlite]
## # Ordered by: desc(mean_cost)
## PRACTICE cost n mean_cost
## <chr> <dbl> <int> <dbl>
## 1 Y00215 144237. 25 5769.
## 2 Y01912 5451. 2 2725.
## 3 Y02774 11869. 6 1978.
## 4 Y03873 23257. 23 1011.
## 5 Y04441 77124. 81 952.
## 6 Y05181 23619. 28 844.
## 7 P81791 14467. 18 804.
## 8 Y02901 15532. 20 777.
## 9 Y03641 21463. 28 767.
## 10 Y04878 41367. 54 766.
## # ℹ more rowsThis operation can be unpicked by running the query in discrete steps and examining the intermediate outputs.
First the grouping and the summary:
tbl(db, "prescriptions") |>
group_by(PRACTICE) |>
summarise(
cost = sum(ACT.COST, na.rm = T),
n = n())Then the creation of a new variable with mutate:
tbl(db, "prescriptions") |>
group_by(PRACTICE) |>
summarise(
cost = sum(ACT.COST, na.rm = T),
n = n()) |>
mutate(mean_cost = cost/n)Before the final ordering by adding the pipe and arrange(desc(mean_cost)) as above. And the observant amongst you will notice that this is the mean and the function `mean could have been used instead!
5.3.3 Final observations
This section has described the different kinds of operations for constructing database queries using the dplyr syntax. These include:
- functions for extracting data (rows / columns) from a database based on pattern matching, logical tests and specific positional references;
- functions for joining or linking database tables together based on some common property or attribute, with consideration of the cardinality of the link, applying the joins first introduced in Practical 2;
- functions for summarising data, with or without grouping, applying different functions applying the
dplyrverbs, also introduced in Practical 2.
These can be used to construct complex queries of filtered records, selected fields and joined database tables. A long and more complex example is in Chapter 4 of Comber and Brunsdon (2021). Using dplyr to do this has a number of advantages, the main one is that dplyr tries to be lazy by never pulling data into R’s memory unless explicitly requested (for example by using the collect function). This is done because dplyr queries create references to the data in the database, and the results that are returned (printed) to the console are just summaries of the query. When collect is used the dplyr query results are retrieved to a local tibble.
However, in some case we may want to pull the data down from the database and perform our database operations. The dbReadTable function does this and returns similar results, but operates in a very different way as indicated by the size of the R objects they create and different times they take to run:
One other aspect of dplyr is that it compiles all of the database commands and translates them to SQL before passing them to the database in one step. This can be illustrated by adding show_query() to the code snippet above:
tbl(db, "prescriptions") |>
group_by(PRACTICE) |>
summarise(mean_cost = mean(ACT.COST, na.rm = T))|>
arrange(desc(mean_cost)) |>
show_query()## <SQL>
## SELECT `PRACTICE`, AVG(`ACT.COST`) AS `mean_cost`
## FROM `prescriptions`
## GROUP BY `PRACTICE`
## ORDER BY `mean_cost` DESCIt also possible to pass SQL code directly to the database using the dbGetQuery function in the DBI package:
dbGetQuery(db,
"SELECT `PRACTICE`, AVG(`ACT.COST`) AS `mean_cost`
FROM `prescriptions`
GROUP BY `PRACTICE`
ORDER BY `mean_cost` DESC") |> as_tibble()SQL is a very powerful language with a standard syntax. Further information on constructing SQL queries can be found at https://www.sqlite.org/queryplanner.html
The database can be closed as follows:
5.4 Summary
This practical has illustrated how database queries can be constructed using the dplyr verbs and joins to wrangle data held in different database tables. These can be used to construct complex queries that integrate and pull data from different tables in databases. In some instances some of the dplyr functions required workarounds, particularly for pattern matching. A key point is that dplyr tries to be as lazy as possible by never pulling data into R unless explicitly requested. In effect all of the dplyr commands are compiled and translated into SQL before they are passed to the database in one step. In this schema, the results of queries (e.g. tbl) create references to the data in the database and the results that are returned and printed to the console are just summaries of the query - the data remain on the database. The dplyr query results can be returned using collect, which retrieves data to a local tibble. The SQL created by dplyr can be examined with the show_query() function.
The examples here used in memory and local on file databases. Of course they connections to remote databases can be made and the dplyr team provide a hypothetical example of the syntax for doing that (see https://db.rstudio.com/dplyr/) - do not run this code!
# do not run this code
con <- DBI::dbConnect(RMySQL::MySQL(),
host = "database.rstudio.com",
user = "hadley",
password = rstudioapi::askForPassword("Database password")
)The dplyr and related packages such as stringr, many of which are loaded with the tidyverse package, contain many functions for manipulating and linking data tables. These wrap SQL and other functions such as pattern matching into a standard format to support data science. However, there are still some gaps in what they can do. In such cases, a bit creative thought can usually overcome the problem.