Chapter 11 Web Scraping
Now we can download data and text from web using url load library
11.1 video
you can find my video! https://youtu.be/4KskacxD0VM
11.2 issue of COVID19 in Korea
Now I want to download Table from https://en.wikipedia.org/wiki/COVID-19_pandemic_by_country_and_territory Please visit the website, url.
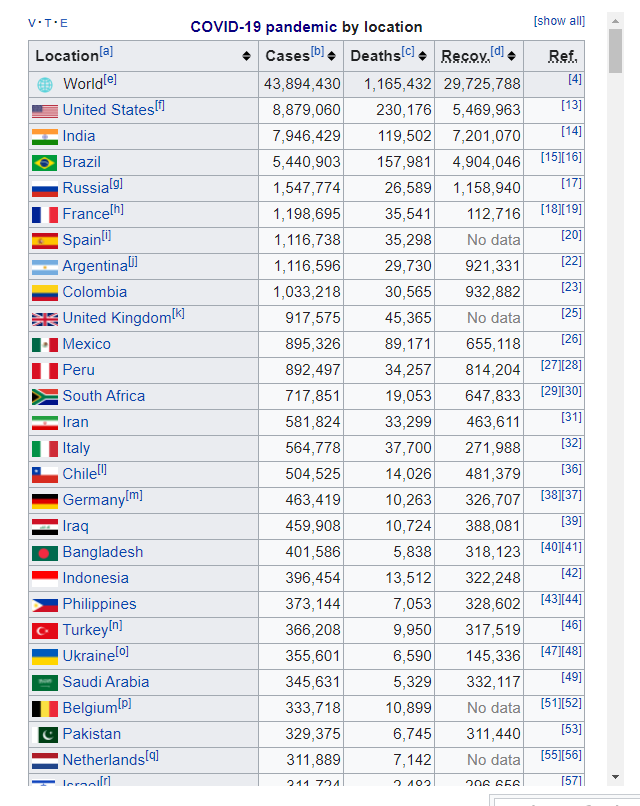
wiki covid
read_html() allows us to read url and its’ contents.
Go to website via chrom and click F12 button. You can see the right window as below. Now I want find table of Covid-19 table. I open the search tab by using ctrl + F. And writing or typing india to find that table.
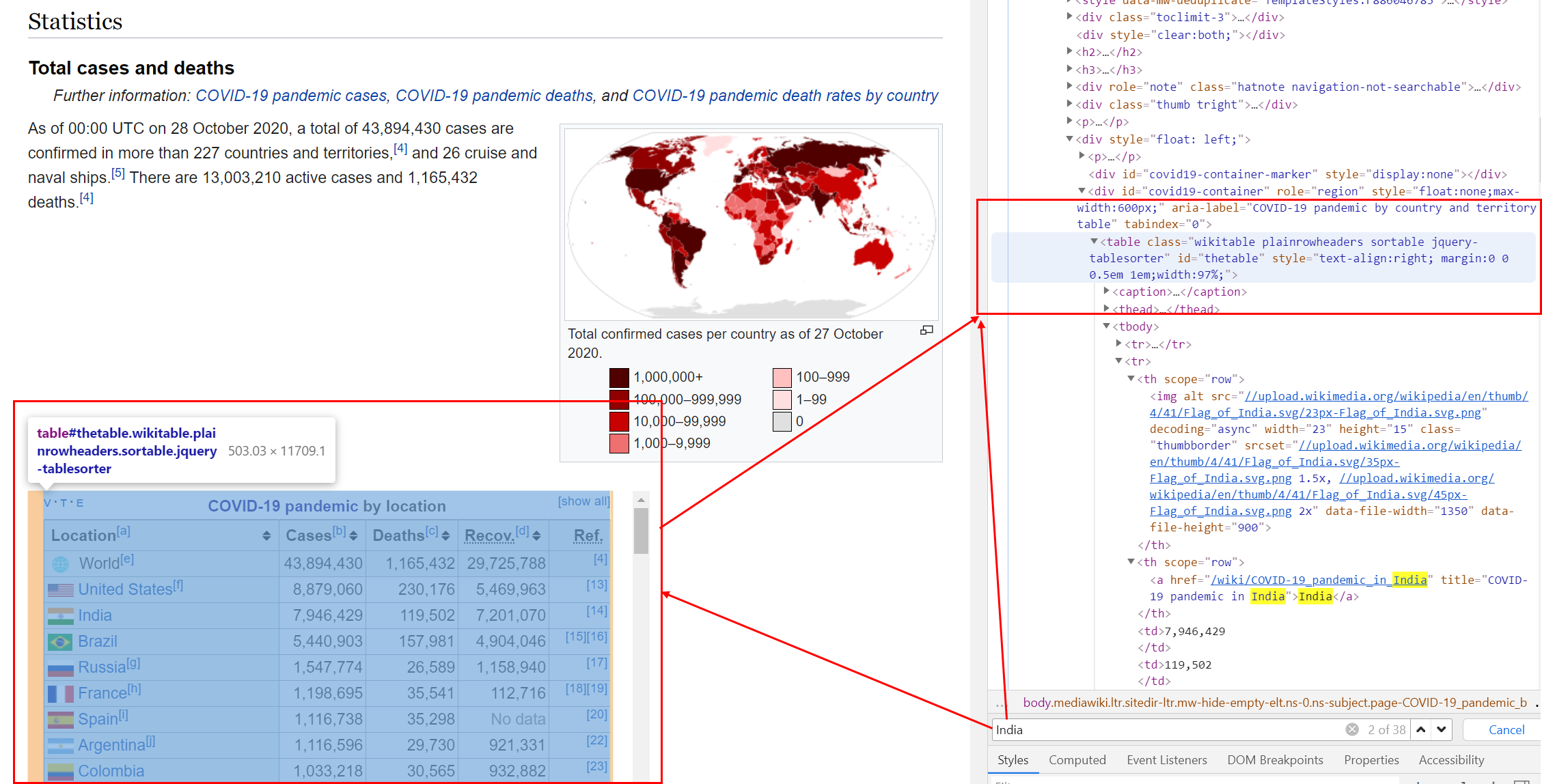
Find source and nodes
Now I try to find table nodes from url
## # A tibble: 234 x 6
## `Location[a]...1` `Location[a]...2` `Cases[b]` `Deaths[c]` `Recov.[d]` Ref.
## <chr> <chr> <chr> <chr> <chr> <chr>
## 1 <NA> World[e] 43,895,968 1,165,455 29,727,057 [4]
## 2 <NA> United States[f] 8,879,060 230,176 5,469,963 [13]
## 3 <NA> India 7,946,429 119,502 7,201,070 [14]
## 4 <NA> Brazil 5,440,903 157,981 4,904,046 [15][~
## 5 <NA> Russia[g] 1,547,774 26,589 1,158,940 [17]
## 6 <NA> France[h] 1,198,695 35,541 112,716 [18][~
## 7 <NA> Spain[i] 1,116,738 35,298 No data [20]
## 8 <NA> Argentina[j] 1,116,596 29,730 921,331 [22]
## 9 <NA> Colombia 1,033,218 30,565 932,882 [23]
## 10 <NA> United Kingdom[k] 917,575 45,365 No data [25]
## # ... with 224 more rowsI remove ‘,’ and macke numeric variables in Cases, Death and Recover.
tab3 <- tab2[ -c(1, 232, 233, 234), -c(1, 6)] %>%
setNames(c("Location", "Cases",
"Death", "Recover"))
tab3 <- tab3 %>%
mutate_at(c('Cases', 'Death', 'Recover'), function(x)(str_replace_all(x, ",", "") %>% as.numeric())) %>%
mutate(Location = str_replace_all(Location, '\\[[:alpha:]]', ""))I used \\[[:alpha:]], \\[ means “[" and [:aplpah:] means any alphabet, and last ] means "]”. So, I try to remove the all character within “[ ]”. Now, Table is.
figs<-tab3 %>%
mutate(mortality = Death /Cases *1000) %>% # mortality per 1 thousnd cases
ggplot(aes(x = Cases, y = mortality, size = Death))+
geom_point() +
scale_x_continuous(trans = 'log')
figs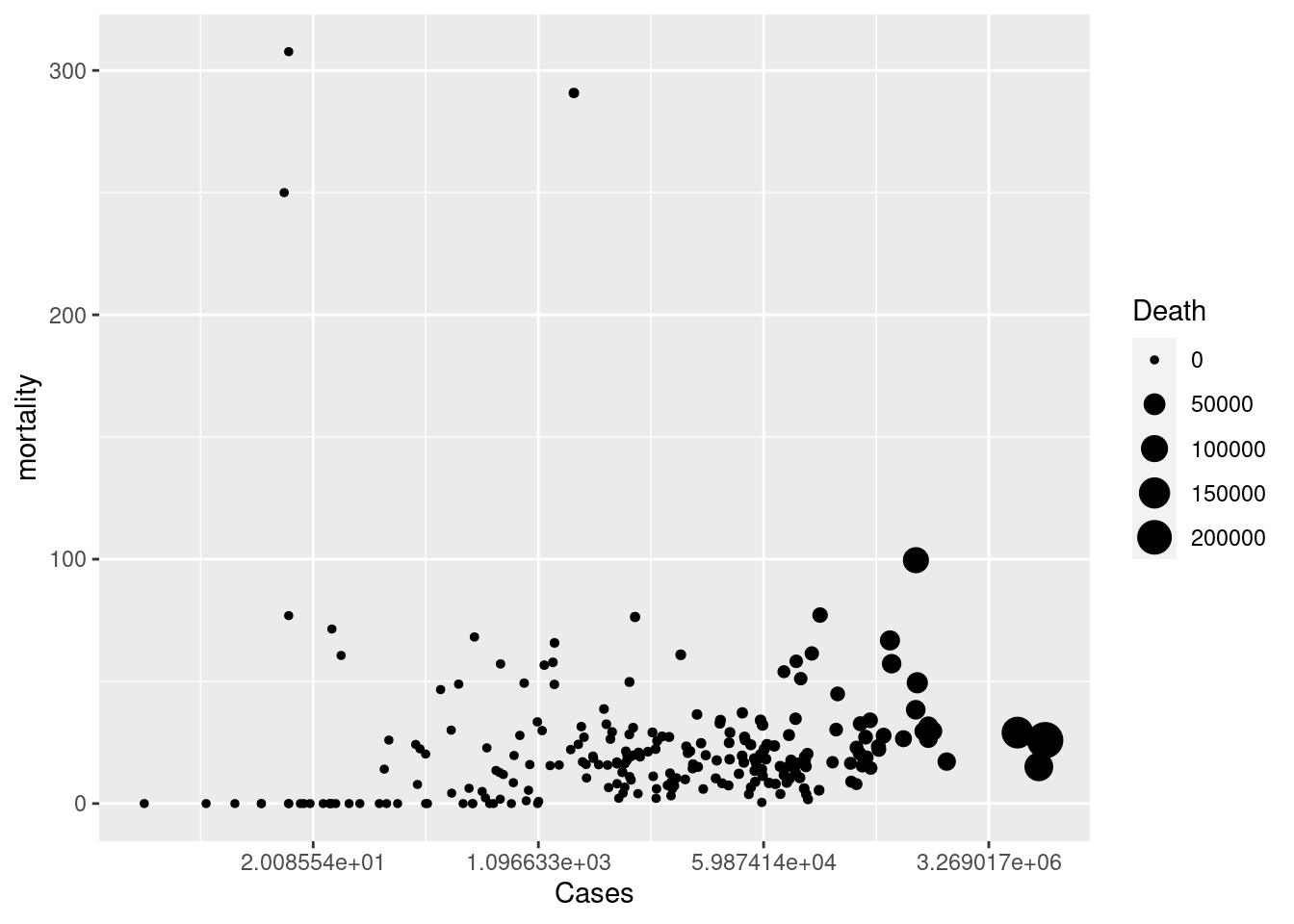
11.3 homework
11.3.1 download Cumulative covid19 death
Download data table from url. You can use tab[[ i ]] code to find cumulative covid19 death. The taret Table in web looks like that.
hint
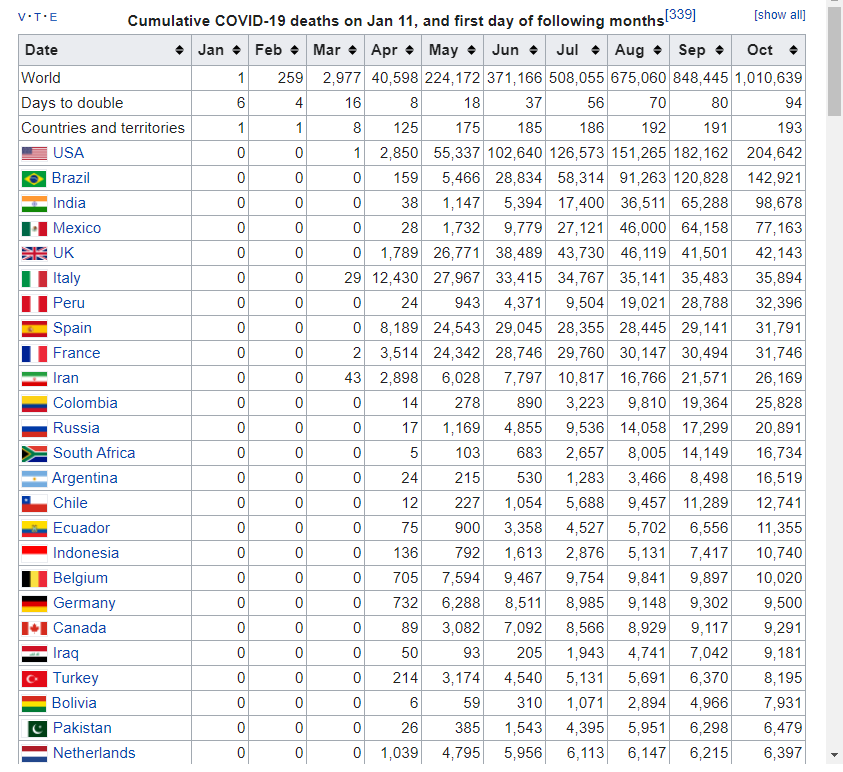 and the table file is
and the table file is
11.3.2 UK, Italy, France, Spain, USA, Canada
select countris of “UK, Italy, France, Spain, USA, Canada” and plot the trends. and upload the final plot in google class
Hint |
|
|---|---|
| step1: | create Month_mortatlity data filter countries names of above |
| step2: | chage character data to numeric data |
| step3: | pivot data to long form |
| step4: | plot the graph! |
Step 1 and 2
step 3
step 4
## [1] "LC_CTYPE=en_US.UTF-8;LC_NUMERIC=C;LC_TIME=en_US.UTF-8;LC_COLLATE=en_US.UTF-8;LC_MONETARY=en_US.UTF-8;LC_MESSAGES=C;LC_PAPER=C;LC_NAME=C;LC_ADDRESS=C;LC_TELEPHONE=C;LC_MEASUREMENT=C;LC_IDENTIFICATION=C"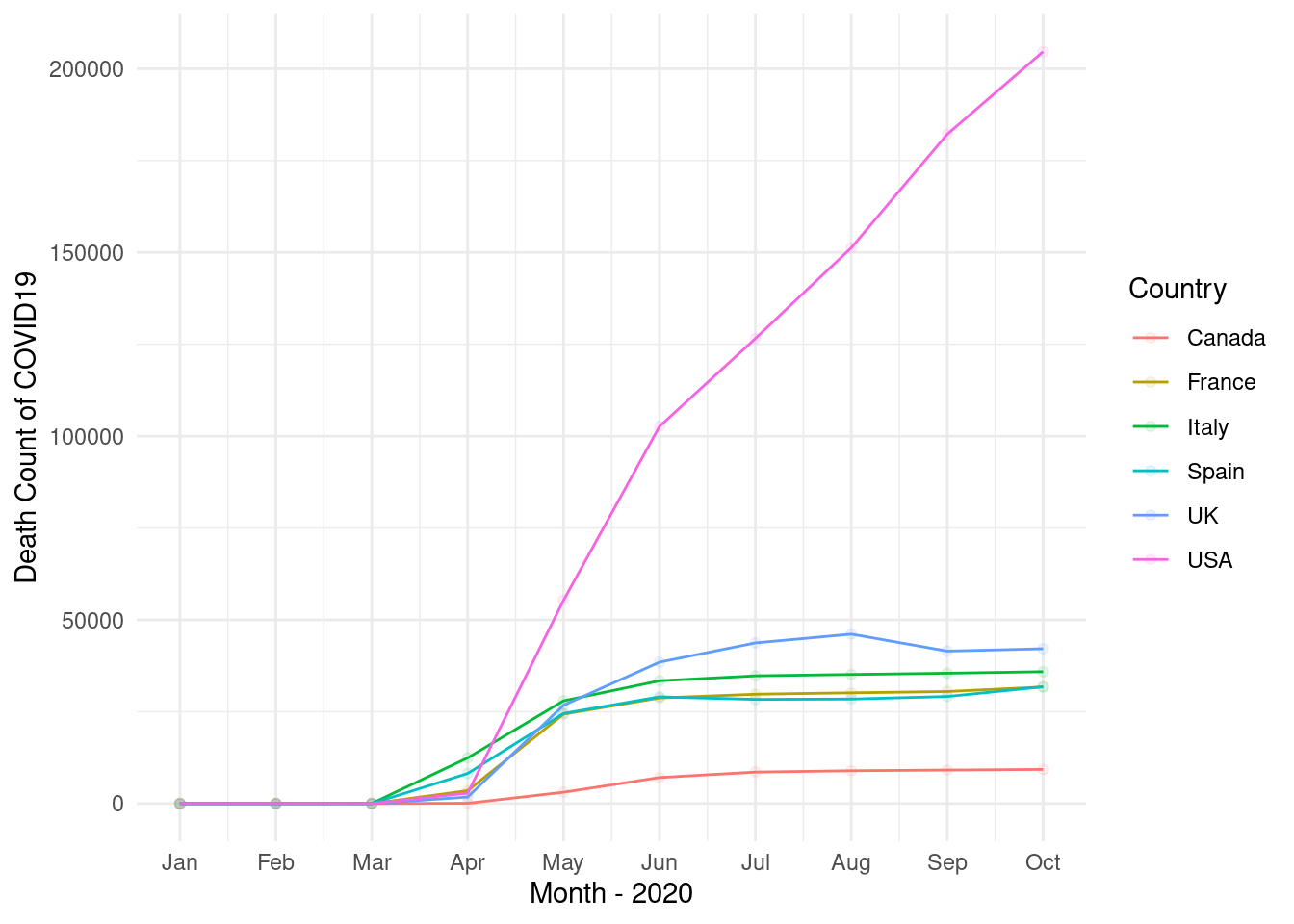
11.4 Review of title from google scholar
my video url is https://youtu.be/oTKEA3IZ7yo
11.4.1 googl scholar
Search the My name of “Jin-Ha Yoon” in google scholar. The url is https://scholar.google.com/citations?hl=en&user=FzE_ZWAAAAAJ&view_op=list_works&sortby=pubdate
url <- "https://scholar.google.com/citations?hl=en&user=FzE_ZWAAAAAJ&view_op=list_works&sortby=pubdate"step1 read the html using url address
step2 filter title using nodes and text, and make data.frame
dat<-gs %>% html_nodes("tbody") %>%
html_nodes("td") %>%
html_nodes("a") %>%
html_text() %>%
data.frame()library(tm)
library(SnowballC)
library(wordcloud)
library(RColorBrewer)
library(dplyr) # for data wrangling
library(tidytext) # for NLP
library(stringr) # to deal with strings
library(knitr) # for tables
library(DT) # for dynamic tables
library(tidyr)step3 split the words (tokenizing) using packages or user own methods.
dat <- dat %>%
setNames(c("titles"))
tokens <-dat %>%
unnest_tokens(word, titles) %>%
count(word, sort = TRUE)%>%
ungroup()
tokens2 <- str_split(dat$titles, " ", simplify = TRUE) %>%
as.data.frame() %>%
mutate(id = row_number()) %>%
pivot_longer(!c(id), names_to = 'Vs', values_to = 'word') %>%
select(-Vs) %>%
filter(!word=="") %>%
count(word, sort = TRUE)%>%
ungroup()step4 import lookup data for removing words
step5 remove stop words and numbers
tokens_clean <- tokens %>%
anti_join(stop_words, by = c("word")) %>%
filter(!str_detect(word, "^[[:digit:]]")) %>%
filter(!str_detect(word, "study|korea"))step6 create word cloud
set.seed(1)
pal <- brewer.pal(12, "Paired")
tokens_clean %>%
with(wordcloud(word, n, random.order = FALSE, colors=pal))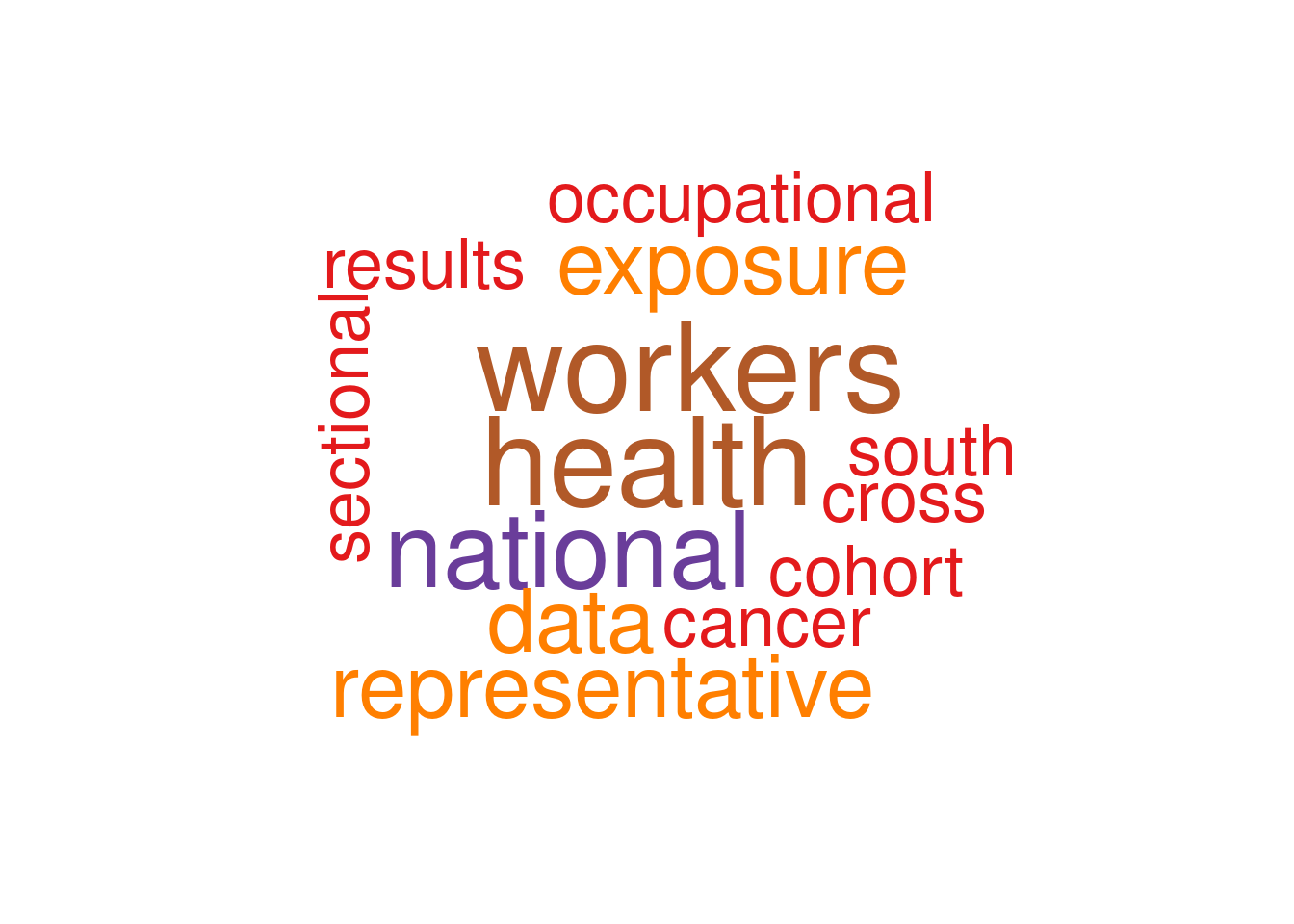
11.5 home work 2
Search you own word in googl scholar. Ror example, You can search “Suicid” or “Hypertension” in googl scholar. And, upload your word cloud to google classroom.